Synthesis and Biological Activity of a VHL-Based PROTAC Specific for p38α
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
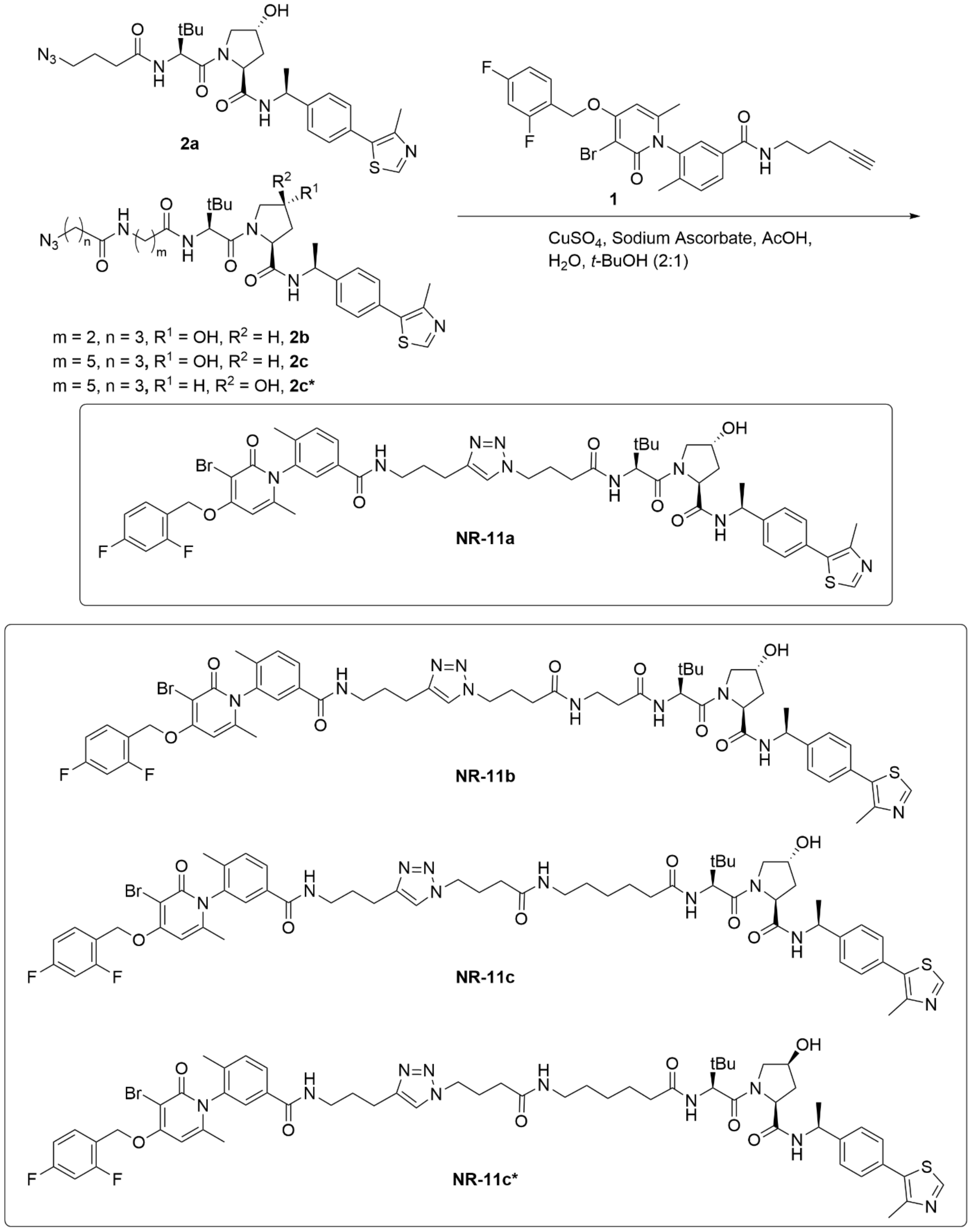
2.1. Chemistry
2.2. General Method CuCAAC
2.2.1. (2S,4R)-1-((S)-2-(4-(4-(2-(3-(3-bromo-4-((2,4-difluorobenzyl)oxy)-6-methyl-2-oxopyridin-1(2H)-yl)-4-methylbenzamido)ethyl)-1H-1,2,3-triazol-1-yl)butanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (NR-11a)
2.2.2. (2S,4R)-1-((S)-2-(3-(4-(4-(2-(3-(3-bromo-4-((2,4-difluorobenzyl)oxy)-6-methyl-2-oxopyridin-1(2H)-yl)-4-methylbenzamido)ethyl)-1H-1,2,3-triazol-1-yl)butanamido)propanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (NR-11b)
2.2.3. (2S,4R)-1-((S)-2-(6-(4-(4-(2-(3-(3-bromo-4-((2,4-difluorobenzyl)oxy)-6-methyl-2-oxopyridin-1(2H)-yl)-4-methylbenzamido)ethyl)-1H-1,2,3-triazol-1-yl)butanamido)hexanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (NR-11c)
2.2.4. (2S,4S)-1-((S)-2-(6-(4-(4-(3-(3-(3-bromo-4-((2,4-difluorobenzyl)oxy)-6-methyl-2-oxopyridin-1(2H)-yl)-4-methylbenzamido)propyl)-1H-1,2,3-triazol-1-yl)butanamido)hexanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (NR-11c*)
2.3. Cell Culture
2.4. Immunoblotting
2.5. Thermal Shift Assays
2.6. RNA Extraction and Gene Expression Analysis
2.7. Treatment of Mice with PROTACs
3. Results
3.1. Design of New VHL-Based PROTACs to Target p38α
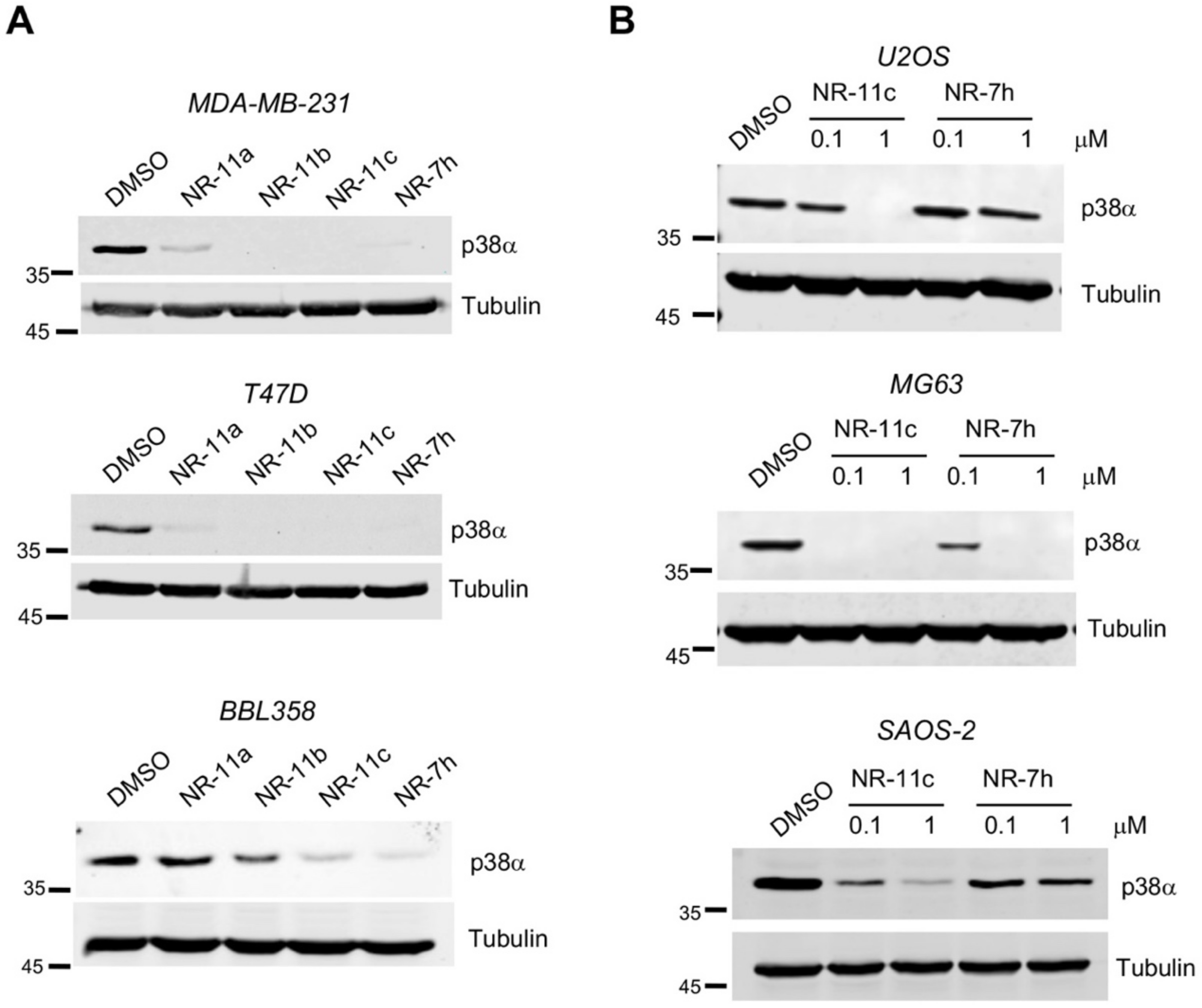
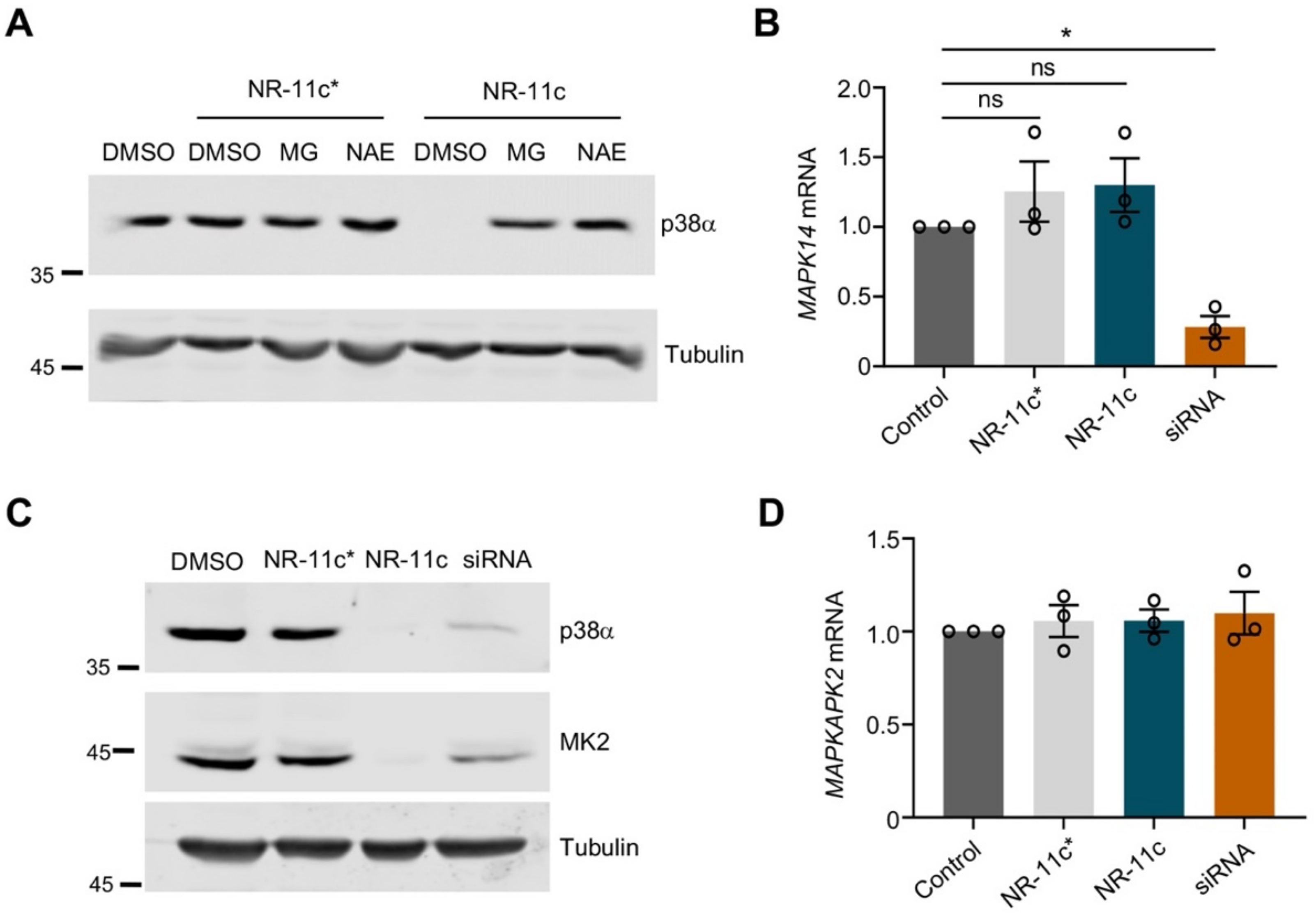
3.2. Characterization of the PROTAC NR-11c
3.3. Effects of the PROTAC NR-11c In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cuadrado, A.; Nebreda, A.R. Mechanisms and functions of p38 MAPK signalling. Biochem. J. 2010, 429, 403–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Wu, J.; Silke, J. An overview of mammalian p38 mitogen-activated protein kinases, central regulators of cell stress and receptor signaling. F1000Research 2020, 9, 1653. