Gene Expression Profiling and Phenotypic Characterization of Circulating Tumor Cells Derived from a Murine Osteosarcoma Model
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
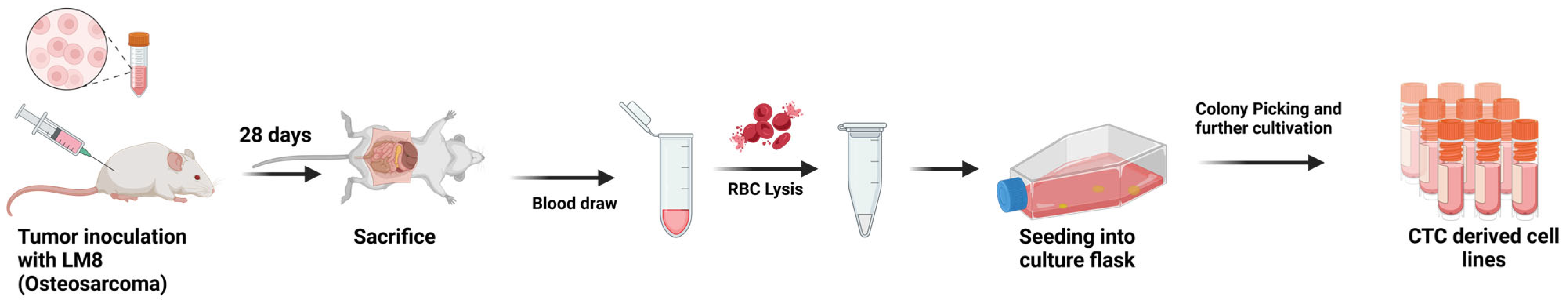
2.1. Cell Culture and Animal Inoculation
2.2. CTC Isolation and Cultivation
2.3. gDNA Isolation and SRY PCR
2.4. Clonogenic Survival Assays After Irradiation
2.5. Immunofluorescence Stainings
2.6. Transwell Migration Assay
2.7. RNA Isolation and mRNA Sequencing
2.8. Bioinformatics
3. Results
3.1. Characterization of CTC-Derived Cell Lines: Confirmation of Origin and Phenotypic Properties
3.2. Transcriptomic Profiling Reveals Distinct Gene Expression Patterns in CTC-Derived Cell Lines
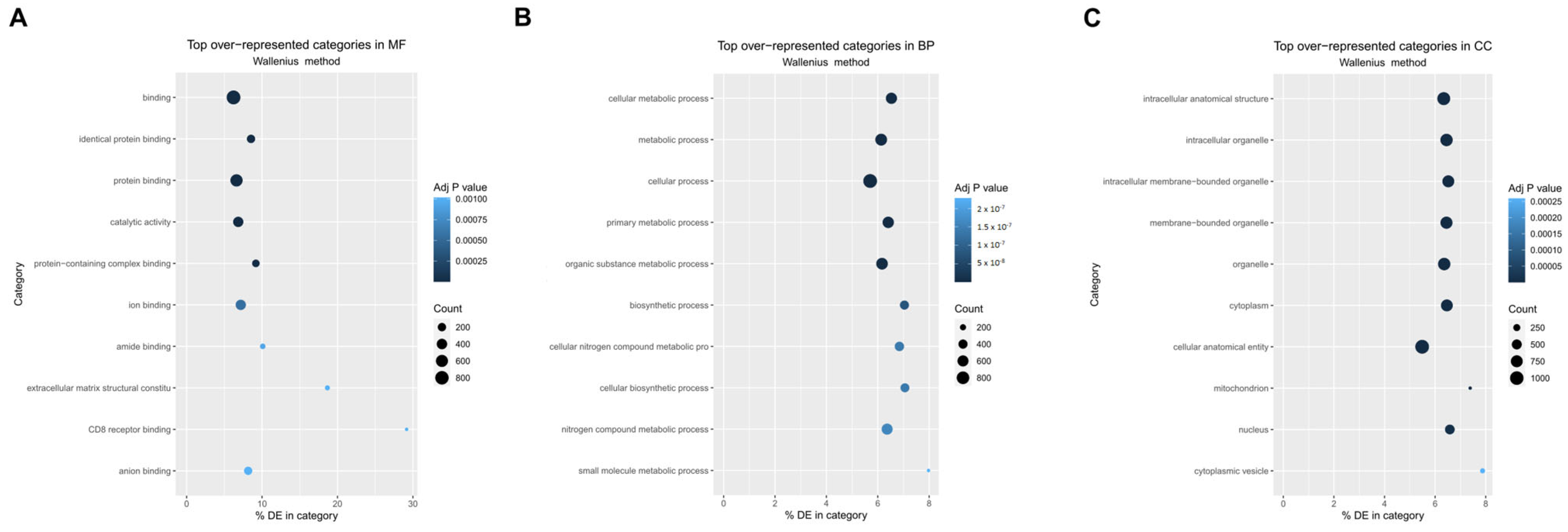
3.3. Gene Ontology and Pathway Enrichment Analysis Reveal Distinct Molecular Signatures in CTC-Derived Cell Lines
4. Discussion
4.1. CD44 as a Marker for OS CTCs and Disease Progression
4.2. Adaptations of OS CTCs: Increased Migration, Nuclear Size Reduction, and VEGFa Expression
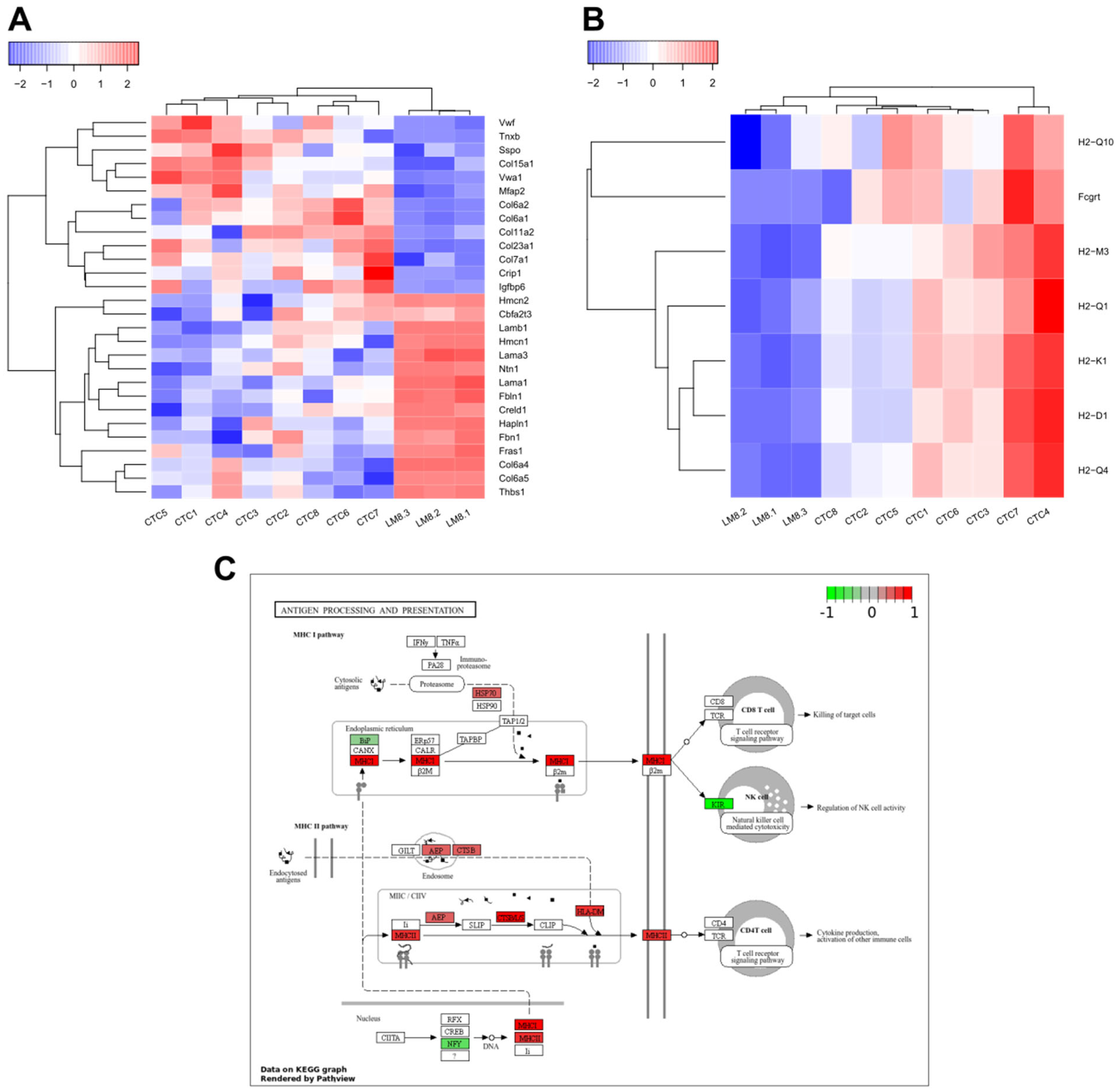
4.3. Gene Expression of LM8-CTCs Differ Significantly from Their Parental Cell Line
4.4. Immune Evasion Strategies of OS CTCs: Platelet Protection and Antigen Presentation
4.5. Extracellular Matrix Remodeling in OS CTCs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BP | Biological processes |
| BSA | Bovine serum albumin |
| CC | Cellular components |
| CD | Cluster of differentiation |
| CTCs | Circulating tumor cells |
| DEGs | Differentially expressed genes |
| DMEM | Dulbeccos Modified Eagle Medium |
| DTCs | Disseminated tumor cells |
| ECM | Extracellular matrix |
| EMT | Epithelial to mesenchymal transition |
| FBS | Fetal bovine serum |
| GO | Gene ontology |
| IF | Immunofluorescence |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MF | Molecular Functions |
