A Novel Gene List Identifies Tumors with a Stromal-Mesenchymal Phenotype and Worse Prognosis in Gastric Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Characteristics and Clinical Samples
2.2. q-RT PCR
2.3. TMA Construction
2.4. Immunohistochemistry
2.5. IHC Scoring and the Determination of MMR Status
2.6. Analysis of Transcriptomic Datasets
2.7. Determination of Genes Constituting the Prognostic Signature
2.8. Gene Set Enrichment and Ingenuity Network Analysis
2.9. Hierarchical Clustering Analysis
2.10. Clustering Based on EMT-Related Gene Expression
2.11. Single-Cell RNA Sequencing Data Analysis
2.12. Statistical Analysis
3. Results
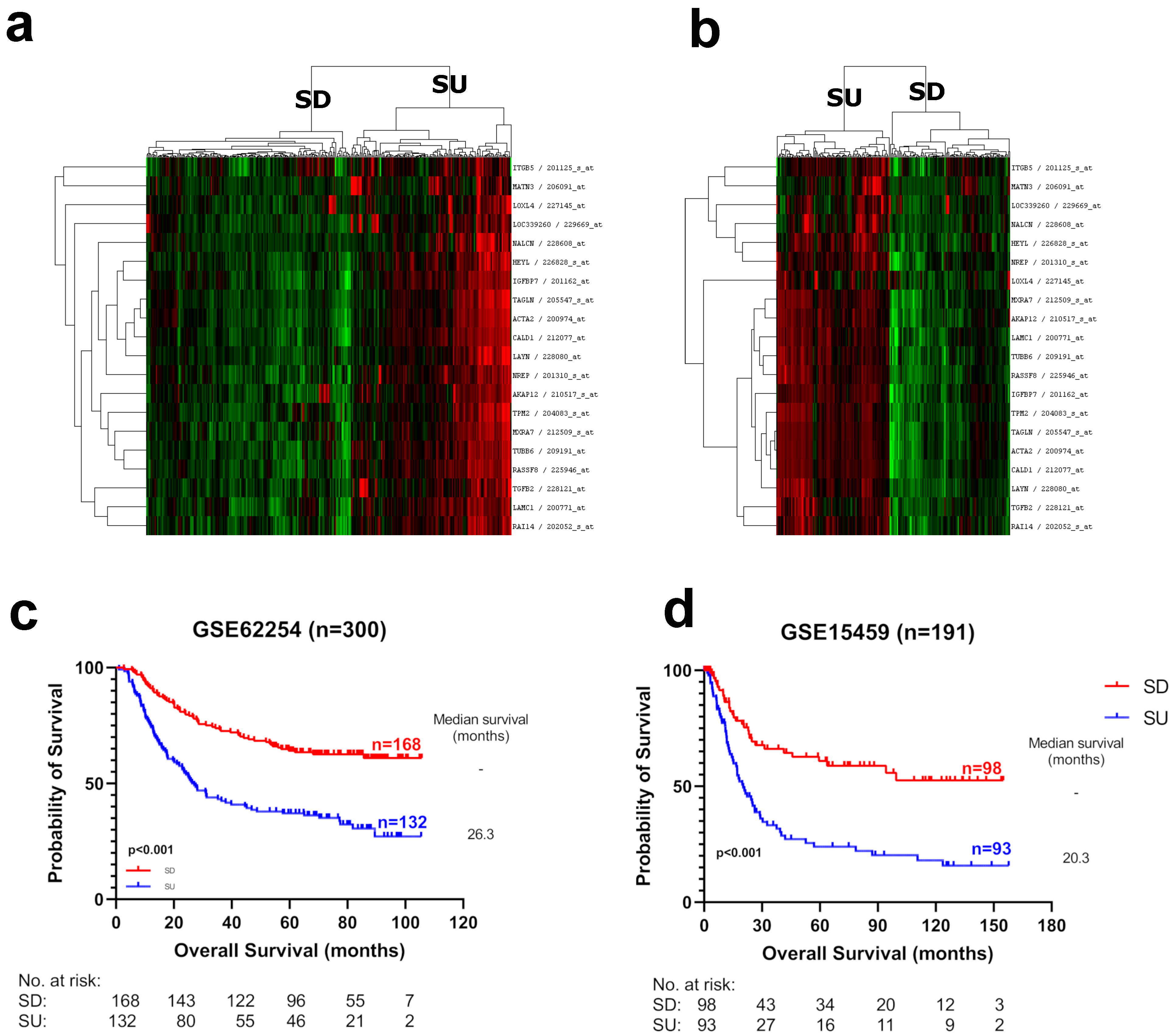
3.1. Identification of Prognostic Markers in Gastric Cancer
3.2. Biological Characteristics of Prognostic Groups
3.3. Expression of Prognostic Genes in Gastric Tumors at the Single-Cell Level
3.4. Evaluation of the SU–SD Signature with Clinical and Biological Subgroups
3.5. SU–SD-Based Classification Is an Independent Prognostic Marker
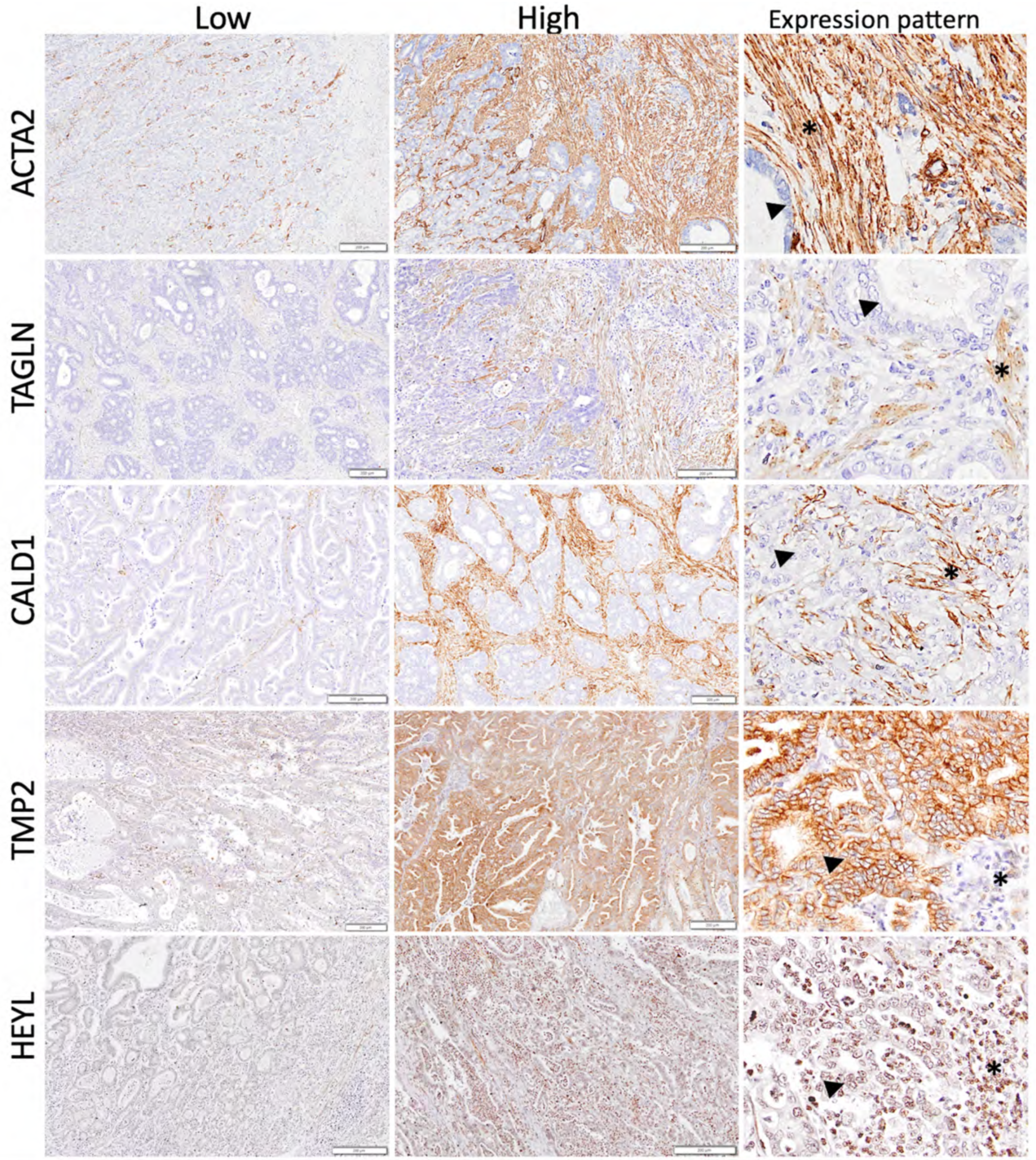
3.6. Validation of the SU–SD Signature Ex Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Rawla, P.; Barsouk, A. Epidemiology of gastric cancer: Global trends, risk factors and prevention. Prz. Gastroenterol. 2019, 14, 26–38. [Google Scholar] [CrossRef]
- Sexton, R.E.; Al Hallak, M.N.; Diab, M.; Azmi, A.S. Gastric cancer: A comprehensive review of current and future treatment strategies. Cancer Metastasis Rev. 2020, 39, 1179–1203. [Google Scholar] [CrossRef]
- Huang, L.; Wu, R.L.; Xu, A.M. Epithelial-mesenchymal transition in gastric cancer. Am. J. Transl. Res. 2015, 7, 2141–2158. [Google Scholar]
- Marano, L.; D’Ignazio, A.; Cammillini, F.; Angotti, R.; Messina, M.; Marrelli, D.; Roviello, F. Comparison between 7th and 8th edition of AJCC TNM staging system for gastric cancer: Old problems and new perspectives. Transl. Gastroenterol. Hepatol. 2019, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Lokuhetty, D.; White, V.A.; Watanabe, R.; Cree, I.A. Digestive System Tumours. World Health Organization Classification of Tumours, 5th ed.; World Health Organization; International Agency for Research on Cancer: Lyon, France, 2019; p. 635. [Google Scholar]
- Marano, L.; Ambrosio, M.R.; Resca, L.; Carbone, L.; Carpineto Samorani, O.; Petrioli, R.; Savelli, V.; Costantini, M.; Malaspina, L.; Polom, K.; et al. The Percentage of Signet Ring Cells Is Inversely Related to Aggressive Behavior and Poor Prognosis in Mixed-Type Gastric Cancer. Front. Oncol. 2022, 12, 897218. [Google Scholar] [CrossRef] [PubMed]
