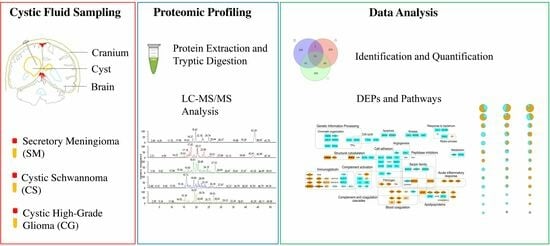
Proteomic Analysis on Sequential Samples of Cystic Fluid Obtained from Human Brain Tumors
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
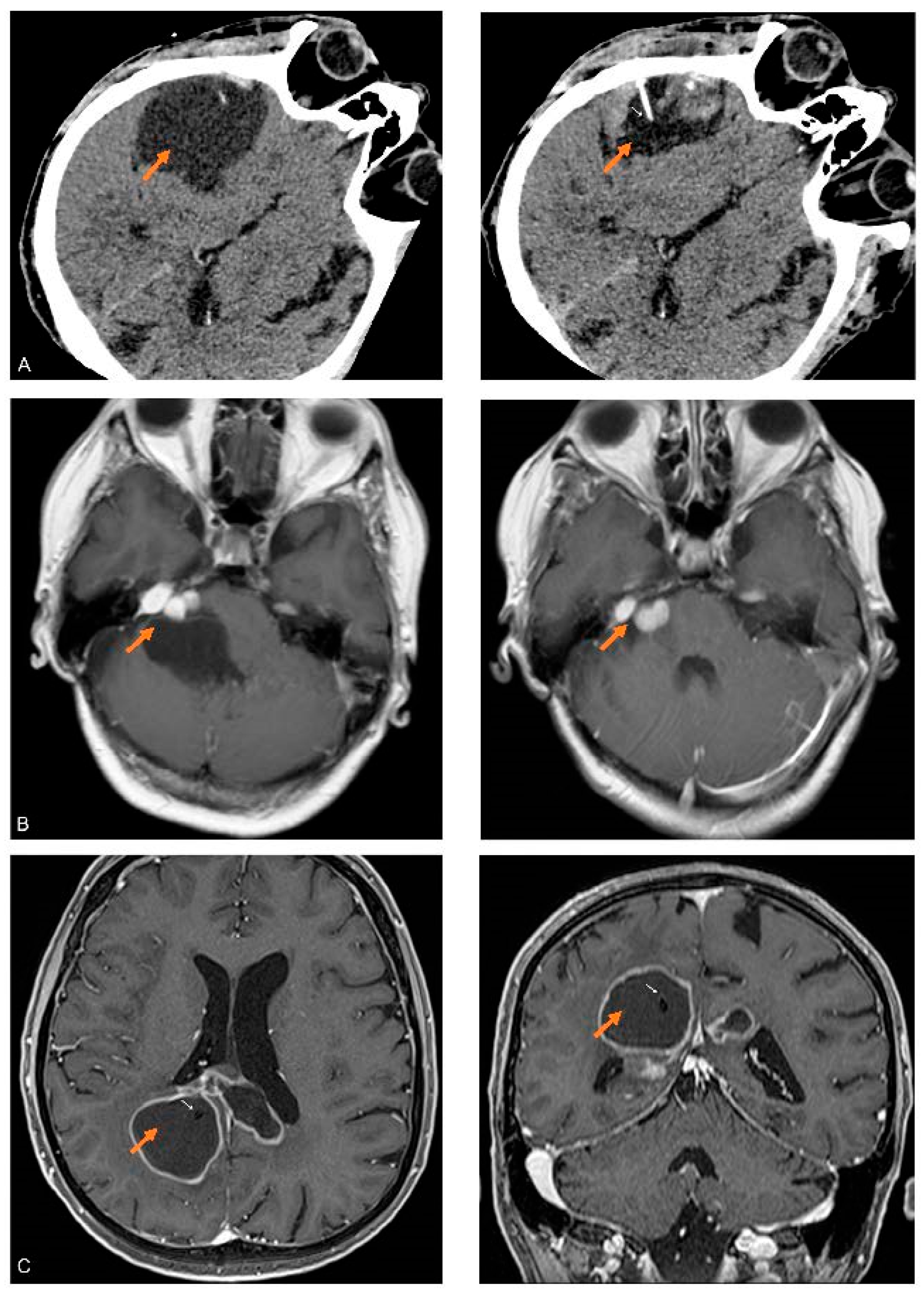
2.1. Patients
2.2. Sample Preparation and LC-MS/MS Analysis
2.3. Extraction of Differentially Expressed Proteins
2.4. Network Analysis
2.5. Overrepresentation Analysis
2.6. Statistical Analysis
3. Results
3.1. Protein Detected and Reproducibility
3.2. Proteins Detected in the Cystic Fluid of All Tumors
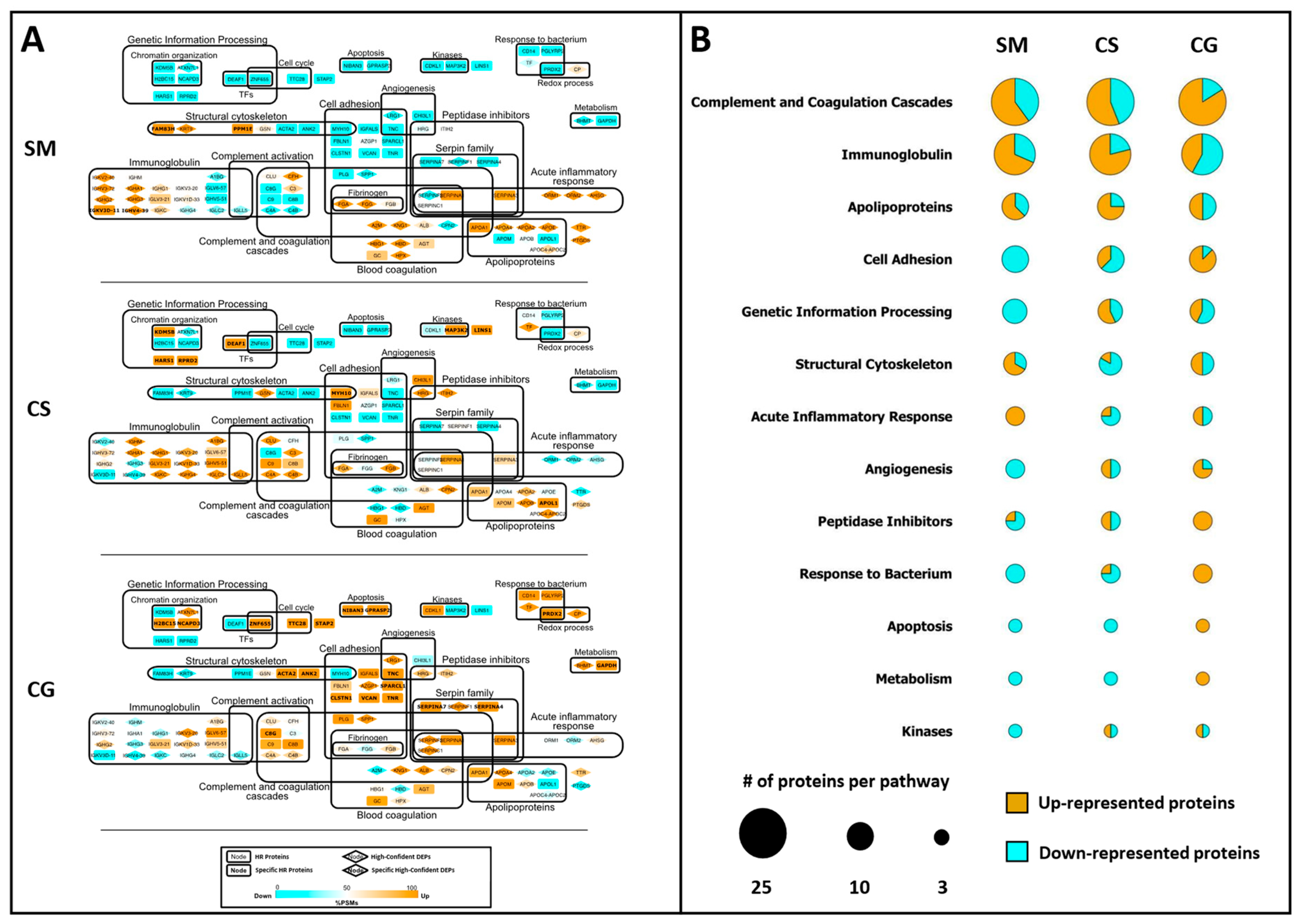
3.3. Label-Free Differential Analysis and Systems Biology
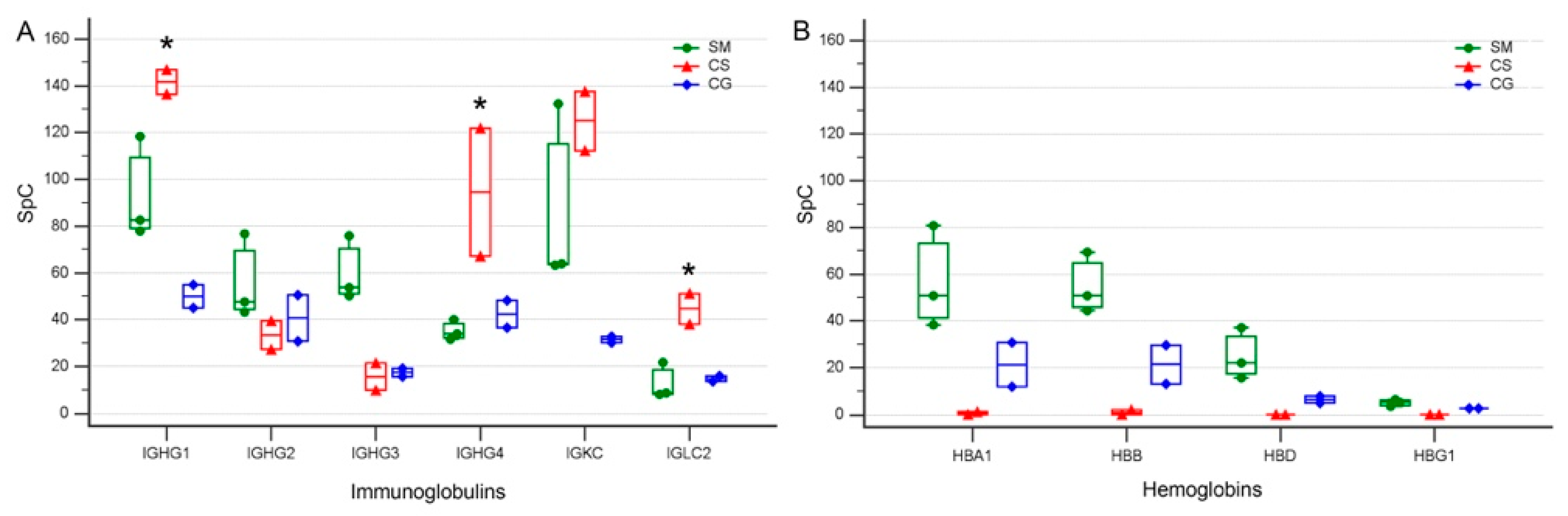
3.4. Levels of Specific Immunoglobulin Are Different in Cystic Fluid from Different Tumors
3.5. Protein Overrepresentation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Curtin, L.; Whitmire, P.; Rickertsen, C.R.; Mazza, G.L.; Canoll, P.; Johnston, S.K.; Mrugala, M.M.; Swanson, K.R.; Hu, L.S. Assessment of Prognostic Value of Cystic Features in Glioblastoma Relative to Sex and Treatment With Standard-of-Care. Front. Oncol. 2020, 10, 580750. [Google Scholar] [CrossRef]
- Westphal, M.; Nausch, H.; Herrmann, H.D. Cyst Fluids of Malignant Human Brain Tumors Contain Substances That Stimulate the Growth of Cultured Human Gliomas of Various Histological Type. Neurosurgery 1989, 25, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Murray, K.J.; Ausman, J.I.; Chou, S.N.; Douglas, S.D. Immunoproteins in Human Brain Tumor Cyst Fluids. J. Neurosurg. 1977, 46, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Lohle, P.N.; Wurzer, H.A.; Seelen, P.J.; Kingma, L.M.; Go, K.G. The Pathogenesis of Cysts Accompanying Intra-Axial Primary and Metastatic Tumors of the Central Nervous System. J. Neurooncol. 1998, 40, 277–285. [Google Scholar] [CrossRef]
- Lohle, P.N.; van Mameren, H.; Zwinderman, K.H.; Teepen, H.L.; Go, K.G.; Wilmink, J.T. On the Pathogenesis of Brain Tumour Cysts: A Volumetric Study of Tumour, Oedema and Cyst. Neuroradiology 2000, 42, 639–642. [Google Scholar] [CrossRef]
- Lonser, R.R.; Vortmeyer, A.O.; Butman, J.A.; Glasker, S.; Finn, M.A.; Ammerman, J.M.; Merrill, M.J.; Edwards, N.A.; Zhuang, Z.; Oldfield, E.H. Edema Is a Precursor to Central Nervous System Peritumoral Cyst Formation. Ann. Neurol. 2005, 58, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Vaquero, J.; Zurita, M.; Oya, S.; Coca, S.; Salas, C. Vascular Permeability Factor Expression in Cerebellar Hemangioblastomas: Correlation with Tumor-Associated Cysts. J. Neurooncol. 1999, 41, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Strugar, J.; Rothbart, D.; Harrington, W.; Criscuolo, G.R. Vascular Permeability Factor in Brain Metastases: Correlation with Vasogenic Brain Edema and Tumor Angiogenesis. J. Neurosurg. 1994, 81, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Strugar, J.G.; Criscuolo, G.R.; Rothbart, D.; Harrington, W.N. Vascular Endothelial Growth/Permeability Factor Expression in Human Glioma Specimens: Correlation with Vasogenic Brain Edema and Tumor-Associated Cysts. J. Neurosurg. 1995, 83, 682–689. [Google Scholar] [CrossRef] [PubMed]
