A Fully Automated Post-Surgical Brain Tumor Segmentation Model for Radiation Treatment Planning and Longitudinal Tracking
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. Related Work
1.2. Purpose
2. Materials and Methods
2.1. Preparation of Training Data
2.2. Segmentation Models and Training
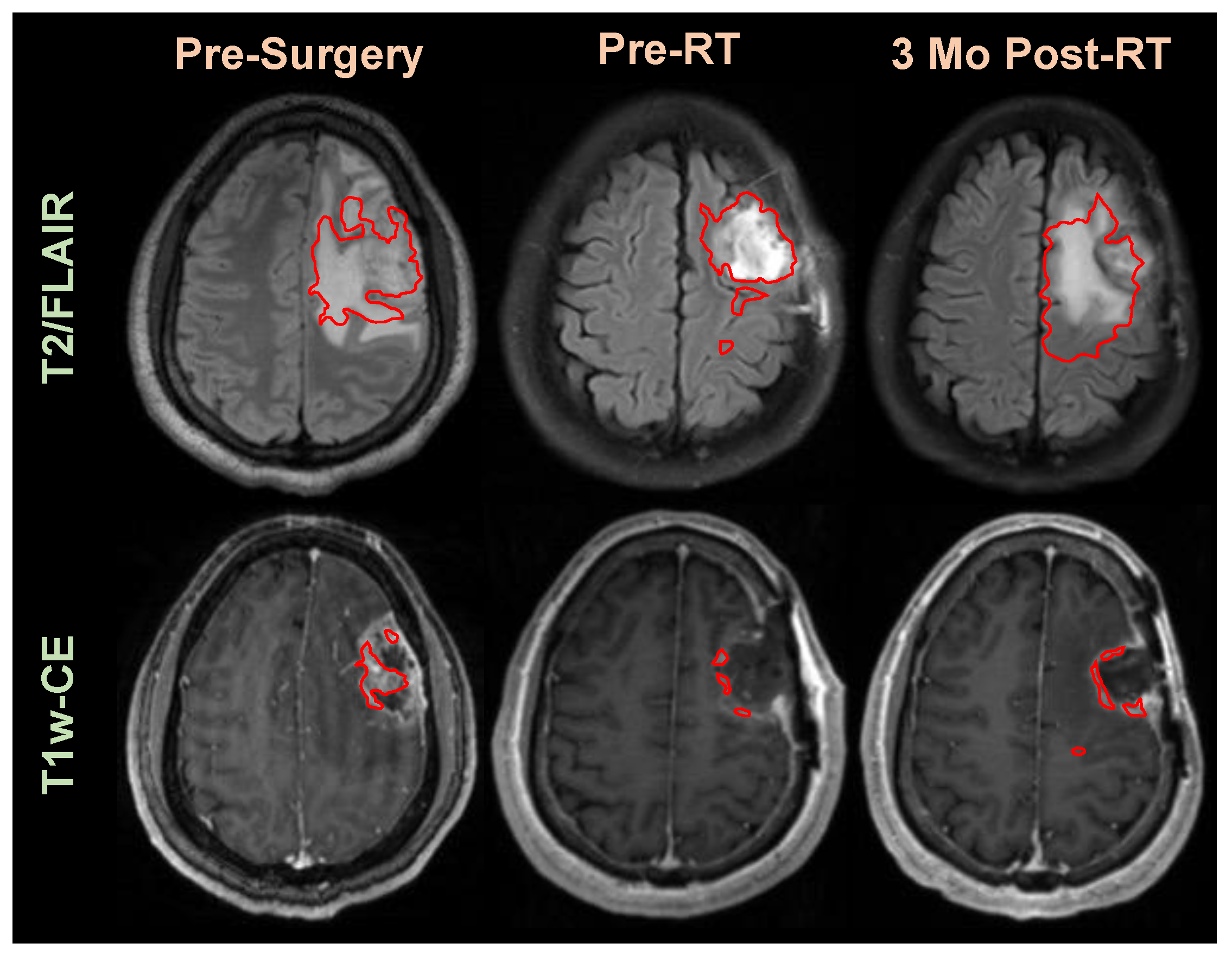
2.3. Application of Segmentation in Longitudinal Tracking
3. Results
3.1. Segmentation for RT Planning
3.2. Application of Segmentation in Longitudinal Tracking
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gilbert, M.R.; Wang, M.; Aldape, K.D.; Stupp, R.; Hegi, M.; Jaeckle, K.A.; Armstrong, T.S.; Wefel, J.S.; Won, M.; Blumenthal, D.T.; et al. RTOG 0525: A randomized phase III trial comparing standard adjuvant temozolomide (TMZ) with a dose-dense (dd) schedule in newly diagnosed glioblastoma (GBM). J. Clin. Oncol. 2011, 29, 2006. [Google Scholar] [CrossRef] [Green Version]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [Green Version]
- Stupp, R.; Taillibert, S.; Kanner, A.; Read, W.; Steinberg, D.; Lhermitte, B.; Toms, S.; Idbaih, A.; Ahluwalia, M.S.; Fink, K.; et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma: A Randomized Clinical Trial. JAMA 2017, 318, 2306–2316. [Google Scholar] [CrossRef] [Green Version]
- Stupp, R.; Taillibert, S.; Kanner, A.A.; Kesari, S.; Steinberg, D.M.; Toms, S.A.; Taylor, L.P.; Lieberman, F.; Silvani, A.; Fink, K.L.; et al. Maintenance Therapy With Tumor-Treating Fields Plus Temozolomide vs Temozolomide Alone for Glioblastoma: A Randomized Clinical Trial. JAMA 2015, 314, 2535–2543. [Google Scholar] [CrossRef] [PubMed]
- Balwant, M.K. A Review on Convolutional Neural Networks for Brain Tumor Segmentation: Methods, Datasets, Libraries, and Future Directions. IRBM 2022, 43, 521–537. [Google Scholar] [CrossRef]
- Ghosh, A.; Thakur, S. Review of Brain Tumor MRI Image Segmentation Methods for BraTS Challenge Dataset. In Proceedings of the 2022 12th International Conference on Cloud Computing, Data Science & Engineering (Confluence), Virtual, 27–28 January 2022; pp. 405–410. [Google Scholar]
- Tillmanns, N.; Lum, A.E.; Cassinelli, G.; Merkaj, S.; Verma, T.; Zeevi, T.; Staib, L.; Subramanian, H.; Bahar, R.C.; Brim, W.; et al. Identifying clinically applicable machine learning algorithms for glioma segmentation: Recent advances and discoveries. Neuro-Oncol. Adv. 2022, 4, vdac093. [Google Scholar] [CrossRef]