- Roviello, F.; Marano, L.; Ambrosio, M.R.; Resca, L.; D’Ignazio, A.; Petrelli, F.; Petrioli, R.; Costantini, M.; Polom, K.; Macchiarelli, R.; et al. Signet ring cell percentage in poorly cohesive gastric cancer patients: A potential novel predictor of survival. Eur. J. Surg. Oncol. 2022, 48, 561–569. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Matsuoka, T.; Yashiro, M. Biomarkers of gastric cancer: Current topics and future perspective. World J. Gastroenterol. 2018, 24, 2818–2832. [Google Scholar] [CrossRef]
- Jing, R.; Cui, M.; Ju, S.; Pan, S. The Changes and Clinical Significance of Preoperative and Postoperative Serum CEA and CA19-9 in Gastric Cancer. Clin. Lab. 2020, 66, 190732. [Google Scholar] [CrossRef] [PubMed]
- Shibata, C.; Nakano, T.; Yasumoto, A.; Mitamura, A.; Sawada, K.; Ogawa, H.; Miura, T.; Ise, I.; Takami, K.; Yamamoto, K.; et al. Comparison of CEA and CA19-9 as a predictive factor for recurrence after curative gastrectomy in gastric cancer. BMC Surg. 2022, 22, 213. [Google Scholar] [CrossRef]
- Feng, F.; Tian, Y.; Xu, G.; Liu, Z.; Liu, S.; Zheng, G.; Guo, M.; Lian, X.; Fan, D.; Zhang, H. Diagnostic and prognostic value of CEA, CA19-9, AFP and CA125 for early gastric cancer. BMC Cancer 2017, 17, 737. [Google Scholar] [CrossRef]
- Shimada, H.; Noie, T.; Ohashi, M.; Oba, K.; Takahashi, Y. Clinical significance of serum tumor markers for gastric cancer: A systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer 2014, 17, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Marrelli, D.; Pinto, E.; De Stefano, A.; Farnetani, M.; Garosi, L.; Roviello, F. Clinical utility of CEA, CA 19-9, and CA 72-4 in the follow-up of patients with resectable gastric cancer. Am. J. Surg. 2001, 181, 16–19. [Google Scholar] [CrossRef] [PubMed]
- Meleth, S.; Reeder-Hayes, K.; Ashok, M.; Clark, R.; Funkhouser, W.; Wines, R.; Hill, C.; Shanahan, E.; McClure, E.; Burson, K.; et al. Technology Assessment of Molecular Pathology Testing for the Estimation of Prognosis for Common Cancers; AHRQ Technology Assessments: Rockville, MD, USA, 2014. [Google Scholar]
- Ajani, J.; Bekaii-Saab, T.; D’Amico, T.A.; Fuchs, C.; Gibson, M.K.; Goldberg, M.; Hayman, J.A.; Ilson, D.H.; Javle, M.; Kelley, S.; et al. Gastric Cancer Clinical Practice Guidelines. J. Natl. Compr. Cancer Netw. 2006, 4, 350–366. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Uner, M.; Isik, A.; Oztop, S.; Karabulut, E.; Demirkol-Canli, S.; Akyol, A. Gastric Carcinoma with Lymphoid Stroma: A Combination of Mismatch Repair Deficient Medullary Type and Epstein-Barr Virus-associated Gastric Carcinomas. Int. J. Surg. Pathol. 