- Vaquero, J.; Zurita, M.; de Oya, S.; Coca, S.; Morales, C.; Salas, C. Expression of Vascular Permeability Factor in Craniopharyngioma. J. Neurosurg. 1999, 91, 831–834. [Google Scholar] [CrossRef]
- Mokhtari, S. Mechanisms of Cyst Formation in Metastatic Lymph Nodes of Head and Neck Squamous Cell Carcinoma. Diagn. Pathol. 2012, 7, 6. [Google Scholar] [CrossRef]
- Huntoon, K.; Wu, T.; Elder, J.B.; Butman, J.A.; Chew, E.Y.; Linehan, W.M.; Oldfield, E.H.; Lonser, R.R. Biological and Clinical Impact of Hemangioblastoma-Associated Peritumoral Cysts in von Hippel-Lindau Disease. J. Neurosurg. 2016, 124, 971–976. [Google Scholar] [CrossRef]
- Villanueva, K.G.; Rea, N.D.; Krieger, M.D. Novel Surgical and Radiologic Risk Factors for Progression or Recurrence of Pediatric Pilocytic Astrocytoma. Pediatr. Neurosurg. 2019, 54, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, O.; Zagzag, D.; Kelly, P.; Golfinos, J.; Levine, P.H. Cytological Diagnosis of Cystic Brain Tumors: A Retrospective Study of 88 Cases. Diagn. Cytopathol. 2004, 31, 221–228. [Google Scholar] [CrossRef]
- Gläsker, S.; Vortmeyer, A.O.; Lonser, R.R.; Lubensky, I.A.; Okamoto, H.; Xia, J.B.; Li, J.; Milne, E.; Kowalak, J.A.; Oldfield, E.H.; et al. Proteomic Analysis of Hemangioblastoma Cyst Fluid. Cancer Biol. Ther. 2006, 5, 549–553. [Google Scholar] [CrossRef]
- Massimi, L.; Martelli, C.; Caldarelli, M.; Castagnola, M.; Desiderio, C. Proteomics in Pediatric Cystic Craniopharyngioma. Brain Pathol. 2017, 27, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Hara, M.; Yokota, H.; Ogashiwa, M.; Takeuchi, K. Biochemical monitoring of postoperative glioma (author’s transl). No Shinkei 1981, 33, 505–511. [Google Scholar]
- Di Silvestre, D.; Brambilla, F.; Mauri, P.L. Multidimensional Protein Identification Technology for Direct-Tissue Proteomics of Heart. In Heart Proteomics; Vivanco, F., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 1005, pp. 25–38. ISBN 978-1-62703-385-5. [Google Scholar]
- Azzalin, A.; Brambilla, F.; Arbustini, E.; Basello, K.; Speciani, A.; Mauri, P.; Bezzi, P.; Magrassi, L. A New Pathway Promotes Adaptation of Human Glioblastoma Cells to Glucose Starvation. Cells 2020, 9, 1249. [Google Scholar] [CrossRef] [PubMed]
- De Palma, A.; Agresta, A.M.; Viglio, S.; Rossi, R.; D’Amato, M.; Di Silvestre, D.; Mauri, P.; Iadarola, P. A Shotgun Proteomic Platform for a Global Mapping of Lymphoblastoid Cells to Gain Insight into Nasu-Hakola Disease. Int. J. Mol. Sci. 2021, 22, 9959. [Google Scholar] [CrossRef] [PubMed]
- Ducret, A.; Van Oostveen, I.; Eng, J.K.; Yates, J.R.; Aebersold, R. High Throughput Protein Characterization by Automated Reverse-Phase Chromatography/Electrospray Tandem Mass Spectrometry. Protein Sci. 1998, 7, 706–719. [Google Scholar] [CrossRef]
- Käll, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-Supervised Learning for Peptide Identification from Shotgun Proteomics Datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef]