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S.; Davis, R.J. Cell Signaling and Stress Responses. Cold Spring Harb. Perspect. Biol. 2016, 8, a006072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuenda, A.; Rousseau, S. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim. Biophys. Acta 2007, 1773, 1358–1375. [Google Scholar] [CrossRef] [Green Version]
- Sanz-Ezquerro, J.J.; Cuenda, A. Molecular Sciences Editorial p38 Signalling Pathway. Int. J. Mol. Sci 2021, 22, 1003. [Google Scholar] [CrossRef]
- Adams, R.H.; Porras, A.; Alonso, G.; Jones, M.; Vintersten, K.; Panelli, S.; Valladares, A.; Perez, L.; Klein, R.; Nebreda, A.R. Essential role of p38alpha MAP kinase in placental but not embryonic cardiovascular development. Mol. Cell 2000, 6, 109–116. [Google Scholar] [CrossRef]
- Mudgett, J.S.; Ding, J.; Guh-Siesel, L.; Chartrain, N.A.; Yang, L.; Gopal, S.; Shen, M.M. Essential role for p38alpha mitogen-activated protein kinase in placental angiogenesis. Proc. Natl. Acad. Sci. USA 2000, 97, 10454–10459. [Google Scholar] [CrossRef] [Green Version]
- Beardmore, V.A.; Hinton, H.J.; Eftychi, C.; Apostolaki, M.; Armaka, M.; Darragh, J.; McIlrath, J.; Carr, J.M.; Armit, L.J.; Clacher, C.; et al. Generation and characterization of p38beta (MAPK11) gene-targeted mice. Mol. Cell. Biol. 2005, 25, 10454–10464. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Bonilla, V.; Perdiguero, E.; Gresh, L.; Serrano, A.L.; Zamora, M.; Sousa-Victor, P.; Jardí, M.; Wagner, E.F.; Muñoz-Cánoves, P. Efficient adult skeletal muscle regeneration in mice deficient in p38beta, p38gamma and p38delta MAP kinases. Cell Cycle 2008, 7, 2208–2214. [Google Scholar] [CrossRef]