| MHC | Major histocompatibility complex |
| MTA1 | Metastasis associated protein 1 |
| NK cells | Natural Killer cells |
| OS | Osteosarcoma |
| PBMCs | Peripheral blood mononuclear cell |
| PBS | Phosphate-buffered saline |
| PCA | Principle component analysis |
| PCR | Polymerase chain reaction |
| Pdl1 | Programmed death-ligand 1 |
| RBCs | Red blood cells |
| SRY | Sex determining region of Y |
| VEGFa | Vascular endothelial growth factor A |
| WBCs | White blood cells |
References
- Ottaviani, G.; Jaffe, N. The Etiology of Osteosarcoma. In Pediatric and Adolescent Osteosarcoma; Jaffe, N., Bruland, O.S., Bielack, S., Eds.; Cancer Treatment and Research; Springer: Boston, MA, USA, 2009; Volume 152, pp. 15–32. ISBN 978-1-4419-0283-2. [Google Scholar]
- Liu, Z.; Xie, Y.; Zhang, C.; Yang, T.; Chen, D. Survival Nomogram for Osteosarcoma Patients: SEER Data Retrospective Analysis with External Validation. Am. J. Cancer Res. 2023, 13, 900–911. [Google Scholar] [PubMed]
- Ritter, J.; Bielack, S.S. Osteosarcoma. Ann. Oncol. 2010, 21, vii320–vii325. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Shen, L.; Luo, M.; Zhang, K.; Li, J.; Yang, Q.; Zhu, F.; Zhou, D.; Zheng, S.; Chen, Y.; et al. Circulating Tumor Cells: Biology and Clinical Significance. Signal Transduct. Target. Ther. 2021, 6, 404. [Google Scholar] [CrossRef]
- Carmona-Ule, N.; González-Conde, M.; Abuín, C.; Cueva, J.F.; Palacios, P.; López-López, R.; Costa, C.; Dávila-Ibáñez, A.B. Short-Term Ex Vivo Culture of CTCs from Advance Breast Cancer Patients: Clinical Implications. Cancers 2021, 13, 2668. [Google Scholar] [CrossRef]
- Fabisiewicz, A.; Szostakowska-Rodzos, M.; Zaczek, A.J.; Grzybowska, E.A. Circulating Tumor Cells in Early and Advanced Breast Cancer; Biology and Prognostic Value. Int. J. Mol. Sci. 2020, 21, 1671. [Google Scholar] [CrossRef]