- Vigani, G.; Di Silvestre, D.; Agresta, A.M.; Donnini, S.; Mauri, P.; Gehl, C.; Bittner, F.; Murgia, I. Molybdenum and Iron Mutually Impact Their Homeostasis in Cucumber (Cucumis sativus) Plants. New Phytol. 2017, 213, 1222–1241. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein–Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING V10: Protein-Protein Interaction Networks, Integrated over the Tree of Life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape Plugin to Assess Overrepresentation of Gene Ontology Categories in Biological Networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed]
- Sereni, L.; Castiello, M.C.; Di Silvestre, D.; Della Valle, P.; Brombin, C.; Ferrua, F.; Cicalese, M.P.; Pozzi, L.; Migliavacca, M.; Bernardo, M.E.; et al. Lentiviral Gene Therapy Corrects Platelet Phenotype and Function in Patients with Wiskott-Aldrich Syndrome. J. Allergy Clin. Immunol. 2019, 144, 825–838. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The Reactome Pathway Knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.; Mi, H. PANTHER: Making Genome-scale Phylogenetics Accessible to All. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef] [PubMed]
- Dey, K.K.; Wang, H.; Niu, M.; Bai, B.; Wang, X.; Li, Y.; Cho, J.-H.; Tan, H.; Mishra, A.; High, A.A.; et al. Deep Undepleted Human Serum Proteome Profiling toward Biomarker Discovery for Alzheimer’s Disease. Clin. Proteom. 2019, 16, 16. [Google Scholar] [CrossRef]
- Bader, J.M.; Geyer, P.E.; Müller, J.B.; Strauss, M.T.; Koch, M.; Leypoldt, F.; Koertvelyessy, P.; Bittner, D.; Schipke, C.G.; Incesoy, E.I.; et al. Proteome Profiling in Cerebrospinal Fluid Reveals Novel Biomarkers of Alzheimer’s Disease. Mol. Syst. Biol. 2020, 16, e9356. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Conradt, H.S.; Gross, G.; Nimtz, M.; Lottspeich, F.; Wurster, U. Purification and Chemical Characterization of Beta-Trace Protein from Human Cerebrospinal Fluid: Its Identification as Prostaglandin D Synthase. J. Neurochem. 1993, 61, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Urade, Y. Biochemical and Structural Characteristics, Gene Regulation, Physiological, Pathological and Clinical Features of Lipocalin-Type Prostaglandin D2 Synthase as a Multifunctional Lipocalin. Front. Physiol. 2021, 12, 718002. [Google Scholar] [CrossRef] [PubMed]
- Ghantasala, S.; Pai, M.G.J.; Biswas, D.; Gahoi, N.; Mukherjee, S.; Kp, M.; Nissa, M.U.; Srivastava, A.; Epari, S.; Shetty, P.; et al. Multiple Reaction Monitoring-Based Targeted Assays for the Validation of Protein Biomarkers in Brain Tumors. Front. Oncol. 2021, 11, 548243. [Google Scholar] [CrossRef] [PubMed]
- Regelsberger, J.; Hagel, C.; Emami, P.; Ries, T.; Heese, O.; Westphal, M. Secretory Meningiomas: A Benign Subgroup Causing Life-Threatening Complications. Neuro Oncol. 2009, 11, 819–824. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, M.; Paetau, A. Mapping of the Keratin Polypeptides in Meningiomas of Different Types: An Immunohistochemical Analysis of 463 Cases. Hum. Pathol. 2002, 33, 590–598. [Google Scholar] [CrossRef]
- Dahlberg, D.; Rummel, J.; Distante, S.; De Souza, G.A.; Stensland, M.E.; Mariussen, E.; Rootwelt, H.; Voie, Ø.; Hassel, B. Glioblastoma Microenvironment Contains Multiple Hormonal and Non-Hormonal Growth-Stimulating Factors. Fluids Barriers CNS 2022, 19, 45. [Google Scholar] [CrossRef]
