Post-Transcriptional Modifications to miRNAs Undergo Widespread Alterations, Creating a Unique Lung Adenocarcinoma IsomiRome
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Sequencing, and Data Processing
2.2. Processing of Small RNA Sequencing Data
2.3. IsomiR and miRNA Quantification and Nomenclature
2.4. Identification of High-Confidence miRNAs and isomiRs
2.5. Computation of Sample-Wide miRNA Editing/Adenylation/Uridylation Rates
2.6. Determination of Significant Differences between ANL and LUAD Samples
2.7. Analysis of Correlations between Variables
2.8. Survival Analyses
2.9. Support Vector Machine and Random Forest Classifiers
2.10. Statistical and Graphical Software
3. Results
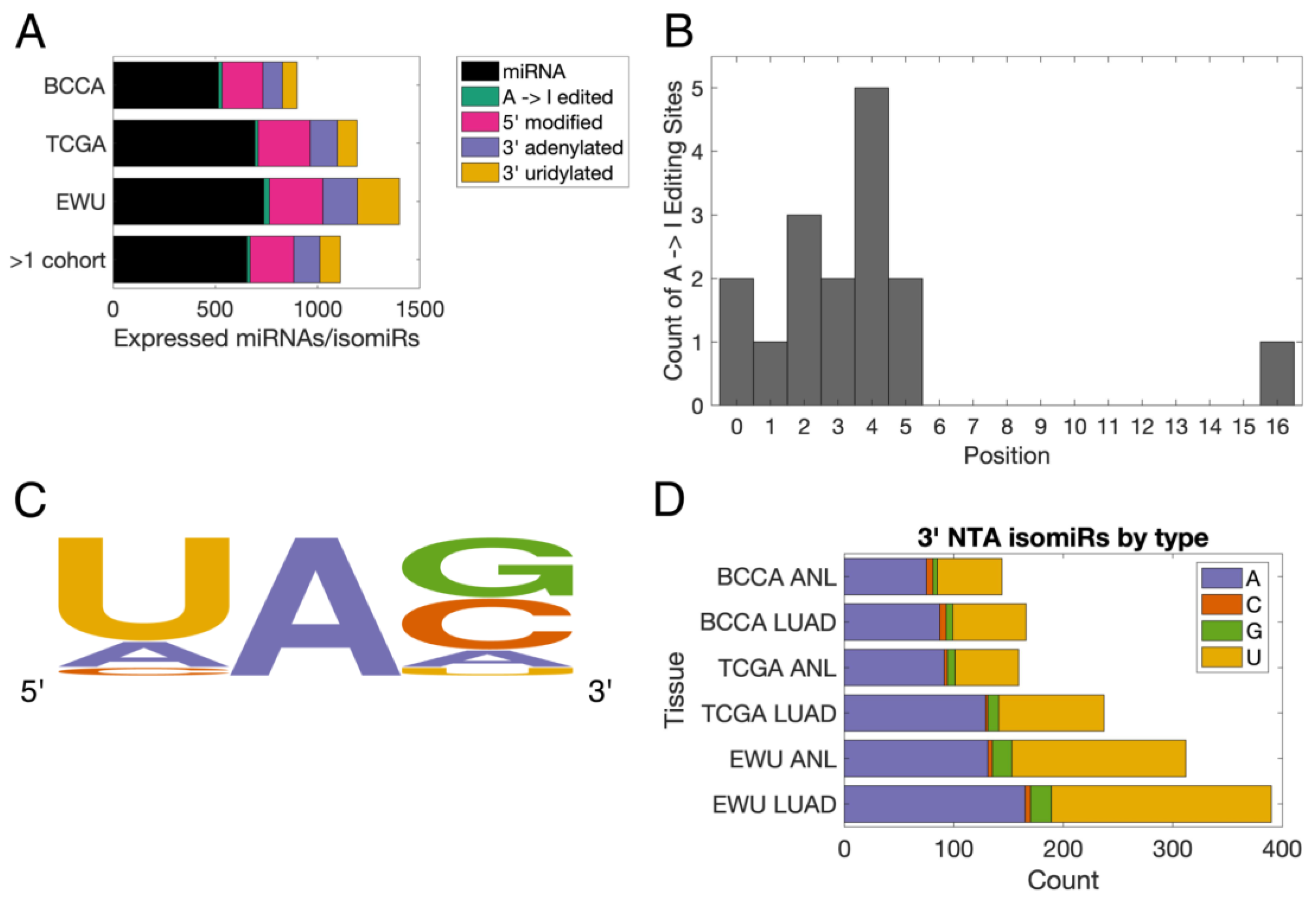
3.1. Characterization of the Lung isomiRome
3.2. The Lung isomiRome Is Widely Altered in LUAD
3.3. miRNA-Modifying Enzymes Are Dysregulated in LUAD
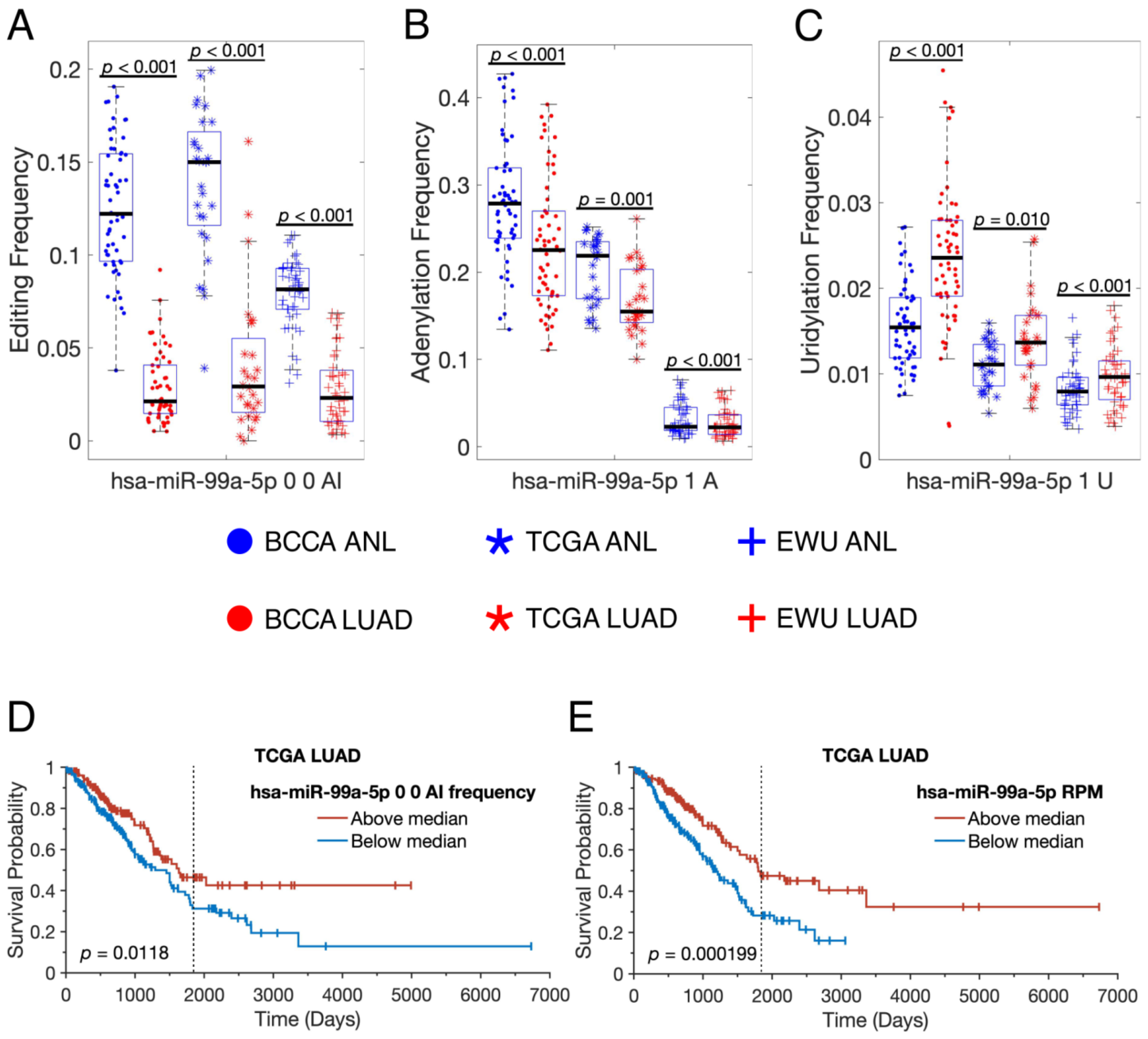
3.4. Individual miRNAs Are Modified in Distinct Fashions in LUAD
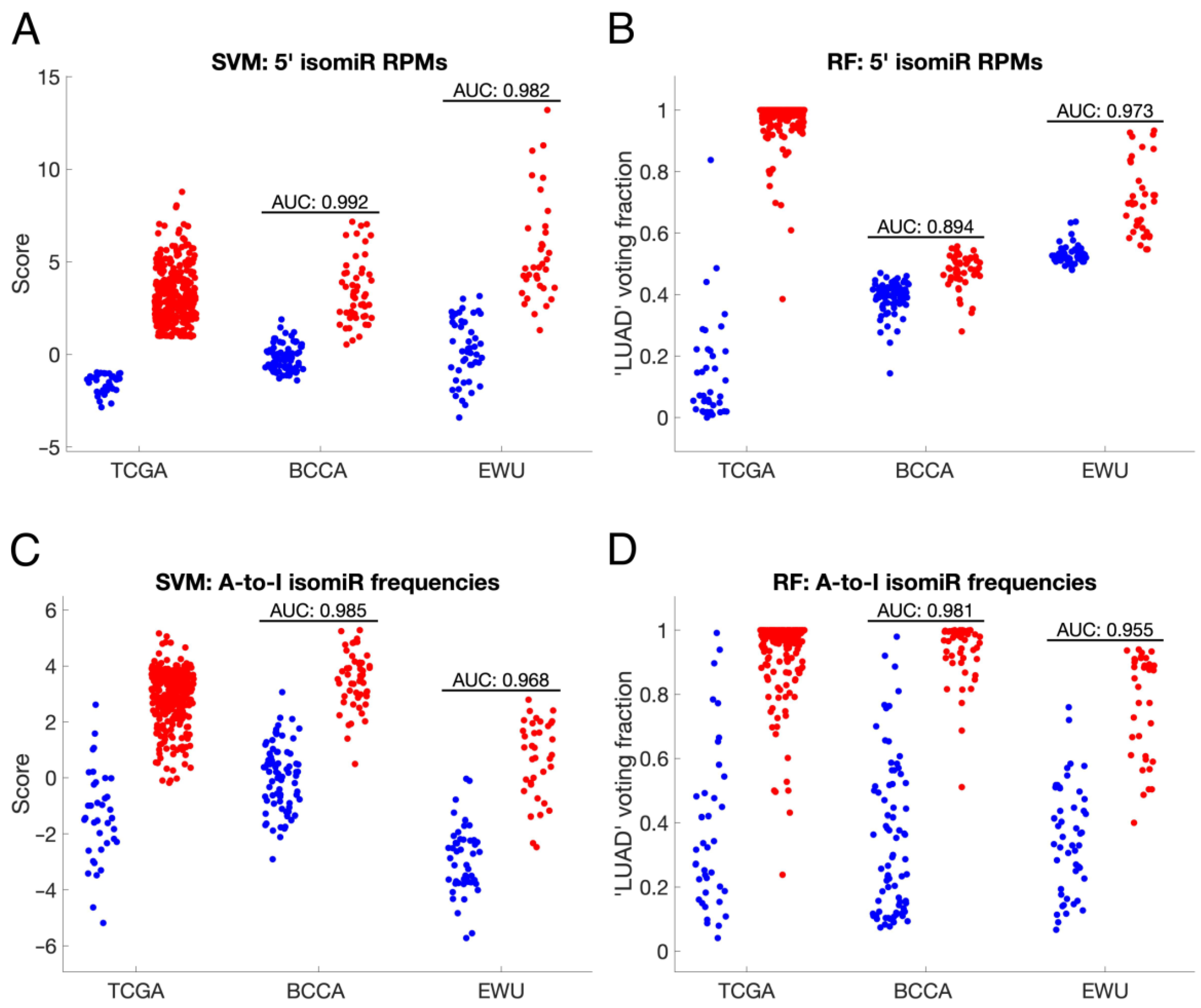
3.5. IsomiR-Based Biomarkers Distinguish ANL and LUAD Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA Function in Animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Wilczynska, A.; Bushell, M. The Complexity of miRNA-Mediated Repression. Cell Death Differ. 2015, 22, 22–33. [Google Scholar] [CrossRef]