- Donato, C.; Kunz, L.; Castro-Giner, F.; Paasinen-Sohns, A.; Strittmatter, K.; Szczerba, B.M.; Scherrer, R.; Di Maggio, N.; Heusermann, W.; Biehlmaier, O.; et al. Hypoxia Triggers the Intravasation of Clustered Circulating Tumor Cells. Cell Rep. 2020, 32, 108105. [Google Scholar] [CrossRef]
- Liu, X.; Li, J.; Cadilha, B.L.; Markota, A.; Voigt, C.; Huang, Z.; Lin, P.P.; Wang, D.D.; Dai, J.; Kranz, G.; et al. Epithelial-Type Systemic Breast Carcinoma Cells with a Restricted Mesenchymal Transition Are a Major Source of Metastasis. Sci. Adv. 2019, 5, eaav4275. [Google Scholar] [CrossRef]
- Mazel, M.; Jacot, W.; Pantel, K.; Bartkowiak, K.; Topart, D.; Cayrefourcq, L.; Rossille, D.; Maudelonde, T.; Fest, T.; Alix-Panabières, C. Frequent Expression of PD-L1 on Circulating Breast Cancer Cells. Mol. Oncol. 2015, 9, 1773–1782. [Google Scholar] [CrossRef]
- Baccelli, I.; Schneeweiss, A.; Riethdorf, S.; Stenzinger, A.; Schillert, A.; Vogel, V.; Klein, C.; Saini, M.; Bäuerle, T.; Wallwiener, M.; et al. Identification of a Population of Blood Circulating Tumor Cells from Breast Cancer Patients That Initiates Metastasis in a Xenograft Assay. Nat. Biotechnol. 2013, 31, 539–544. [Google Scholar] [CrossRef]
- Lim, M.; Park, S.; Jeong, H.-O.; Park, S.H.; Kumar, S.; Jang, A.; Lee, S.; Kim, D.U.; Cho, Y.-K. Circulating Tumor Cell Clusters Are Cloaked with Platelets and Correlate with Poor Prognosis in Unresectable Pancreatic Cancer. Cancers 2021, 13, 5272. [Google Scholar] [CrossRef]
- Ting, D.T.; Wittner, B.S.; Ligorio, M.; Vincent Jordan, N.; Shah, A.M.; Miyamoto, D.T.; Aceto, N.; Bersani, F.; Brannigan, B.W.; Xega, K.; et al. Single-Cell RNA Sequencing Identifies Extracellular Matrix Gene Expression by Pancreatic Circulating Tumor Cells. Cell Rep. 2014, 8, 1905–1918. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.-J.; Tan, J.-C.; Qin, X.; Liu, B.; Yuan, Z.