2022, 30, 623–633. [Google Scholar] [CrossRef]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Ooi, C.H.; Ivanova, T.; Wu, J.; Lee, M.; Tan, I.B.; Tao, J.; Ward, L.; Koo, J.H.; Gopalakrishnan, V.; Zhu, Y.; et al. Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLoS Genet. 2009, 5, e1000676. [Google Scholar] [CrossRef]
- Wang, G.; Hu, N.; Yang, H.H.; Wang, L.; Su, H.; Wang, C.; Clifford, R.; Dawsey, E.M.; Li, J.M.; Ding, T.; et al. Comparison of global gene expression of gastric cardia and noncardia cancers from a high-risk population in china. PLoS ONE 2013, 8, e63826. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Choi, I.J.; Kim, C.G.; Kim, H.S.; Oshima, A.; Michalowski, A.; Green, J.E. A gene expression signature of acquired chemoresistance to cisplatin and fluorouracil combination chemotherapy in gastric cancer patients. PLoS ONE 2011, 6, e16694. [Google Scholar] [CrossRef] [PubMed]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martinez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Trevino, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.J.; Park, J.; Shin, Y.; Choi, Y.; Park, S.W.; Kang, S.G.; Son, H.Y.; Huh, Y.M. Deconvolution of diffuse gastric cancer and the suppression of CD34 on the BALB/c nude mice model. BMC Cancer 2020, 20, 314. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.Y.; Lim, J.Y.; Cheong, J.H.; Park, Y.Y.; Yoon, S.L.; Kim, S.M.; Kim, S.B.; Kim, H.; Hong, S.W.; Park, Y.N.; et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin. Cancer Res. 2011, 17, 1850–1857. [Google Scholar] [CrossRef]
- Oh, S.C.; Sohn, B.H.; Cheong, J.H.; Kim, S.B.; Lee, J.E.; Park, K.C.; Lee, S.H.; Park, J.L.; Park, Y.Y.; Lee, H.S.; et al. Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat. Commun. 2018, 9, 1777. [Google Scholar] [CrossRef]
- Terry, M.; Therneau, P.M.G. Modeling Survival Data: Extending the Cox Model; Springer: New York, NY, USA, 2000. [Google Scholar]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Tan, T.Z.; Miow, Q.H.; Miki, Y.; Noda, T.; Mori, S.; Huang, R.Y.; Thiery, J.P. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol. Med. 2014, 6, 1279–1293. [Google Scholar] [CrossRef]
- Kumar, V.; Ramnarayanan, K.; Sundar, R.; Padmanabhan, N.; Srivastava, S.; Koiwa, M.; Yasuda, T.; Koh, V.; Huang, K.K.; Tay, S.T.; et al. Single-Cell Atlas of Lineage States, Tumor Microenvironment, and Subtype-Specific Expression Programs in Gastric Cancer. Cancer Discov. 2022, 12, 670–691. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef]