- Iqbal, M.A.; Arora, S.; Prakasam, G.; Calin, G.A.; Syed, M.A. MicroRNA in Lung Cancer: Role, Mechanisms, Pathways and Therapeutic Relevance. Mol. Aspects Med. 2019, 70, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Chen, Y.; Chen, H.; Fei, S.; Chen, D.; Cai, X.; Liu, L.; Lin, B.; Su, H.; Zhao, L.; et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin. Cancer Res. 2017, 23, 5311–5319. [Google Scholar] [CrossRef] [PubMed]
- Sholl, L.M.; Aisner, D.L.; Varella-Garcia, M.; Berry, L.D.; Dias-Santagata, D.; Wistuba, I.I.; Chen, H.; Fujimoto, J.; Kugler, K.; Franklin, W.A.; et al. Multi-Institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. J. Thorac. Oncol. 2015, 10, 768–777. [Google Scholar] [CrossRef]
- Tomasello, L.; Distefano, R.; Nigita, G.; Croce, C.M. The MicroRNA Family Gets Wider: The IsomiRs Classification and Role. Front. Cell Dev. Biol. 2021, 9, 668648. [Google Scholar] [CrossRef]
- Cloonan, N.; Wani, S.; Xu, Q.; Gu, J.; Lea, K.; Heater, S.; Barbacioru, C.; Steptoe, A.L.; Martin, H.C.; Nourbakhsh, E.; et al. MicroRNAs and Their isomiRs Function Cooperatively to Target Common Biological Pathways. Genome Biol. 2011, 12, R126. [Google Scholar] [CrossRef]
- Tan, G.C.; Chan, E.; Molnar, A.; Sarkar, R.; Alexieva, D.; Isa, I.M.; Robinson, S.; Zhang, S.; Ellis, P.; Langford, C.F.; et al. 5′ isomiR Variation Is of Functional and Evolutionary Importance. Nucleic Acids Res. 2014, 42, 9424–9435. [Google Scholar] [CrossRef]