-C. Significance of Circulating Tumor Cells in Osteosarcoma Patients Treated by Neoadjuvant Chemotherapy and Surgery. CMAR 2018, 10, 3333–3339. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.; Zuo, D.; Chen, J.; Liu, Z.; Wang, Z.; Yang, L.; Shi, Q.; Hua, Y. Detection and Surveillance of Circulating Tumor Cells in Osteosarcoma for Predicting Therapy Response and Prognosis. Cancer Biol. Med. 2022, 19, 1397–1409. [Google Scholar] [CrossRef]
- Li, M.; Lu, Y.; Long, Z.; Li, M.; Kong, J.; Chen, G.; Wang, Z. Prognostic and Clinicopathological Significance of Circulating Tumor Cells in Osteosarcoma. J. Bone Oncol. 2019, 16, 100236. [Google Scholar] [CrossRef]
- Chalopin, A.; Tellez-Gabriel, M.; Brown, H.K.; Vallette, F.; Heymann, M.-F.; Gouin, F.; Heymann, D. Isolation of Circulating Tumor Cells in a Preclinical Model of Osteosarcoma: Effect of Chemotherapy. J. Bone Oncol. 2018, 12, 83–90. [Google Scholar] [CrossRef]
- Green, D.; Eyre, H.; Singh, A.; Taylor, J.T.; Chu, J.; Jeys, L.; Sumathi, V.; Coonar, A.; Rassl, D.; Babur, M.; et al. Targeting the MAPK7/MMP9 Axis for Metastasis in Primary Bone Cancer. Oncogene 2020, 39, 5553–5569. [Google Scholar] [CrossRef]
- Asai, T.; Ueda, T.; Itoh, K.; Yoshioka, K.; Aoki, Y.; Mori, S.; Yoshikawa, H. Establishment and Characterization of a Murine Osteosarcoma Cell Line (LM8) with High Metastatic Potential to the Lung. Int. J. Cancer 1998, 76, 418–422. [Google Scholar] [CrossRef]
- Tanaka, T.; Yui, Y.; Naka, N.; Wakamatsu, T.; Yoshioka, K.; Araki, N.; Yoshikawa, H.; Itoh, K. Dynamic Analysis of Lung Metastasis by Mouse Osteosarcoma LM8: VEGF Is a Candidate for Anti-Metastasis Therapy. Clin. Exp. Metastasis 2013, 30, 369–379. [Google Scholar] [CrossRef]
- Vitacchio, T. Figure 1: CTC Isolation and Cultivation. Available online: https://BioRender.com/s18m011 (accessed on 19 March 2025).