- Wang, F.; Jiang, R.; Zheng, L.; Meng, C.; Biswal, B. 3D U-Net Based Brain Tumor Segmentation and Survival Days Prediction. In Proceedings of the Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Cham, Switzerland, 4 October 2020; pp. 131–141. [Google Scholar]
- Hatamizadeh, A.; Nath, V.; Tang, Y.; Yang, D.; Roth, H.R.; Xu, D. Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images. In Proceedings of the Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Cham, Switzerland, 4 October 2022; pp. 272–284. [Google Scholar]
- Shusharina, N.; Söderberg, J.; Edmunds, D.; Löfman, F.; Shih, H.; Bortfeld, T. Automated delineation of the clinical target volume using anatomically constrained 3D expansion of the gross tumor volume. Radiother. Oncol. 2020, 146, 37–43. [Google Scholar] [CrossRef]
- Sadeghi, S.; Farzin, M.; Gholami, S. Fully automated clinical target volume segmentation for glioblastoma radiotherapy using a deep convolutional neural network. Pol. J. Radiol. 2023, 88, 31–40. [Google Scholar] [CrossRef]
- Ermiş, E.; Jungo, A.; Poel, R.; Blatti-Moreno, M.; Meier, R.; Knecht, U.; Aebersold, D.M.; Fix, M.K.; Manser, P.; Reyes, M.; et al. Fully automated brain resection cavity delineation for radiation target volume definition in glioblastoma patients using deep learning. Radiat. Oncol. 2020, 15, 100. [Google Scholar] [CrossRef]
- Ramesh, K.; Gurbani, S.S.; Mellon, E.A.; Huang, V.; Goryawala, M.; Barker, P.B.; Kleinberg, L.; Shu, H.G.; Shim, H.; Weinberg, B.D. The Longitudinal Imaging Tracker (BrICS-LIT):A Cloud Platform for Monitoring Treatment Response in Glioblastoma Patients. Tomography 2020, 6, 93–100. [Google Scholar] [CrossRef]
- Chukwueke, U.N.; Wen, P.Y. Use of the Response Assessment in Neuro-Oncology (RANO) criteria in clinical trials and clinical practice. CNS Oncol. 2019, 8, CNS28. [Google Scholar] [CrossRef] [Green Version]
- Kickingereder, P.; Isensee, F.; Tursunova, I.; Petersen, J.; Neuberger, U.; Bonekamp, D.; Brugnara, G.; Schell, M.; Kessler, T.; Foltyn, M.; et al. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: A multicentre, retrospective study. Lancet Oncol. 2019, 20, 728–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinberg, B.; Ramesh, K.; Gurbani, S.; Schreibmann, E.; Kleinberg, L.; Shu, H.-K.; Shim, H. NIMG-23. Brain tumor reporting and data system (BT-rads) and quantitative tools to guide its implementation. Neuro Oncol. 2019, 21, vi166. [Google Scholar] [CrossRef]