- Xu, X.; Wang, Y.; Mojumdar, K.; Zhou, Z.; Jeong, K.J.; Mangala, L.S.; Yu, S.; Tsang, Y.H.; Rodriguez-Aguayo, C.; Lu, Y.; et al. A-to-I-Edited miRNA-379-5p Inhibits Cancer Cell Proliferation through CD97-Induced Apoptosis. J. Clin. Investig. 2019, 129, 5343–5356. [Google Scholar] [CrossRef]
- Shoshan, E.; Mobley, A.K.; Braeuer, R.R.; Kamiya, T.; Huang, L.; Vasquez, M.E.; Salameh, A.; Lee, H.J.; Kim, S.J.; Ivan, C.; et al. Reduced Adenosine-to-Inosine miR-455-5p Editing Promotes Melanoma Growth and Metastasis. Nat. Cell Biol. 2015, 17, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zheng, Z.; Chen, P.; Wu, M. Tumor Classification and Biomarker Discovery Based on the 5′isomiR Expression Level. BMC Cancer 2019, 19, 127. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Fang, S.; Di, Y.; Ying, W.; Tan, Y.; Gu, W. Modulation of NF-κB/miR-21/PTEN Pathway Sensitizes Non-Small Cell Lung Cancer to Cisplatin. PLoS ONE 2015, 10, e0121547. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, J.; Zhao, F.; Liu, Q.; Jiang, K.; Yang, G. MicroRNA-21 (miR-21) Represses Tumor Suppressor PTEN and Promotes Growth and Invasion in Non-Small Cell Lung Cancer (NSCLC). Clin. Chim. Acta 2010, 411, 846–852. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Chen, Q.; Wang, J.; Mao, Q.; Dong, G.; Shi, R.; Zheng, Y.; Xu, L.; Jiang, F. DNA Methylation Mediated Silencing of microRNA-145 Is a Potential Prognostic Marker in Patients with Lung Adenocarcinoma. Sci. Rep. 2015, 5, 16901. [Google Scholar] [CrossRef]