- Che, L.H.; Liu, J.W.; Huo, J.P.; Luo, R.; Xu, R.M.; He, C.; Li, Y.Q.; Zhou, A.J.; Huang, P.; Chen, Y.Y.; et al. A single-cell atlas of liver metastases of colorectal cancer reveals reprogramming of the tumor microenvironment in response to preoperative chemotherapy. Cell Discov. 2021, 7, 80. [Google Scholar] [CrossRef]
- Demirkol, S.; Gomceli, I.; Isbilen, M.; Dayanc, B.E.; Tez, M.; Bostanci, E.B.; Turhan, N.; Akoglu, M.; Ozyerli, E.; Durdu, S.; et al. A Combined ULBP2 and SEMA5A Expression Signature as a Prognostic and Predictive Biomarker for Colon Cancer. J. Cancer 2017, 8, 1113–1122. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, O.; Naumann, M. NF-kappaB Signaling in Gastric Cancer. Toxins 2017, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Rodriquenz, M.G.; Roviello, G.; D’Angelo, A.; Lavacchi, D.; Roviello, F.; Polom, K. MSI and EBV Positive Gastric Cancer’s Subgroups and Their Link With Novel Immunotherapy. J. Clin. Med. 2020, 9, 1427. [Google Scholar] [CrossRef]
- Marttila, M.; Lemola, E.; Wallefeld, W.; Memo, M.; Donner, K.; Laing, N.G.; Marston, S.; Gronholm, M.; Wallgren-Pettersson, C. Abnormal actin binding of aberrant beta-tropomyosins is a molecular cause of muscle weakness in TPM2-related nemaline and cap myopathy. Biochem. J. 2012, 442, 231–239. [Google Scholar] [CrossRef]
- Tajsharghi, H.; Ohlsson, M.; Palm, L.; Oldfors, A. Myopathies associated with beta-tropomyosin mutations. Neuromuscul. Disord. 2012, 22, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Khachigian, L.M.; Black, B.L.; Ferdinandy, P.; De Caterina, R.; Madonna, R.; Geng, Y.J. Transcriptional regulation of vascular smooth muscle cell proliferation, differentiation and senescence: Novel targets for therapy. Vascul. Pharmacol. 2022, 146, 107091. [Google Scholar] [CrossRef] [PubMed]
- Tawfik, O.; Rao, D.; Nothnick, W.B.; Graham, A.; Mau, B.; Fan, F. Transgelin, a Novel Marker of Smooth Muscle Differentiation, Effectively Distinguishes Endometrial Stromal Tumors from Uterine Smooth Muscle Tumors. Int. J. Gynecol. Obstet. Reprod. Med. Res. 2014, 1, 26–31. [Google Scholar]
- Mayanagi, T.; Sobue, K. Diversification of caldesmon-linked actin cytoskeleton in cell motility. Cell Adhes. Migr. 2011, 5, 150–159. [Google Scholar] [CrossRef]
- Li, D.; Yin, Y.; He, M.; Wang, J. Identification of Potential Biomarkers Associated with Prognosis in Gastric Cancer via Bioinformatics Analysis. Med. Sci. Monit. 2021, 27, e929104. [Google Scholar] [CrossRef]