- Gore, A.; Hoch, M.J.; Shu, H.G.; Olson, J.J.; Voloschin, A.D.; Weinberg, B.D. Institutional Implementation of a Structured Reporting System: Our Experience with the Brain Tumor Reporting and Data System. Acad. Radiol. 2019, 26, 974–980. [Google Scholar] [CrossRef]
- Kim, S.; Hoch, M.J.; Cooper, M.E.; Gore, A.; Weinberg, B.D. Using a Website to Teach a Structured Reporting System, the Brain Tumor Reporting and Data System. Curr. Probl. Diagn. Radiol. 2021, 50, 356–361. [Google Scholar] [CrossRef]
- Lee, S.J.; Weinberg, B.D.; Gore, A.; Banerjee, I. A Scalable Natural Language Processing for Inferring BT-RADS Categorization from Unstructured Brain Magnetic Resonance Reports. J. Digit. Imaging 2020, 33, 1393–1400. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Weinberg, B.D.; Hu, R.; Saindane, A.; Mullins, M.; Allen, J.; Hoch, M.J. Quantitative Improvement in Brain Tumor MRI Through Structured Reporting (BT-RADS). Acad. Radiol. 2020, 27, 780–784. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-Like Pure Transformer for Medical Image Segmentation. In Proceedings of the Computer Vision—ECCV 2022 Workshops, Cham, Switzerland, 23–27 October 2022; pp. 205–218. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Yang, C.; Guo, X.; Wang, T.; Yang, Y.; Ji, N.; Li, D.; Lv, H.; Ma, T. Automatic Brain Tumor Segmentation Method Based on Modified Convolutional Neural Network. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Ann ual International Conference, Berlin, Germany, 23–27 July 2019; pp. 998–1001. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference, Athens, Greece, 17–21 October 2016; pp. 424–432. [Google Scholar]
- Fushiki, T. Estimation of prediction error by using K-fold cross-validation. Stat. Comput. 2011, 21, 137–146. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Barragán-Montero, A.; Javaid, U.; Valdés, G.; Nguyen, D.; Desbordes, P.; Macq, B.; Willems, S.; Vandewinckele, L.; Holmström, M.; Löfman, F.; et al. Artificial intelligence and machine learning for medical imaging: A technology review. Phys. Medica 2021, 83, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Giansanti, D.; Di Basilio, F. The Artificial Intelligence in Digital Radiology: Part 1: The Challenges, Acceptance and Consensus. Healthcare 2022, 10, 509. [Google Scholar] [CrossRef]
- Saw, S.N.; Ng, K.H. Current challenges of implementing artificial intelligence in medical imaging. Phys. Med. 2022, 100, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Martín-Noguerol, T.; Paulano-Godino, F.; López-Ortega, R.; Górriz, J.M.; Riascos, R.F.; Luna, A. Artificial intelligence in radiology: Relevance of collaborative work between radiologists and engineers for building a multidisciplinary team. Clin. Radiol. 2021, 76, 317–324. [Google Scholar] [CrossRef]