- Chen, Z.; Zeng, H.; Guo, Y.; Liu, P.; Pan, H.; Deng, A.; Hu, J. miRNA-145 Inhibits Non-Small Cell Lung Cancer Cell Proliferation by Targeting c-Myc. J. Exp. Clin. Cancer Res. 2010, 29, 151. [Google Scholar] [CrossRef]
- Grenda, A.; Krawczyk, P.; Błach, J.; Chmielewska, I.; Kubiatowski, T.; Kieszko, S.; Wojas-Krawczyk, K.; Kucharczyk, T.; Jarosz, B.; Paśnik, I.; et al. Tissue MicroRNA Expression as a Predictor of Response to Immunotherapy in NSCLC Patients. Front. Oncol. 2020, 10, 563613. [Google Scholar] [CrossRef]
- Wang, S.; Su, X.; Bai, H.; Zhao, J.; Duan, J.; An, T.; Zhuo, M.; Wang, Z.; Wu, M.; Li, Z.; et al. Identification of Plasma microRNA Profiles for Primary Resistance to EGFR-TKIs in Advanced Non-Small Cell Lung Cancer (NSCLC) Patients with EGFR Activating Mutation. J. Hematol. Oncol. 2015, 8, 127. [Google Scholar] [CrossRef]
- Teng, P.-C.; Liang, Y.; Yarmishyn, A.A.; Hsiao, Y.-J.; Lin, T.-Y.; Lin, T.-W.; Teng, Y.-C.; Yang, Y.-P.; Wang, M.-L.; Chien, C.-S.; et al. RNA Modifications and Epigenetics in Modulation of Lung Cancer and Pulmonary Diseases. Int. J. Mol. Sci. 2021, 22, 10592. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Program (TCGA)—NCI. Available online: https://www.cancer.gov/ccg/research/genome-sequencing/tcga (accessed on 12 August 2024).