- Evangelou, P.; Groll, M.; Oppermann, H.; Gaunitz, F.; Eisenlöffel, C.; Müller, W.; Eschrich, K.; Schänzer, A.; Nestler, U. Assessment of ApoC1, LuzP6, C12orf75 and OCC-1 in Cystic Glioblastoma Using MALDI-TOF Mass Spectrometry, Immunohistochemistry and QRT-PCR. Med. Mol. Morphol. 2019, 52, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Dziegielewska, K.M.; Saunders, N.R.; Schejter, E.J.; Zakut, H.; Zevin-Sonkin, D.; Zisling, R.; Soreq, H. Synthesis of Plasma Proteins in Fetal, Adult, and Neoplastic Human Brain Tissue. Dev. Biol. 1986, 115, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Qiu, F.; Huang, Y.; Saunders, N.R.; Habgood, M.D.; Dziegielewska, K.M. Age Dependent Contribution of Entry via the CSF to the Overall Brain Entry of Small and Large Hydrophilic Markers. Fluids Barriers CNS 2022, 19, 90. [Google Scholar] [CrossRef] [PubMed]
- Saunders, N.R.; Dziegielewska, K.M.; Møllgård, K.; Habgood, M.D. Physiology and Molecular Biology of Barrier Mechanisms in the Fetal and Neonatal Brain. J. Physiol. 2018, 596, 5723–5756. [Google Scholar] [CrossRef]
- Liddelow, S.A.; Dzięgielewska, K.M.; Møllgård, K.; Whish, S.C.; Noor, N.M.; Wheaton, B.J.; Gehwolf, R.; Wagner, A.; Traweger, A.; Bauer, H.; et al. Cellular Specificity of the Blood-CSF Barrier for Albumin Transfer across the Choroid Plexus Epithelium. PLoS ONE 2014, 9, e106592. [Google Scholar] [CrossRef] [PubMed]
- Jha, M.K.; Kim, J.-H.; Song, G.J.; Lee, W.-H.; Lee, I.-K.; Lee, H.-W.; An, S.S.A.; Kim, S.; Suk, K. Functional Dissection of Astrocyte-Secreted Proteins: Implications in Brain Health and Diseases. Prog. Neurobiol. 2018, 162, 37–69. [Google Scholar] [CrossRef] [PubMed]
- Rooprai, H.K.; Martin, A.J.; King, A.; Appadu, U.D.; Gullan, R.W.; Thomas, N.W.M.; Pilkington, G.J. Lack of Correlation Between Immunohistochemical Expression of SPARC and Invasion in Different Grades of Meningiomas. Anticancer. Res. 2020, 40, 3081–3089. [Google Scholar] [CrossRef] [PubMed]
- Enault, S.; Muñoz, D.; Simion, P.; Ventéo, S.; Sire, J.-Y.; Marcellini, S.; Debiais-Thibaud, M. Evolution of Dental Tissue Mineralization: An Analysis of the Jawed Vertebrate SPARC and SPARC-L Families. BMC Evol. Biol. 2018, 18, 127. [Google Scholar] [CrossRef] [PubMed]
- Turtoi, A.; Musmeci, D.; Naccarato, A.G.; Scatena, C.; Ortenzi, V.; Kiss, R.; Murtas, D.; Patsos, G.; Mazzucchelli, G.; De Pauw, E.; et al. Sparc-Like Protein 1 Is a New Marker of Human Glioma Progression. J. Proteome Res. 2012, 11, 5011–5021. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, F.; Narayanan, A.; Gallotti, A.L.; Pieri, V.; Mazzoleni, S.; Cominelli, M.; Rezzola, S.; Corsini, M.; Brugnara, G.; Altabella, L.; et al. Enhanced SPARCL1 Expression in Cancer Stem Cells Improves Preclinical Modeling of Glioblastoma by Promoting Both Tumor Infiltration and Angiogenesis. Neurobiol. Dis. 2020, 134, 104705. [Google Scholar] [CrossRef]
- Cosgrove, D.; Madison, J. Molecular and Cellular Mechanisms Underlying the Initiation and Progression of Alport Glomerular Pathology. Front. Med. 2022, 9, 846152. [Google Scholar] [CrossRef] [PubMed]
- Bose, S.; Pathireddy, S.; Baradhi, K.M.; Aeddula, N.R. Alport’s Syndrome and Intracranial Aneurysm: Mere Coincidence or Undiscovered Causal Relationship. BMJ Case Rep. 2019, 12, e228175. [Google Scholar] [CrossRef]