- Morgan, E.; Arnold, M.; Camargo, M.C.; Gini, A.; Kunzmann, A.T.; Matsuda, T.; Meheus, F.; Verhoeven, R.H.A.; Vignat, J.; Laversanne, M.; et al. The current and future incidence and mortality of gastric cancer in 185 countries, 2020–2040: A population-based modelling study. EClinicalMedicine 2022, 47, 101404. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Taube, J.M.; Anders, R.A.; Pardoll, D.M. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer 2016, 16, 275–287. [Google Scholar] [CrossRef] [PubMed]
- De Rosa, S.; Sahnane, N.; Tibiletti, M.G.; Magnoli, F.; Vanoli, A.; Sessa, F.; Chiaravalli, A.M. EBV(+) and MSI Gastric Cancers Harbor High PD-L1/PD-1 Expression and High CD8(+) Intratumoral Lymphocytes. Cancers 2018, 10, 102. [Google Scholar] [CrossRef]
- Ferrasi, A.C.; Pinheiro, N.A.; Rabenhorst, S.H.; Caballero, O.L.; Rodrigues, M.A.; de Carvalho, F.; Leite, C.V.; Ferreira, M.V.; Barros, M.A.; Pardini, M.I. Helicobacter pylori and EBV in gastric carcinomas: Methylation status and microsatellite instability. World J. Gastroenterol. 2010, 16, 312–319. [Google Scholar] [CrossRef]
- Sun, K.; Jia, K.; Lv, H.; Wang, S.Q.; Wu, Y.; Lei, H.; Chen, X. EBV-Positive Gastric Cancer: Current Knowledge and Future Perspectives. Front. Oncol. 2020, 10, 583463. [Google Scholar] [CrossRef] [PubMed]
- Boccardi, V.; Marano, L.; Rossetti, R.R.; Rizzo, M.R.; di Martino, N.; Paolisso, G. Serum CD26 levels in patients with gastric cancer: A novel potential diagnostic marker. BMC Cancer 2015, 15, 703. [Google Scholar] [CrossRef]
- Zhou, L.; Lu, H.; Zeng, F.; Zhou, Q.; Li, S.; Wu, Y.; Yuan, Y.; Xin, L. Constructing a new prognostic signature of gastric cancer based on multiple data sets. Bioengineered 2021, 12, 2820–2835. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ni, S.; Wang, H.; Zhang, Q.; Weng, W. Charactering tumor microenvironment reveals stromal-related transcription factors promote tumor carcinogenesis in gastric cancer. Cancer Med. 2020, 9, 5247–5257. [Google Scholar] [CrossRef]
- Eiro, N.; Fernandez-Gomez, J.M.; Gonzalez-Ruiz de Leon, C.; Fraile, M.; Gonzalez-Suarez, J.; Lobo-Rodriguez, B.; Garcia-Rodriguez, J.; Escaf, S.; Vizoso, F.J. Gene Expression Profile of Stromal Factors in Cancer-Associated Fibroblasts from Prostate Cancer. Diagnostics 2022, 12, 1605. [Google Scholar] [CrossRef]
- Rao, K.B.; Malathi, N.; Narashiman, S.; Rajan, S.T. Evaluation of myofibroblasts by expression of alpha smooth muscle actin: A marker in fibrosis, dysplasia and carcinoma. J. Clin. Diagn. Res. 2014, 8, ZC14–ZC17. [Google Scholar] [CrossRef]