- Pinto, Y.; Buchumenski, I.; Levanon, E.Y.; Eisenberg, E. Human Cancer Tissues Exhibit Reduced A-to-I Editing of miRNAs Coupled with Elevated Editing of Their Targets. Nucleic Acids Res. 2018, 46, 71–82. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, X.; Yu, S.; Jeong, K.J.; Zhou, Z.; Han, L.; Tsang, Y.H.; Li, J.; Chen, H.; Mangala, L.S.; et al. Systematic Characterization of A-to-I RNA Editing Hotspots in microRNAs across Human Cancers. Genome Res. 2017, 27, 1112–1125. [Google Scholar] [CrossRef] [PubMed]
- Nigita, G.; Distefano, R.; Veneziano, D.; Romano, G.; Rahman, M.; Wang, K.; Pass, H.; Croce, C.M.; Acunzo, M.; Nana-Sinkam, P. Tissue and Exosomal miRNA Editing in Non-Small Cell Lung Cancer. Sci. Rep. 2018, 8, 10222. [Google Scholar] [CrossRef] [PubMed]
- Maemura, K.; Watanabe, K.; Ando, T.; Hiyama, N.; Sakatani, T.; Amano, Y.; Kage, H.; Nakajima, J.; Yatomi, Y.; Nagase, T.; et al. Altered Editing Level of microRNAs Is a Potential Biomarker in Lung Adenocarcinoma. Cancer Sci. 2018, 109, 3326–3335. [Google Scholar] [CrossRef] [PubMed]
- Distefano, R.; Nigita, G.; Le, P.; Romano, G.; Acunzo, M.; Nana-Sinkam, P. Disparities in Lung Cancer: miRNA Isoform Characterization in Lung Adenocarcinoma. Cancers 2022, 14, 773. [Google Scholar] [CrossRef]
- Salem, O.; Erdem, N.; Jung, J.; Münstermann, E.; Wörner, A.; Wilhelm, H.; Wiemann, S.; Körner, C. The Highly Expressed 5′isomiR of Hsa-miR-140-3p Contributes to the Tumor-Suppressive Effects of miR-140 by Reducing Breast Cancer Proliferation and Migration. BMC Genom. 2016, 17, 566. [Google Scholar] [CrossRef] [PubMed]
- D’Ambrogio, A.; Gu, W.; Udagawa, T.; Mello, C.C.; Richter, J.D. Specific miRNA Stabilization by Gld2-Catalyzed Monoadenylation. Cell Rep. 2012, 2, 1537–1545. [Google Scholar] [CrossRef]
- Boele, J.; Persson, H.; Shin, J.W.; Ishizu, Y.; Newie, I.S.; Søkilde, R.; Hawkins, S.M.; Coarfa, C.; Ikeda, K.; Takayama, K.; et al. PAPD5-Mediated 3′ Adenylation and Subsequent Degradation of miR-21 Is Disrupted in Proliferative Disease. Proc. Natl. Acad. Sci. USA 2014, 111, 11467–11472. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Ando, Y.; de Hoon, M.J.L.; Tomaru, Y.; Nishibu, T.; Ukekawa, R.; Funakoshi, T.; Kurokawa, T.; Suzuki, H.; Hayashizaki, Y.; et al. A Comprehensive Survey of 3′ Animal miRNA Modification Events and a Possible Role for 3′ Adenylation in Modulating miRNA Targeting Effectiveness. Genome Res. 2010, 20, 1398–1410. [Google Scholar] [CrossRef] [PubMed]
- Enfield, K.S.S.; Marshall, E.A.; Anderson, C.; Ng, K.W.; Rahmati, S.; Xu, Z.; Fuller, M.; Milne, K.; Lu, D.; Shi, R.; et al. Epithelial Tumor Suppressor ELF3 Is a Lineage-Specific Amplified Oncogene in Lung Adenocarcinoma. Nat. Commun. 2019, 10, 5438. [Google Scholar] [CrossRef]
- Vucic, E.A.; Thu, K.L.; Pikor, L.A.; Enfield, K.S.S.; Yee, J.; English, J.C.; MacAulay, C.E.; Lam, S.; Jurisica, I.; Lam, W.L. Smoking Status Impacts microRNA Mediated Prognosis and Lung Adenocarcinoma Biology. BMC Cancer 2014, 14, 778. [Google Scholar] [CrossRef]
- Cohn, D.E.; Barros-Filho, M.C.; Minatel, B.C.; Pewarchuk, M.E.; Marshall, E.A.; Vucic, E.A.; Sage, A.P.; Telkar, N.; Stewart, G.L.; Jurisica, I.; et al. Reactivation of Multiple Fetal miRNAs in Lung Adenocarcinoma. Cancers 2021, 13, 2686. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 2005, 15, 3439–3440. [Google Scholar] [CrossRef]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef]