- Boethius, J.; Lefvert, A.K.; Sidén, A. Evidence of a Local Immune Activation in Cystic Brain Tumors. J. Neurosurg. 1990, 73, 933–935. [Google Scholar] [CrossRef]
- Abbritti, R.V.; Polito, F.; Cucinotta, M.; Lo Giudice, C.; Caffo, M.; Tomasello, C.; Germanò, A.; Aguennouz, M. Meningiomas and Proteomics: Focus on New Potential Biomarkers and Molecular Pathways. Cancer Genom. Proteom. 2016, 13, 369–379. [Google Scholar]
- Zhang, J.; Furuta, T.; Sabit, H.; Tamai, S.; Jiapaer, S.; Dong, Y.; Kinoshita, M.; Uchida, Y.; Ohtsuki, S.; Terasaki, T.; et al. Gelsolin Inhibits Malignant Phenotype of Glioblastoma and Is Regulated by MiR-654-5p and MiR-450b-5p. Cancer Sci. 2020, 111, 2413–2422. [Google Scholar] [CrossRef]
- Edvardsson Rasmussen, J.; Laurell, G.; Rask-Andersen, H.; Bergquist, J.; Eriksson, P.O. The Proteome of Perilymph in Patients with Vestibular Schwannoma. A Possibility to Identify Biomarkers for Tumor Associated Hearing Loss? PLoS ONE 2018, 13, e0198442. [Google Scholar] [CrossRef]
- Lassaletta, L.; Calvino, M.; Morales-Puebla, J.M.; Lapunzina, P.; Rodriguez-de la Rosa, L.; Varela-Nieto, I.; Martinez-Glez, V. Biomarkers in Vestibular Schwannoma–Associated Hearing Loss. Front. Neurol. 2019, 10, 978. [Google Scholar] [CrossRef] [PubMed]
- Nangami, G.N.; Sakwe, A.M.; Izban, M.G.; Rana, T.; Lammers, P.E.; Thomas, P.; Chen, Z.; Ochieng, J. Fetuin-A (Alpha 2HS Glycoprotein) Modulates Growth, Motility, Invasion, and Senescence in High-Grade Astrocytomas. Cancer Med. 2016, 5, 3532–3543. [Google Scholar] [CrossRef] [PubMed]
- Martelli, C.; Serra, R.; Inserra, I.; Rossetti, D.V.; Iavarone, F.; Vincenzoni, F.; Castagnola, M.; Urbani, A.; Tamburrini, G.; Caldarelli, M.; et al. Investigating the Protein Signature of Adamantinomatous Craniopharyngioma Pediatric Brain Tumor Tissue: Towards the Comprehension of Its Aggressive Behavior. Dis. Markers 2019, 2019, 3609789. [Google Scholar] [CrossRef] [PubMed]
- Martelli, C.; Iavarone, F.; Vincenzoni, F.; Rossetti, D.V.; D’Angelo, L.; Tamburrini, G.; Caldarelli, M.; Di Rocco, C.; Messana, I.; Castagnola, M.; et al. Proteomic Characterization of Pediatric Craniopharyngioma Intracystic Fluid by LC-MS Top-down/Bottom-up Integrated Approaches. Electrophoresis 2014, 35, 2172–2183. [Google Scholar] [CrossRef] [PubMed]
- Müller, H.L.; Merchant, T.E.; Warmuth-Metz, M.; Martinez-Barbera, J.-P.; Puget, S. Craniopharyngioma. Nat. Rev. Dis. Primers 2019, 5, 75. [Google Scholar] [CrossRef] [PubMed]
- Guttenplan, K.A.; Weigel, M.K.; Prakash, P.; Wijewardhane, P.R.; Hasel, P.; Rufen-Blanchette, U.; Münch, A.E.; Blum, J.A.; Fine, J.; Neal, M.C.; et al. Neurotoxic Reactive Astrocytes Induce Cell Death via Saturated Lipids. Nature 2021, 599, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Hok-A-Hin, Y.S.; Dijkstra, A.A.; Rábano, A.; Hoozemans, J.J.; Castillo, L.; Seelaar, H.; van Swieten, J.C.; Pijnenburg, Y.A.L.; Teunissen, C.E.; Del Campo, M. Apolipoprotein L1 Is Increased in Frontotemporal Lobar Degeneration Post-Mortem Brain but Not in Ante-Mortem Cerebrospinal Fluid. Neurobiol. Dis. 2022, 172, 105813. [Google Scholar] [CrossRef] [PubMed]