- Del Barco Barrantes, I.; Coya, J.M.; Maina, F.; Arthurd, J.S.C.; Nebreda, A.R. Genetic analysis of specific and redundant roles for p38alpha and p38beta MAPKs during mouse development. Proc. Natl. Acad. Sci. USA 2011, 108, 12764–12769. [Google Scholar] [CrossRef]
- Igea, A.; Nebreda, A.R. The Stress Kinase p38a as a Target for Cancer Therapy. Can. Res. 2015, 75, 3997–4002. [Google Scholar] [CrossRef] [Green Version]
- Canovas, B.; Nebreda, A.R. Diversity and versatility of p38 kinase signalling in health and disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 346–366. [Google Scholar] [CrossRef]
- Martínez-Limón, A.; Joaquin, M.; Caballero, M.; Posas, F.; de Nadal, E. The p38 Pathway: From Biology to Cancer Therapy. Int. J. Mol. Sci. 2020, 21, 1913. [Google Scholar] [CrossRef] [Green Version]
- Haller, V.; Nahidino, P.; Forster, M.; Laufer, S.A. An updated patent review of p38 MAP kinase inhibitors (2014–2019). Expert Opin. Ther. Pat. 2020, 30, 453–466. [Google Scholar] [CrossRef]
- Burslem, G.M.; Crews, C.M. Proteolysis-Targeting Chimeras as Therapeutics and Tools for Biological Discovery. Cell 2020, 181, 102–114. [Google Scholar] [CrossRef]
- Nalawansha, D.A.; Crews, C.M. PROTACs: An Emerging Therapeutic Modality in Precision Medicine. Cell Chem. Biol. 2020, 27, 998–1014. [Google Scholar] [CrossRef]
- Ishida, T.; Ciulli, A. E3 Ligase Ligands for PROTACs: How They Were Found and How to Discover New Ones. SLAS Discov. 2021, 26, 484–502. [Google Scholar] [CrossRef]
- Donoghue, C.; Cubillos-Rojas, M.; Gutierrez-Prat, N.; Sanchez-Zarzalejo, C.; Verdaguer, X.; Riera, A.; Nebreda, A.R. Optimal linker length for small molecule PROTACs that selectively target p38α and p38β for degradation. Eur. J. Med. Chem. 2020, 201, 112451. [Google Scholar] [CrossRef]