- Chu, A.; Robertson, G.; Brooks, D.; Mungall, A.J.; Birol, I.; Coope, R.; Ma, Y.; Jones, S.; Marra, M.A. Large-Scale Profiling of microRNAs for The Cancer Genome Atlas. Nucleic Acids Res. 2016, 44, e3. [Google Scholar] [CrossRef]
- Yu, N.; Yong, S.; Kim, H.K.; Choi, Y.-L.; Jung, Y.; Kim, D.; Seo, J.; Lee, Y.E.; Baek, D.; Lee, J.; et al. Identification of Tumor Suppressor miRNAs by Integrative miRNA and mRNA Sequencing of Matched Tumor-Normal Samples in Lung Adenocarcinoma. Mol. Oncol. 2019, 13, 1356–1368. [Google Scholar] [CrossRef]
- Fehlmann, T.; Kern, F.; Laham, O.; Backes, C.; Solomon, J.; Hirsch, P.; Volz, C.; Müller, R.; Keller, A. miRMaster 2.0: Multi-Species Non-Coding RNA Sequencing Analyses at Scale. Nucleic Acids Res. 2021, 49, W397–W408. [Google Scholar] [CrossRef] [PubMed]
- Alon, S.; Mor, E.; Vigneault, F.; Church, G.M.; Locatelli, F.; Galeano, F.; Gallo, A.; Shomron, N.; Eisenberg, E. Systematic Identification of Edited microRNAs in the Human Brain. Genome Res. 2012, 22, 1533–1540. [Google Scholar] [CrossRef]
- Liu, C.-J.; Fu, X.; Xia, M.; Zhang, Q.; Gu, Z.; Guo, A.-Y. miRNASNP-v3: A Comprehensive Database for SNPs and Disease-Related Variations in miRNAs and miRNA Targets. Nucleic Acids Res. 2021, 49, D1276–D1281. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Ibing, S.; Michels, B.E.; Mosdzien, M.; Meyer, H.R.; Feuerbach, L.; Körner, C. On the Impact of Batch Effect Correction in TCGA isomiR Expression Data. NAR Cancer 2021, 3, zcab007. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.-M. CCA Addition to tRNA: Implications for tRNA Quality Control. IUBMB Life 2010, 62, 251–260. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- McCall, M.N.; Kim, M.-S.; Adil, M.; Patil, A.H.; Lu, Y.; Mitchell, C.J.; Leal-Rojas, P.; Xu, J.; Kumar, M.; Dawson, V.L.; et al. Toward the Human Cellular microRNAome. Genome Res. 2017, 27, 1769–1781. [Google Scholar] [CrossRef] [PubMed]
- Creed, J.; Gerke, T.; Berglund, A. MatSurv: Survival Analysis and Visualization in MATLAB. J. Open Source Softw. 2020, 5, 1830. [Google Scholar] [CrossRef]
- Amin, E.M.; Liu, Y.; Deng, S.; Tan, K.S.; Chudgar, N.; Mayo, M.W.; Sanchez-Vega, F.; Adusumilli, P.S.; Schultz, N.; Jones, D.R. The RNA-Editing Enzyme ADAR Promotes Lung Adenocarcinoma Migration and Invasion by Stabilizing FAK. Sci. Signal 2017, 10, eaah3941. [Google Scholar] [CrossRef]
- Mizuno, K.; Tanigawa, K.; Nohata, N.; Misono, S.; Okada, R.; Asai, S.; Moriya, S.; Suetsugu, T.; Inoue, H.; Seki, N. FAM64A: A Novel Oncogenic Target of Lung Adenocarcinoma Regulated by Both Strands of miR-99a (miR-99a-5p and miR-99a-3p). Cells 2020, 9, 2083. [Google Scholar] [CrossRef]
- Chan, T.H.M.; Qamra, A.; Tan, K.T.; Guo, J.; Yang, H.; Qi, L.; Lin, J.S.; Ng, V.H.E.; Song, Y.; Hong, H.; et al. ADAR-Mediated RNA Editing Predicts Progression and Prognosis of Gastric Cancer. Gastroenterology 2016, 151, 637–650.e10. [Google Scholar] [CrossRef]