- Lambert, J.-F.; Benoit, B.O.; Colvin, G.A.; Carlson, J.; Delville, Y.; Quesenberry, P.J. Quick Sex Determination of Mouse Fetuses. J. Neurosci. Methods 2000, 95, 127–132. [Google Scholar] [CrossRef]
- Franken, N.A.P.; Rodermond, H.M.; Stap, J.; Haveman, J.; Van Bree, C. Clonogenic Assay of Cells In Vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef]
- Batut, B.; Freeberg, M.; Heydarian, M.; Erxleben, A.; Videm, P.; Blank, C.; Doyle, M.; Soranzo, N.; van Heusden, P.; Delisle, L. Reference-Based RNA-Seq Data Analysis (Galaxy Training Materials). Available online: https://training.galaxyproject.org/training-material/topics/transcriptomics/tutorials/ref-based/tutorial.html#citing-this-tutorial (accessed on 7 March 2025).
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor Package for Pathway-Based Data Integration and Visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
- Artigas, N.; Ureña, C.; Rodríguez-Carballo, E.; Rosa, J.L.; Ventura, F. Mitogen-Activated Protein Kinase (MAPK)-Regulated Interactions between Osterix and Runx2 Are Critical for the Transcriptional Osteogenic Program. J. Biol. Chem. 2014, 289, 27105–27117. [Google Scholar] [CrossRef]
- Zoch, M.L.; Clemens, T.L.; Riddle, R.C. New Insights into the Biology of Osteocalcin. Bone 2016, 82, 42–49. [Google Scholar] [CrossRef]
- Helm, A.; Tinganelli, W.; Simoniello, P.; Kurosawa, F.; Fournier, C.; Shimokawa, T.; Durante, M. Reduction of Lung Metastases in a Mouse Osteosarcoma Model Treated With Carbon Ions and Immune Checkpoint Inhibitors. Int. J. Radiat. Oncol. Biol. Phys. 2021, 109, 594–602. [Google Scholar] [CrossRef]
- Tinganelli, W.; Weber, U.; Puspitasari, A.; Simoniello, P.; Abdollahi, A.; Oppermann, J.; Schuy, C.; Horst, F.; Helm, A.; Fournier, C.; et al. FLASH with Carbon Ions: Tumor Control, Normal Tissue Sparing, and Distal Metastasis in a Mouse Osteosarcoma Model. Radiother. Oncol. 2022, 175, 185–190. [Google Scholar] [CrossRef]
- Loreth, D.; Schuette, M.; Zinke, J.; Mohme, M.; Piffko, A.; Schneegans, S.; Stadler, J.; Janning, M.; Loges, S.; Joosse, S.A.; et al. CD74 and CD44 Expression on CTCs in Cancer Patients with Brain Metastasis. IJMS 2021, 22, 6993. [Google Scholar] [CrossRef]
- Liu, X.; Taftaf, R.; Kawaguchi, M.; Chang, Y.-F.; Chen, W.; Entenberg, D.; Zhang, Y.; Gerratana, L.; Huang, S.; Patel, D.B.; et al. Homophilic CD44 Interactions Mediate Tumor Cell Aggregation and Polyclonal Metastasis in Patient-Derived Breast Cancer Models. Cancer Discov. 2019, 9, 96–113. [Google Scholar] [CrossRef] [PubMed]
- Gerardo-Ramírez, M.; Keggenhoff, F.L.; Giam, V.; Becker, D.; Groth, M.; Hartmann, N.; Straub, B.K.; Morrison, H.; Galle, P.R.; Marquardt, J.U.; et al. CD44 Contributes to the Regulation of MDR1 Protein and Doxorubicin Chemoresistance in Osteosarcoma. Int. J. Mol. Sci. 2022, 23, 8616. [Google Scholar] [CrossRef]