- Corraliza-Gomez, M.; Del Caño-Espinel, M.; Sanchez, D.; Ganfornina, M.D. The Neuroprotective Lipocalin Apolipoprotein D Stably Interacts with Specific Subtypes of Detergent-Resistant Membrane Domains in a Basigin-Independent Manner. Mol. Neurobiol. 2022, 59, 4015–4029. [Google Scholar] [CrossRef]
- Kuga, T.; Inoue, N.; Sometani, K.; Murataka, S.; Saraya, M.; Sugita, R.; Mikami, T.; Takeda, Y.; Taniguchi, M.; Nishida, K.; et al. The Conserved C-Terminal Residues of FAM83H Are Required for the Recruitment of Casein Kinase 1 to the Keratin Cytoskeleton. Sci. Rep. 2022, 12, 11819. [Google Scholar] [CrossRef] [PubMed]
- Haslund-Vinding, J.; Møller, J.R.; Ziebell, M.; Vilhardt, F.; Mathiesen, T. The Role of Systemic Inflammatory Cells in Meningiomas. Neurosurg. Rev. 2022, 45, 1205–1215. [Google Scholar] [CrossRef]
- Vandooren, J.; Itoh, Y. Alpha-2-Macroglobulin in Inflammation, Immunity and Infections. Front. Immunol. 2021, 12, 803244. [Google Scholar] [CrossRef]
- Sharma, S.; Ray, S.; Moiyadi, A.; Sridhar, E.; Srivastava, S. Quantitative Proteomic Analysis of Meningiomas for the Identification of Surrogate Protein Markers. Sci. Rep. 2014, 4, 7140. [Google Scholar] [CrossRef] [PubMed]
- Torres-Martin, M.; Lassaletta, L.; San-Roman-Montero, J.; De Campos, J.M.; Isla, A.; Gavilan, J.; Melendez, B.; Pinto, G.R.; Burbano, R.R.; Castresana, J.S.; et al. Microarray Analysis of Gene Expression in Vestibular Schwannomas Reveals SPP1/MET Signaling Pathway and Androgen Receptor Deregulation. Int. J. Oncol. 2013, 42, 848–862. [Google Scholar] [CrossRef] [PubMed]
- Vetrano, I.G.; Dei Cas, M.; Nazzi, V.; Eoli, M.; Innocenti, N.; Saletti, V.; Potenza, A.; Carrozzini, T.; Pollaci, G.; Gorla, G.; et al. The Lipid Asset Is Unbalanced in Peripheral Nerve Sheath Tumors. Int. J. Mol. Sci. 2021, 23, 61. [Google Scholar] [CrossRef] [PubMed]
- Graca, F.A.; Sheffield, N.; Puppa, M.; Finkelstein, D.; Hunt, L.C.; Demontis, F. A Large-Scale Transgenic RNAi Screen Identifies Transcription Factors That Modulate Myofiber Size in Drosophila. PLoS Genet. 2021, 17, e1009926. [Google Scholar] [CrossRef]
- Dertschnig, S.; Evans, P.; Santos e Sousa, P.; Manzo, T.; Ferrer, I.R.; Stauss, H.J.; Bennett, C.L.; Chakraverty, R. Graft-versus-Host Disease Reduces Lymph Node Display of Tissue-Restricted Self-Antigens and Promotes Autoimmunity. J. Clin. Investig. 2020, 130, 1896–1911. [Google Scholar] [CrossRef]
- Pietras, A.; Katz, A.M.; Ekström, E.J.; Wee, B.; Halliday, J.J.; Pitter, K.L.; Werbeck, J.L.; Amankulor, N.M.; Huse, J.T.; Holland, E.C. Osteopontin-CD44 Signaling in the Glioma Perivascular Niche Enhances Cancer Stem Cell Phenotypes and Promotes Aggressive Tumor Growth. Cell Stem Cell 2014, 14, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Akawi, N.A.; Al-Jasmi, F.; Al-Shamsi, A.M.; Ali, B.R.; Al-Gazali, L. LINS, a Modulator of the WNT Signaling Pathway, Is Involved in Human Cognition. Orphanet J. Rare Dis. 2013, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Kwon, M.; Jung, J.; Chae, H.B.; Lee, J.; Yoon, Y.-J.; Moon, I.S.; Lee, H.K.; Namkung, W.; Stankovic, K.M.; et al. Celastrol Suppresses the Growth of Vestibular Schwannoma in Mice by Promoting the Degradation of β-Catenin. Acta Pharmacol. Sin. 2022, 43, 2993–3001. [Google Scholar] [CrossRef] [PubMed]