- Gutiérrez-Vázquez, C.; Enright, A.J.; Rodríguez-Galán, A.; Pérez-García, A.; Collier, P.; Jones, M.R.; Benes, V.; Mizgerd, J.P.; Mittelbrunn, M.; Ramiro, A.R.; et al. 3′ Uridylation Controls Mature microRNA Turnover during CD4 T-Cell Activation. RNA 2017, 23, 882–891. [Google Scholar] [CrossRef]
- Wyman, S.K.; Knouf, E.C.; Parkin, R.K.; Fritz, B.R.; Lin, D.W.; Dennis, L.M.; Krouse, M.A.; Webster, P.J.; Tewari, M. Post-Transcriptional Generation of miRNA Variants by Multiple Nucleotidyl Transferases Contributes to miRNA Transcriptome Complexity. Genome Res. 2011, 21, 1450–1461. [Google Scholar] [CrossRef] [PubMed]
- Koppers-Lalic, D.; Hackenberg, M.; de Menezes, R.; Misovic, B.; Wachalska, M.; Geldof, A.; Zini, N.; de Reijke, T.; Wurdinger, T.; Vis, A.; et al. Non-invasive Prostate Cancer Detection by Measuring miRNA Variants (isomiRs) in Urine Extracellular Vesicles. Oncotarget 2016, 7, 22566–22578. [Google Scholar] [CrossRef] [PubMed]
- Koi, Y.; Tsutani, Y.; Nishiyama, Y.; Ueda, D.; Ibuki, Y.; Sasada, S.; Akita, T.; Masumoto, N.; Kadoya, T.; Yamamoto, Y.; et al. Predicting the Presence of Breast Cancer Using Circulating Small RNAs, Including Those in the Extracellular Vesicles. Cancer Sci. 2020, 111, 2104–2115. [Google Scholar] [CrossRef] [PubMed]




| Characteristic | EWU (n = 48) | TCGA (n = 389) | BCCA (n = 63) |
|---|---|---|---|
| Median Age (Range) | 59.5 (37–78) | 66 (39–88) | 70 (45–86) |
| Sex | |||
| Male | 0 (0%) | 173 (44%) | 19 (30%) |
| Female | 48 (100%) | 216 (56%) | 44 (70%) |
| Smoking History | |||
| Current or Former | 7 (15%) | 307 (79%) | 40 (63%) |
| Never | 41 (85%) | 64 (16%) | 23 (37%) |
| Stage | |||
| IA | 23 (48%) | 102 (26%) | 23 (37%) |
| IB | 7 (15%) | 103 (26%) | 18 (29%) |
| IIA | 5 (10%) | 43 (11%) | 2 (3%) |
| IIB | 1 (2%) | 52 (13%) | 11 (17%) |
| IIIA | 12 (25%) | 57 (15%) | 4 (6%) |
| IIIB | 0 (0%) | 6 (2%) | 1 (2%) |
| IV | 0 (0%) | 17 (4%) | 1 (2%) |
| miRNA | 5′ Shift | Edited Position | Adjacent Nucleotides * |
|---|---|---|---|
| hsa-let-7d-3p | 0 | 4 | UAC |
| hsa-miR-151a-3p | −2 | 2 | UAG |
| hsa-miR-200b-3p | 0 | 4 | UAC |
| hsa-miR-376c-3p | 0 | 5 | UAG |
| hsa-miR-379-5p | 0 | 4 | UAG |
| hsa-miR-381-3p | −1 | 3 | UAC |
| hsa-miR-381-3p | 0 | 3 | UAC |
| hsa-miR-411-5p | −1 | 4 | UAG |
| hsa-miR-411-5p | 0 | 4 | UAG |
| hsa-miR-455-5p | 0 | 16 | UAC |
| hsa-miR-4662a-5p | 0 | 2 | UAG |
| hsa-miR-497-5p | 0 | 1 | CAG |
| hsa-miR-589-3p | 0 | 5 | AAC |
| hsa-miR-9903 | 0 | 2 | UAU |
| hsa-miR-99a-5p | −1 | 0 | AAA |
| hsa-miR-99a-5p | 0 | 0 | AAA |
| Edited isomiR | ADAR ρ Value | ADAR p-Value | ADARB1 ρ Value | ADARB1 p-Value | Group * |
|---|---|---|---|---|---|
| hsa-let-7d-3p 0 4 AI | 0.079 | 0.124 | 0.198 | 9.38 × 10−5 | ADARB1 |
| hsa-miR-151a-3p -2 2 AI | 0.267 | 1.12 × 10−7 | 0.067 | 0.192 | ADAR |
| hsa-miR-200b-3p 0 4 AI | 0.452 | 9.65 × 10−21 | −0.040 | 0.436 | ADAR |
| hsa-miR-376c-3p 0 5 AI | −0.031 | 0.547 | 0.030 | 0.559 | Neither |
| hsa-miR-379-5p 0 4 AI | −0.030 | 0.561 | 0.249 | 7.63 × 10−7 | ADARB1 |
| hsa-miR-381-3p -1 3 AI | −0.070 | 0.170 | 0.006 | 0.904 | Neither |
| hsa-miR-381-3p 0 3 AI | 0.061 | 0.233 | −0.118 | 0.021 | Neither |
| hsa-miR-411-5p -1 4 AI | −0.047 | 0.357 | 0.103 | 0.044 | Neither |
| hsa-miR-411-5p 0 4 AI | 0.008 | 0.874 | 0.171 | 7.97 × 10−4 | ADARB1 |
| hsa-miR-455-5p 0 16 AI | −0.030 | 0.559 | 0.268 | 9.75 × 10−8 | ADARB1 |