- Ji, H.; Kong, L.; Wang, Y.; Hou, Z.; Kong, W.; Qi, J.; Jin, Y. CD44 Expression Is Correlated with Osteosarcoma Cell Progression and Immune Infiltration and Affects the Wnt/β-Catenin Signaling Pathway. J. Bone Oncol. 2023, 41, 100487. [Google Scholar] [CrossRef]
- Liu, T.; Ma, Q.; Zhang, Y.; Wang, X.; Xu, K.; Yan, K.; Dong, W.; Fan, Q.; Zhang, Y.; Qiu, X. Self-Seeding Circulating Tumor Cells Promote the Proliferation and Metastasis of Human Osteosarcoma by Upregulating Interleukin-8. Cell Death Dis. 2019, 10, 575. [Google Scholar] [CrossRef]
- Chen, J.-Y.; Chou, H.-H.; Lim, S.C.; Huang, Y.-J.; Lai, K.-C.; Guo, C.-L.; Tung, C.-Y.; Su, C.-T.; Wang, J.; Liu, E.; et al. Multiomic Characterization and Drug Testing Establish Circulating Tumor Cells as an Ex Vivo Tool for Personalized Medicine. iScience 2022, 25, 105081. [Google Scholar] [CrossRef]
- Mendelaar, P.A.J.; Kraan, J.; Van, M.; Zeune, L.L.; Terstappen, L.W.M.M.; Oomen-de Hoop, E.; Martens, J.W.M.; Sleijfer, S. Defining the Dimensions of Circulating Tumor Cells in a Large Series of Breast, Prostate, Colon, and Bladder Cancer Patients. Mol. Oncol. 2021, 15, 116–125. [Google Scholar] [CrossRef]
- McGregor, A.L.; Hsia, C.-R.; Lammerding, J. Squish and Squeeze—The Nucleus as a Physical Barrier during Migration in Confined Environments. Curr. Opin. Cell Biol. 2016, 40, 32–40. [Google Scholar] [CrossRef]
- Yang, Y.; Cao, Y. The Impact of VEGF on Cancer Metastasis and Systemic Disease. Semin. Cancer Biol. 2022, 86, 251–261. [Google Scholar] [CrossRef]
- Kallergi, G.; Markomanolaki, H.; Giannoukaraki, V.; Papadaki, M.A.; Strati, A.; Lianidou, E.S.; Georgoulias, V.; Mavroudis, D.; Agelaki, S. Hypoxia-Inducible Factor-1α and Vascular Endothelial Growth Factor Expression in Circulating Tumor Cells of Breast Cancer Patients. Breast Cancer Res. 2009, 11, R84. [Google Scholar] [CrossRef]
- Rath, B.; Plangger, A.; Klameth, L.; Hochmair, M.; Ulsperger, E.; Boeckx, B.; Neumayer, C.; Hamilton, G. Small Cell Lung Cancer: Circulating Tumor Cell Lines and Expression of Mediators of Angiogenesis and Coagulation. Explor. Target. Anti-Tumor Ther. 2023, 4, 355–365. [Google Scholar] [CrossRef]
- Agnoletto, C.; Corrà, F.; Minotti, L.; Baldassari, F.; Crudele, F.; Cook, W.J.J.; Di Leva, G.; d’Adamo, A.P.; Gasparini, P.; Volinia, S. Heterogeneity in Circulating Tumor Cells: The Relevance of the Stem-Cell Subset. Cancers 2019, 11, 483. [Google Scholar] [CrossRef]
- Yu, T.; Wang, C.; Xie, M.; Zhu, C.; Shu, Y.; Tang, J.; Guan, X. Heterogeneity of CTC Contributes to the Organotropism of Breast Cancer. Biomed. Pharmacother. 2021, 137, 111314. [Google Scholar] [CrossRef]
- Dai, S.; Shao, X.; Wei, Q.; Du, S.; Hou, C.; Li, H.; Jin, D. Association of circulating tumor cells and IMP3 expression with metastasis of osteosarcoma. Front. Oncol. 2023, 13, 819357. [Google Scholar] [CrossRef]