- Prunotto, M.; Bruschi, M.; Gunning, P.; Gabbiani, G.; Weibel, F.; Ghiggeri, G.M.; Petretto, A.; Scaloni, A.; Bonello, T.; Schevzov, G.; et al. Stable incorporation of alpha-smooth muscle actin into stress fibers is dependent on specific tropomyosin isoforms. Cytoskeleton 2015, 72, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.J. Regulation of heterogeneous cancer-associated fibroblasts: The molecular pathology of activated signaling pathways. J. Exp. Clin. Cancer Res. 2020, 39, 112. [Google Scholar] [CrossRef]
- Cheong, J.H.; Wang, S.C.; Park, S.; Porembka, M.R.; Christie, A.L.; Kim, H.; Kim, H.S.; Zhu, H.; Hyung, W.J.; Noh, S.H.; et al. Development and validation of a prognostic and predictive 32-gene signature for gastric cancer. Nat. Commun. 2022, 13, 774. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Bai, J.; Shi, R.; Shao, X.; Yang, Y.; Jin, Y.; Che, X.; Zhang, Y.; Qu, X.; Liu, Y.; et al. TGFB2 serves as a link between epithelial-mesenchymal transition and tumor mutation burden in gastric cancer. Int. Immunopharmacol. 2020, 84, 106532. [Google Scholar] [CrossRef]
- Ko, Y.C.; Lai, T.Y.; Hsu, S.C.; Wang, F.H.; Su, S.Y.; Chen, Y.L.; Tsai, M.L.; Wu, C.C.; Hsiao, J.R.; Chang, J.Y.; et al. Index of Cancer-Associated Fibroblasts Is Superior to the Epithelial-Mesenchymal Transition Score in Prognosis Prediction. Cancers 2020, 12, 1718. [Google Scholar] [CrossRef]
- Tang, Y.A.; Chen, Y.F.; Bao, Y.; Mahara, S.; Yatim, S.; Oguz, G.; Lee, P.L.; Feng, M.; Cai, Y.; Tan, E.Y.; et al. Hypoxic tumor microenvironment activates GLI2 via HIF-1alpha and TGF-beta2 to promote chemoresistance in colorectal cancer. Proc. Natl. Acad. Sci. USA 2018, 115, E5990–E5999. [Google Scholar] [CrossRef]
- Liu, Y.J.; Zeng, S.H.; Hu, Y.D.; Zhang, Y.H.; Li, J.P. Overexpression of NREP Promotes Migration and Invasion in Gastric Cancer Through Facilitating Epithelial-Mesenchymal Transition. Front. Cell Dev. Biol. 2021, 9, 746194. [Google Scholar] [CrossRef]
- Brown, K.M.; Xue, A.; Smith, R.C.; Samra, J.S.; Gill, A.J.; Hugh, T.J. Cancer-associated stroma reveals prognostic biomarkers and novel insights into the tumour microenvironment of colorectal cancer and colorectal liver metastases. Cancer Med. 2022, 11, 492–506. [Google Scholar] [CrossRef]
- Rupp, C.; Scherzer, M.; Rudisch, A.; Unger, C.; Haslinger, C.; Schweifer, N.; Artaker, M.; Nivarthi, H.; Moriggl, R.; Hengstschlager, M.; et al. IGFBP7, a novel tumor stroma marker, with growth-promoting effects in colon cancer through a paracrine tumor-stroma interaction. Oncogene 2015, 34, 815–825. [Google Scholar] [CrossRef]