- Loren, G.; Espuny, I.; Llorente, A.; Donoghue, C.; Verdaguer, X.; Gomis, R.R.; Riera, A. Design and optimization of oestrogen receptor PROTACs based on 4-hydroxytamoxifen. Eur. J. Med. Chem. 2022, 243, 114770. [Google Scholar] [CrossRef]
- Cánovas, B.; Igea, A.; Sartori, A.A.; Gomis, R.R.; Paull, T.T.; Isoda, M.; Pérez-Montoyo, H.; Serra, V.; González-Suárez, E.; Stracker, T.H.; et al. Targeting p38α Increases DNA Damage, Chromosome Instability, and the Anti-tumoral Response to Taxanes in Breast Cancer Cells. Cancer Cell 2018, 33, 1094–1110.e1098. [Google Scholar] [CrossRef]
- Cecchini, C.; Pannilunghi, S.; Tardy, S.; Scapozza, L. From Conception to Development: Investigating PROTACs Features for Improved Cell Permeability and Successful Protein Degradation. Front. Chem. 2021, 9, 672267. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Prat, N.; Cubillos-Rojas, M.; Cánovas, B.; Kuzmanic, A.; Gupta, J.; Igea, A.; Llonch, E.; Gaestel, M.; Nebreda, A.R. MK2 degradation as a sensor of signal intensity that controls stress-induced cell fate. Proc. Natl. Acad. Sci. USA. 2021, 118, e2024562118. [Google Scholar] [CrossRef] [PubMed]
- White, A.; Pargellis, C.A.; Studts, J.M.; Werneburg, B.G.; Farmer, B.T. Molecular basis of MAPK-activated protein kinase 2:p38 assembly. Proc. Natl. Acad. Sci. USA 2007, 104, 6353–6358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saokham, P.; Muankaew, C.; Jansook, P.; Loftsson, T. Solubility of Cyclodextrins and Drug/Cyclodextrin Complexes. Molecules 2018, 23, 1161. [Google Scholar] [CrossRef] [Green Version]