- El Ayachi, I.; Baeza, N.; Fernandez, C.; Colin, C.; Scavarda, D.; Pesheva, P.; Figarella-Branger, D. KIAA0510, the 3’-Untranslated Region of the Tenascin-R Gene, and Tenascin-R Are Overexpressed in Pilocytic Astrocytomas. Neuropathol. Appl. Neurobiol. 2010, 36, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Bi, B.; Li, F.; Guo, J.; Li, C.; Jing, R.; Lv, X.; Chen, X.; Wang, F.; Azadzoi, K.M.; Wang, L.; et al. Label-Free Quantitative Proteomics Unravels the Importance of RNA Processing in Glioma Malignancy. Neuroscience 2017, 351, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, M.; Loiselle, M.; Vaganay, C.; Pons, S.; Letavernier, E.; Demonchy, J.; Fodil, S.; Nouacer, M.; Placier, S.; Frère, P.; et al. Tumor Lysis Syndrome and AKI: Beyond Crystal Mechanisms. J. Am. Soc. Nephrol. 2022, 33, 1154–1171. [Google Scholar] [CrossRef]
- Nacev, B.A.; Feng, L.; Bagert, J.D.; Lemiesz, A.E.; Gao, J.; Soshnev, A.A.; Kundra, R.; Schultz, N.; Muir, T.W.; Allis, C.D. The Expanding Landscape of “oncohistone” Mutations in Human Cancers. Nature 2019, 567, 473–478. [Google Scholar] [CrossRef]
- Sahu, V.; Lu, C. Oncohistones: Hijacking the Histone Code. Annu. Rev. Cancer Biol. 2022, 6, 293–312. [Google Scholar] [CrossRef] [PubMed]
- Mena, H.A.; Carestia, A.; Scotti, L.; Parborell, F.; Schattner, M.; Negrotto, S. Extracellular Histones Reduce Survival and Angiogenic Responses of Late Outgrowth Progenitor and Mature Endothelial Cells. J. Thromb. Haemost. 2016, 14, 397–410. [Google Scholar] [CrossRef]
- Lim, C.H.; Adav, S.S.; Sze, S.K.; Choong, Y.K.; Saravanan, R.; Schmidtchen, A. Thrombin and Plasmin Alter the Proteome of Neutrophil Extracellular Traps. Front. Immunol. 2018, 9, 1554. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.A.; Lyon, M.; Simpson, D.; Mason, D.; Beynon, R.J.; Moots, R.J.; Wright, H.L. Caught in a Trap? Proteomic Analysis of Neutrophil Extracellular Traps in Rheumatoid Arthritis and Systemic Lupus Erythematosus. Front. Immunol. 2019, 10, 423. [Google Scholar] [CrossRef]
- Papayannopoulos, V. Neutrophil Extracellular Traps in Immunity and Disease. Nat. Rev. Immunol. 2018, 18, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Slowinski, J.; Mazurek, U.; Bierzynska-Macyszyn, G.; Widel, M.; Latocha, M.; Glogowska-Ligus, J.; Stomal, M.; Mrowka, R. Cell Proliferative Activity Estimated by Histone H2B MRNA Level Correlates with Cytogenetic Damage Induced by Radiation in Human Glioblastoma Cell Lines. J. Neurooncol. 2005, 71, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Kostrikina, I.A.; Buneva, V.N.; Granieri, E.; Nevinsky, G.A. Extreme Diversity of IgGs Against Histones, DNA, and Myelin Basic Protein in the Cerebrospinal Fluid and Blood of Patients with Multiple Sclerosis. Biomolecules 2020, 10, E630. [Google Scholar] [CrossRef] [PubMed]


| Tu | Accession | Gene | Protein | MW (KDa) | pI |
|---|---|---|---|---|---|
| SM | A0A494C1T9 | FAM83H | Protein FAM83H | 149 | 6.71 |
| SM | P01824 | IGHV4-39 | Immunoglobulin heavy variable 4–39 | 13.9 | 9.26 |
| SM | A0A0A0MRZ8 | IGKV3D-11 | Immunoglobulin kappa variable 3D-11 | 12.6 | 5.29 |