- Shen, X.J.; Zhang, Y.W.; Deng, H.; Chen, L.L.; Chen, P.S. [Fibroblasts-colorectal cancer cells interaction induces the expression of IGFBP7]. Zhejiang Da Xue Xue Bao Yi Xue Ban 2009, 38, 151–157. [Google Scholar]
- Calon, A.; Lonardo, E.; Berenguer-Llergo, A.; Espinet, E.; Hernando-Momblona, X.; Iglesias, M.; Sevillano, M.; Palomo-Ponce, S.; Tauriello, D.V.; Byrom, D.; et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 2015, 47, 320–329. [Google Scholar] [CrossRef]
- Liu, D.; Liu, S.; Fang, Y.; Liu, L.; Hu, K. Comprehensive Analysis of the Expression and Prognosis for ITGBs: Identification of ITGB5 as a Biomarker of Poor Prognosis and Correlated with Immune Infiltrates in Gastric Cancer. Front. Cell Dev. Biol. 2021, 9, 816230. [Google Scholar] [CrossRef] [PubMed]
- Michael, G.; Walker, W.V. Cell adhesion and matrix remodeling genes identified by co-expression analysis. Gene Funct. Dis. 2003, 3, 109–112. [Google Scholar] [CrossRef]
- Yu, B.; Chen, X.; Li, J.; Qu, Y.; Su, L.; Peng, Y.; Huang, J.; Yan, J.; Yu, Y.; Gu, Q.; et al. Stromal fibroblasts in the microenvironment of gastric carcinomas promote tumor metastasis via upregulating TAGLN expression. BMC Cell Biol. 2013, 14, 17. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, K.; Sun, J.; Tang, J.; Zhou, J. Development and Validation of an Autophagy-Stroma-Based Microenvironment Gene Signature for Risk Stratification in Colorectal Cancer. OncoTargets Ther. 2021, 14, 3503–3515. [Google Scholar] [CrossRef] [PubMed]
- Rho, J.H.; Roehrl, M.H.; Wang, J.Y. Tissue proteomics reveals differential and compartment-specific expression of the homologs transgelin and transgelin-2 in lung adenocarcinoma and its stroma. J. Proteome Res. 2009, 8, 5610–5618. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Bian, S.; Zhou, X.; Cui, Y.; Wang, W.; Wen, L.; Guo, L.; Fu, W.; Tang, F. Single-Cell Multiomics Sequencing Reveals Prevalent Genomic Alterations in Tumor Stromal Cells of Human Colorectal Cancer. Cancer Cell 2020, 38, 818–828.e5. [Google Scholar] [CrossRef]
- Xue, R.; Hua, L.; Xu, W.; Gao, Y.; Pang, Y.; Hao, J. Derivation and Validation of the Potential Core Genes in Pancreatic Cancer for Tumor-Stroma Crosstalk. Biomed. Res. Int. 2018, 2018, 4283673. [Google Scholar] [CrossRef]