- Choi, Y.; Choi, M.; Kim, M.; Ha, J.-W.; Kim, S.; Choo, J. Stargan: Unified generative adversarial networks for multi-domain image-to-image translation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8789–8797. [Google Scholar]
- Xin, B.; Hu, Y.; Zheng, Y.; Liao, H. Multi-modality generative adversarial networks with tumor consistency loss for brain mr image synthesis. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 4–7 April 2020; pp. 1803–1807. [Google Scholar]
- Isola, P.; Zhu, J.-Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]






| Brain Lesion Segmentation | Strengths | Weaknesses |
|---|---|---|
| Previous Efforts |
|
|
| Proposed Effort |
|
|
| Model | GTV1 | GTV2 | ||||||
|---|---|---|---|---|---|---|---|---|
| Train (Dice) | Test (Dice) | Test (Hausdorff) | Test (Jaccard) | Train (Dice) | Test (Dice) | Test (Hausdorff) | Test (Jaccard) | |
| 2D Unet | 0.93 | 0.43 | 78.50 | 0.19 | 0.92 | 0.56 | 75.41 | 0.34 |
| 2D Resunet | 0.93 | 0.58 | 58.50 | 0.36 | 0.91 | 0.57 | 35.57 | 0.35 |
| 2D Swin-Unet | 0.89 | 0.64 | 60.71 | 0.44 | 0.86 | 0.51 | 35.63 | 0.31 |
| 3D Unet | 0.77 | 0.72 | 12.77 | 0.51 | 0.79 | 0.73 | 10.75 | 0.58 |
| 3D Swin-UNETR | 0.64 | 0.60 | 38.32 | 0.36 | 0.65 | 0.64 | 23.33 | 0.44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramesh, K.K.; Xu, K.M.; Trivedi, A.G.; Huang, V.; Sharghi, V.K.; Kleinberg, L.R.; Mellon, E.A.; Shu, H.-K.G.; Shim, H.; Weinberg, B.D. A Fully Automated Post-Surgical Brain Tumor Segmentation Model for Radiation Treatment Planning and Longitudinal Tracking. Cancers 2023, 15, 3956. https://doi.org/10.3390/cancers15153956
Ramesh KK, Xu KM, Trivedi AG, Huang V, Sharghi VK, Kleinberg LR, Mellon EA, Shu H-KG, Shim H, Weinberg BD. A Fully Automated Post-Surgical Brain Tumor Segmentation Model for Radiation Treatment Planning and Longitudinal Tracking. Cancers. 2023; 15(15):3956. https://doi.org/10.3390/cancers15153956
Chicago/Turabian StyleRamesh, Karthik K., Karen M. Xu, Anuradha G. Trivedi, Vicki Huang, Vahid Khalilzad Sharghi, Lawrence R. Kleinberg, Eric A. Mellon, Hui-Kuo G. Shu, Hyunsuk Shim, and Brent D. Weinberg. 2023. "A Fully Automated Post-Surgical Brain Tumor Segmentation Model for Radiation Treatment Planning and Longitudinal Tracking" Cancers 15, no. 15: 3956. https://doi.org/10.3390/cancers15153956