- Brewster, M.E.; Loftsson, T. Cyclodextrins as pharmaceutical solubilizers. Adv. Drug Deliv. Rev. 2007, 59, 645–666. [Google Scholar] [CrossRef]
- Li, X.; Pu, W.; Zheng, Q.; Ai, M.; Chen, S.; Peng, Y. Proteolysis-targeting chimeras (PROTACs) in cancer therapy. Mol. Cancer 2022, 21, 99. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Y.; Wang, J.; Xing, D. VHL-based PROTACs as potential therapeutic agents: Recent progress and perspectives. Eur. J. Med. Chem. 2022, 227, 113906. [Google Scholar] [CrossRef]
- Békés, M.; Langley, D.R.; Crews, C.M. PROTAC targeted protein degraders: The past is prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef]
- Pike, A.; Williamson, B.; Harlfinger, S.; Martin, S.; McGinnity, D.F. Optimising proteolysis-targeting chimeras (PROTACs) for oral drug delivery: A drug metabolism and pharmacokinetics perspective. Drug Discov. Today 2020, 25, 1793–1800. [Google Scholar] [CrossRef]
- Kofink, C.; Trainor, N.; Mair, B.; Wohrle, S.; Wurm, M.; Mischerikow, N.; Roy, M.J.; Bader, G.; Greb, P.; Garavel, G.; et al. A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo. Nat. Commun. 2022, 13, 5969. [Google Scholar] [CrossRef]
- Luo, G.; Li, Z.; Lin, X.; Li, X.; Chen, Y.; Xi, K.; Xiao, M.; Wei, H.; Zhu, L.; Xiang, H. Discovery of an orally active VHL-recruiting PROTAC that achieves robust HMGCR degradation and potent hypolipidemic activity in vivo. Acta Pharm. Sin. B 2021, 11, 1300–1314. [Google Scholar] [CrossRef]
- Diehl, C.J.; Ciulli, A. Discovery of small molecule ligands for the von Hippel-Lindau (VHL) E3 ligase and their use as inhibitors and PROTAC degraders. Chem. Soc. Rev. 2022, 51, 8216–8257. [Google Scholar] [CrossRef]