| hsa-miR-4662a-5p 0 2 AI | −0.017 | 0.745 | 0.224 | 9.28 × 10−6 | ADARB1 |
| hsa-miR-497-5p 0 1 AI | −0.046 | 0.374 | 0.233 | 3.80 × 10−6 | ADARB1 |
| hsa-miR-589-3p 0 5 AI | 0.319 | 1.52 × 10−10 | −0.027 | 0.592 | ADAR |
| hsa-miR-9903 0 2 AI | −0.052 | 0.343 | −0.095 | 0.081 | Neither |
| hsa-miR-99a-5p -1 0 AI | −0.135 | 0.008 | 0.116 | 0.023 | ADARB1 |
| hsa-miR-99a-5p 0 0 AI | −0.081 | 0.114 | 0.310 | 5.48 × 10−10 | ADARB1 |
| RPM-Based Classifiers | Frequency-Based Classifiers | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| miRNA | 3′ U | 3′ A | A-to-I | 5′ | 3′ U | 3′ A | A-to-I | 5′ | ||
| Mean classification error in 10-fold CV | BCCA SVM | 1.56% | 1.56% | 0.78% | 3.13% | 2.34% | 0.78% | 0.00% | 4.69% | 3.13% |
| TCGA SVM | 1.45% | 1.16% | 2.03% | 1.45% | 0.58% | 1.16% | 2.62% | 1.74% | 1.16% | |
| EWU SVM | 2.38% | 2.38% | 2.38% | 2.38% | 2.38% | 1.19% | 3.57% | 11.90% | 3.57% | |
| Mean | 1.80% | 1.70% | 1.73% | 2.32% | 1.77% | 1.04% | 2.06% | 6.11% | 2.62% | |
| Out of bag error | BCCA RF | 0.78% | 0.78% | 0.00% | 1.56% | 0.78% | 4.69% | 1.56% | 3.91% | 4.69% |
| TCGA RF | 1.45% | 0.58% | 0.87% | 1.16% | 0.87% | 1.45% | 2.62% | 2.62% | 1.74% | |
| EWU RF | 1.19% | 0.00% | 0.00% | 1.19% | 0.00% | 8.33% | 14.29% | 3.57% | 5.95% | |
| Mean | 1.14% | 0.45% | 0.29% | 1.30% | 0.55% | 4.82% | 6.16% | 3.37% | 4.13% | |
| RPM-Based Classifiers | Frequency-Based Classifiers | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| miRNA | 3′ U | 3′ A | A-to-I | 5′ | 3′ U | 3′ A | A-to-I | 5′ | ||
| Test Cohort AUCs | Features | 368 | 37 | 41 | 7 | 115 | 17 | 34 | 7 | 36 |
| BCCA SVM | 0.944 | 0.998 | 0.962 | 0.997 | 0.992 | 0.663 | 0.476 | 0.985 | 0.716 | |
| EWU SVM | 0.951 | 0.995 | 0.884 | 0.934 | 0.982 | 0.582 | 0.752 | 0.968 | 0.893 | |
| BCCA RF | 0.901 | 0.891 | 0.855 | 0.990 | 0.894 | 0.634 | 0.756 | 0.981 | 0.889 | |
| EWU RF | 0.988 | 0.910 | 0.995 | 0.988 | 0.973 | 0.572 | 0.704 | 0.955 | 0.606 | |
| Mean | 0.946 | 0.949 | 0.924 | 0.977 | 0.960 | 0.613 | 0.672 | 0.972 | 0.776 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cohn, D.E.; Souza, V.G.P.; Forder, A.; Telkar, N.; Stewart, G.L.; Lam, W.L. Post-Transcriptional Modifications to miRNAs Undergo Widespread Alterations, Creating a Unique Lung Adenocarcinoma IsomiRome. Cancers 2024, 16, 3322. https://doi.org/10.3390/cancers16193322
Cohn DE, Souza VGP, Forder A, Telkar N, Stewart GL, Lam WL. Post-Transcriptional Modifications to miRNAs Undergo Widespread Alterations, Creating a Unique Lung Adenocarcinoma IsomiRome. Cancers. 2024; 16(19):3322. https://doi.org/10.3390/cancers16193322
Chicago/Turabian StyleCohn, David E., Vanessa G. P. Souza, Aisling Forder, Nikita Telkar, Greg L. Stewart, and Wan L. Lam. 2024. "Post-Transcriptional Modifications to miRNAs Undergo Widespread Alterations, Creating a Unique Lung Adenocarcinoma IsomiRome" Cancers 16, no. 19: 3322. https://doi.org/10.3390/cancers16193322
APA StyleCohn, D. E., Souza, V. G. P., Forder, A., Telkar, N., Stewart, G. L., & Lam, W. L. (2024). Post-Transcriptional Modifications to miRNAs Undergo Widespread Alterations, Creating a Unique Lung Adenocarcinoma IsomiRome. Cancers, 16(19), 3322. https://doi.org/10.3390/cancers16193322