- Li, L.; Wang, X. Identification of gastric cancer subtypes based on pathway clustering. NPJ Precis. Oncol. 2021, 5, 46. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.P.; Sima, Z.H.; Wang, H.C.; Zhang, J.Y.; Sun, L.S.; Chen, F.; Li, T.J. Identification of the involvement of LOXL4 in generation of keratocystic odontogenic tumors by RNA-Seq analysis. Int. J. Oral Sci. 2014, 6, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Li, L.; Li, M.; Wang, X. HIF1A expression correlates with increased tumor immune and stromal signatures and aggressive phenotypes in human cancers. Cell Oncol. 2020, 43, 877–888. [Google Scholar] [CrossRef]
- Kitajima, Y.; Miyazaki, K. The Critical Impact of HIF-1a on Gastric Cancer Biology. Cancers 2013, 5, 15–26. [Google Scholar] [CrossRef]
- Zheng, H.; Liu, H.; Li, H.; Dou, W.; Wang, X. Weighted Gene Co-expression Network Analysis Identifies a Cancer-Associated Fibroblast Signature for Predicting Prognosis and Therapeutic Responses in Gastric Cancer. Front. Mol. Biosci. 2021, 8, 744677. [Google Scholar] [CrossRef] [PubMed]
- Mak, T.K.; Li, X.; Huang, H.; Wu, K.; Huang, Z.; He, Y.; Zhang, C. The cancer-associated fibroblast-related signature predicts prognosis and indicates immune microenvironment infiltration in gastric cancer. Front. Immunol. 2022, 13, 951214. [Google Scholar] [CrossRef] [PubMed]
- Demirkol, S. Prediction of Prognosis and Chemosensitivity in Gastrointestinal Cancers. Ph.D. Thesis, Bilkent University, Ankara, Turkey, 2017. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demirkol Canli, S.; Uner, M.; Kucukkaraduman, B.; Karaoglu, D.A.; Isik, A.; Turhan, N.; Akyol, A.; Gomceli, I.; Gure, A.O. A Novel Gene List Identifies Tumors with a Stromal-Mesenchymal Phenotype and Worse Prognosis in Gastric Cancer. Cancers 2023, 15, 3035. https://doi.org/10.3390/cancers15113035
Demirkol Canli S, Uner M, Kucukkaraduman B, Karaoglu DA, Isik A, Turhan N, Akyol A, Gomceli I, Gure AO. A Novel Gene List Identifies Tumors with a Stromal-Mesenchymal Phenotype and Worse Prognosis in Gastric Cancer. Cancers. 2023; 15(11):3035. https://doi.org/10.3390/cancers15113035
Chicago/Turabian StyleDemirkol Canli, Secil, Meral Uner, Baris Kucukkaraduman, Diren Arda Karaoglu, Aynur Isik, Nesrin Turhan, Aytekin Akyol, Ismail Gomceli, and Ali Osmay Gure. 2023. "A Novel Gene List Identifies Tumors with a Stromal-Mesenchymal Phenotype and Worse Prognosis in Gastric Cancer" Cancers 15, no. 11: 3035. https://doi.org/10.3390/cancers15113035
APA StyleDemirkol Canli, S., Uner, M., Kucukkaraduman, B., Karaoglu, D. A., Isik, A., Turhan, N., Akyol, A., Gomceli, I., & Gure, A. O. (2023). A Novel Gene List Identifies Tumors with a Stromal-Mesenchymal Phenotype and Worse Prognosis in Gastric Cancer. Cancers, 15(11), 3035. https://doi.org/10.3390/cancers15113035