- Liu, X.; Kalogeropulou, A.F.; Domingos, S.; Makukhin, N.; Nirujogi, R.S.; Singh, F.; Shpiro, N.; Saalfrank, A.; Sammler, E.; Ganley, I.G.; et al. Discovery of XL01126: A Potent, Fast, Cooperative, Selective, Orally Bioavailable and Blood Brain Barrier Penetrant Prote-olysis Targeting Chimera Degrader of Leucine Rich Repeat Kinase 2. J. Am. Chem. Soc. 2022, 144, 16930–16952. [Google Scholar] [CrossRef]
- Luo, X.; Archibeque, I.; Dellamaggiore, K.; Smither, K.; Homann, O.; Lipford, J.R.; Mohl, D. Profiling of diverse tumor types establishes the broad utility of VHL-based ProTaCs and triages candidate ubiquitin ligases. iScience 2022, 25, 103985. [Google Scholar] [CrossRef]
- Melero, I.; Castanon, E.; Alvarez, M.; Champiat, S.; Marabelle, A. Intratumoural administration and tumour tissue targeting of cancer immunotherapies. Nat. Rev. Clin. Oncol. 2021, 18, 558–576. [Google Scholar] [CrossRef]
- Smith, B.E.; Wang, S.L.; Jaime-Figueroa, S.; Harbin, A.; Wang, J.; Hamman, B.D.; Crews, C.M. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat. Commun. 2019, 10, 131. [Google Scholar] [CrossRef] [Green Version]
- Bondeson, D.P.; Smith, B.E.; Burslem, G.M.; Buhimschi, A.D.; Hines, J.; Jaime-Figueroa, S.; Wang, J.; Hamman, B.D.; Ishchenko, A.; Crews, C.M. Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead. Cell Chem. Biol. 2018, 25, 78–87.e75. [Google Scholar] [CrossRef] [Green Version]
- Remillard, D.; Buckley, D.L.; Paulk, J.; Brien, G.L.; Sonnett, M.; Seo, H.S.; Dastjerdi, S.; Wühr, M.; Dhe-Paganon, S.; Armstrong, S.A.; et al. Degradation of the BAF Complex Factor BRD9 by Heterobifunctional Ligands. Angew. Chem. Int. Ed. Engl. 2017, 56, 5738. [Google Scholar] [CrossRef] [Green Version]
- Zengerle, M.; Chan, K.-H.; Ciulli, A. Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS Chem. Biol. 2015, 10, 2. [Google Scholar] [CrossRef] [Green Version]