| SM | Q8WY54 | PPM1E | Protein phosphatase 1E | 83.9 | 5.01 |
| CS | O14791 | APOL1 | Apolipoprotein L1 | 43.9 | 5.81 |
| CS | A0A0J9YWD6 | DEAF1 | Deformed epidermal autoregulatory factor 1 homolog (Frag.) | 17.7 | 7.11 |
| CS | E7ETE2 | HARS1 | Histidyl-tRNA synthetase 1 | 57.8 | 5.88 |
| CS | Q9UGL1 | KDM5B | Lysine-specific demethylase 5B | 175.5 | 6.7 |
| CS | Q8NG48 | LINS1 | Protein Lines homolog 1 | 85.8 | 6.52 |
| CS | Q9Y2U5 | MAP3K2 | Mitogen-activated protein kinase kinase kinase 2 | 69.7 | 8 |
| CS | P35580 | MYH10 | Myosin-10 | 228.9 | 5.54 |
| CS | Q5VT52 | RPRD2 | Regulation of nuclear pre-mRNA domain-containing protein 2 | 155.9 | 7.42 |
| CG | P62736 | ACTA2 | Actin, aortic smooth muscle | 42 | 5.39 |
| CG | A0A5F9ZHS1 | ANK2 | Ankyrin-2 | 436.8 | 5.16 |
| CG | P07360 | C8G | Complement component C8 gamma chain | 22.3 | 8.31 |
| CG | O94985 | CLSTN1 | Calsyntenin-1 | 109.7 | 4.91 |
| CG | P04406 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 36 | 8.46 |
| CG | Q96D09 | GPRASP2 | G-protein coupled receptor-associated sorting protein 2 | 93.7 | 5.01 |
| CG | U3KQK0 | H2BC15 | Histone H2B | 18.8 | 10.54 |
| CG | P42695 | NCAPD3 | Condensin-2 complex subunit D3 | 168.8 | 7.5 |
| CG | Q86XR2 | NIBAN3 | Protein Niban 3 | 77.4 | 8.63 |
| CG | P32119 | PRDX2 | Peroxiredoxin-2 | 21.9 | 5.97 |
| CG | P29622 | SERPINA4 | Kallistatin | 48.5 | 7.75 |
| CG | P05543 | SERPINA7 | Thyroxine-binding globulin | 46.3 | 6.3 |
| CG | Q14515 | SPARCL1 | SPARC-like protein 1 | 75.2 | 4.81 |
| CG | Q9UGK3 | STAP2 | Signal-transducing adaptor protein 2 | 44.9 | 8.16 |
| CG | P24821 | TNC | Tenascin | 240.7 | 4.89 |
| CG | Q92752 | TNR | Tenascin-R | 149.5 | 4.82 |
| CG | Q96AY4 | TTC28 | Tetratricopeptide repeat protein 28 | 270.7 | 6.89 |
| CG | P13611 | VCAN | Versican core protein | 372.6 | 4.51 |
| CG | Q8N720 | ZNF655 | Zinc-finger protein 655 | 57.4 | 7.14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magrassi, L.; Brambilla, F.; Viganò, R.; Di Silvestre, D.; Benazzi, L.; Bellantoni, G.; Danesino, G.M.; Comincini, S.; Mauri, P. Proteomic Analysis on Sequential Samples of Cystic Fluid Obtained from Human Brain Tumors. Cancers 2023, 15, 4070. https://doi.org/10.3390/cancers15164070
Magrassi L, Brambilla F, Viganò R, Di Silvestre D, Benazzi L, Bellantoni G, Danesino GM, Comincini S, Mauri P. Proteomic Analysis on Sequential Samples of Cystic Fluid Obtained from Human Brain Tumors. Cancers. 2023; 15(16):4070. https://doi.org/10.3390/cancers15164070
Chicago/Turabian StyleMagrassi, Lorenzo, Francesca Brambilla, Raffaello Viganò, Dario Di Silvestre, Louise Benazzi, Giuseppe Bellantoni, Gian Marco Danesino, Sergio Comincini, and Pierluigi Mauri. 2023. "Proteomic Analysis on Sequential Samples of Cystic Fluid Obtained from Human Brain Tumors" Cancers 15, no. 16: 4070. https://doi.org/10.3390/cancers15164070
APA StyleMagrassi, L., Brambilla, F., Viganò, R., Di Silvestre, D., Benazzi, L., Bellantoni, G., Danesino, G. M., Comincini, S., & Mauri, P. (2023). Proteomic Analysis on Sequential Samples of Cystic Fluid Obtained from Human Brain Tumors. Cancers, 15(16), 4070. https://doi.org/10.3390/cancers15164070