- Lai, A.C.; Toure, M.; Hellerschmied, D.; Salami, J.; Jaime-Figueroa, S.; Ko, E.; Hines, J.; Crews, C.M. Modular PROTAC Design for the Degradation of Oncogenic BCR-ABL. Angew. Chem. Int. Ed. Engl. 2016, 55, 807. [Google Scholar] [CrossRef]
- Goracci, L.; Desantis, J.; Valeri, A.; Castellani, B.; Eleuteri, M.; Cruciani, G. Understanding the Metabolism of Proteolysis Targeting Chimeras (PROTACs): The Next Step toward Pharmaceutical Applications. J. Med. Chem. 2020, 63, 11615–11638. [Google Scholar] [CrossRef] [PubMed]
- Watt, G.F.; Scott-Stevens, P.; Gaohua, L. Targeted protein degradation in vivo with Proteolysis Targeting Chimeras: Current status and future considerations. Drug Discov. Today Technol. 2019, 31, 69–80. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cubillos-Rojas, M.; Loren, G.; Hakim, Y.Z.; Verdaguer, X.; Riera, A.; Nebreda, A.R. Synthesis and Biological Activity of a VHL-Based PROTAC Specific for p38α. Cancers 2023, 15, 611. https://doi.org/10.3390/cancers15030611
Cubillos-Rojas M, Loren G, Hakim YZ, Verdaguer X, Riera A, Nebreda AR. Synthesis and Biological Activity of a VHL-Based PROTAC Specific for p38α. Cancers. 2023; 15(3):611. https://doi.org/10.3390/cancers15030611
Chicago/Turabian StyleCubillos-Rojas, Mónica, Guillem Loren, Yusuf Z. Hakim, Xavier Verdaguer, Antoni Riera, and Angel R. Nebreda. 2023. "Synthesis and Biological Activity of a VHL-Based PROTAC Specific for p38α" Cancers 15, no. 3: 611. https://doi.org/10.3390/cancers15030611
APA StyleCubillos-Rojas, M., Loren, G., Hakim, Y. Z., Verdaguer, X., Riera, A., & Nebreda, A. R. (2023). Synthesis and Biological Activity of a VHL-Based PROTAC Specific for p38α. Cancers, 15(3), 611. https://doi.org/10.3390/cancers15030611