- Gu, X.; Wei, S.; Lv, X. Circulating Tumor Cells: From New Biological Insights to Clinical Practice. Signal Transduct. Target. Ther. 2024, 9, 226. [Google Scholar] [CrossRef]
- Paul, S.; Lal, G. The Molecular Mechanism of Natural Killer Cells Function and Its Importance in Cancer Immunotherapy. Front. Immunol. 2017, 8, 1124. [Google Scholar] [CrossRef]
- Labelle, M.; Hynes, R.O. The Initial Hours of Metastasis: The Importance of Cooperative Host–Tumor Cell Interactions during Hematogenous Dissemination. Cancer Discov. 2012, 2, 1091–1099. [Google Scholar] [CrossRef]
- Bryckaert, M.; Rosa, J.-P.; Denis, C.V.; Lenting, P.J. Of von Willebrand Factor and Platelets. Cell. Mol. Life Sci. 2015, 72, 307–326. [Google Scholar] [CrossRef]
- Rodriguez-Martinez, A.; Simon-Saez, I.; Perales, S.; Garrido-Navas, C.; Russo, A.; De Miguel-Perez, D.; Puche-Sanz, I.; Alaminos, C.; Ceron, J.; Lorente, J.A.; et al. Exchange of Cellular Components between Platelets and Tumor Cells: Impact on Tumor Cells Behavior. Theranostics 2022, 12, 2150–2161. [Google Scholar] [CrossRef]
- Placke, T.; Örgel, M.; Schaller, M.; Jung, G.; Rammensee, H.-G.; Kopp, H.-G.; Salih, H.R. Platelet-Derived MHC Class I Confers a Pseudonormal Phenotype to Cancer Cells That Subverts the Antitumor Reactivity of Natural Killer Immune Cells. Cancer Res. 2012, 72, 440–448. [Google Scholar] [CrossRef]
- Lei, P.-J.; Pereira, E.R.; Andersson, P.; Amoozgar, Z.; Van Wijnbergen, J.W.; O’Melia, M.J.; Zhou, H.; Chatterjee, S.; Ho, W.W.; Posada, J.M.; et al. Cancer Cell Plasticity and MHC-II–Mediated Immune Tolerance Promote Breast Cancer Metastasis to Lymph Nodes. J. Exp. Med. 2023, 220, e20221847. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of Extracellular Matrix Remodelling in Tumour Progression and Metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Z.; Yang, X.; Lu, W.; Chen, Y.; Lin, Y.; Wang, J.; Lin, S.; Yun, J.-P. H3K27 Acetylation Activated-COL6A1 Promotes Osteosarcoma Lung Metastasis by Repressing STAT1 and Activating Pulmonary Cancer-Associated Fibroblasts. Theranostics 2021, 11, 1473–1492. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benje, M.; Vitacchio, T.; Fritsche, D.; Tinganelli, W. Gene Expression Profiling and Phenotypic Characterization of Circulating Tumor Cells Derived from a Murine Osteosarcoma Model. Cancers 2025, 17, 1210. https://doi.org/10.3390/cancers17071210
Benje M, Vitacchio T, Fritsche D, Tinganelli W. Gene Expression Profiling and Phenotypic Characterization of Circulating Tumor Cells Derived from a Murine Osteosarcoma Model. Cancers. 2025; 17(7):1210. https://doi.org/10.3390/cancers17071210
Chicago/Turabian StyleBenje, Malte, Tamara Vitacchio, Dennis Fritsche, and Walter Tinganelli. 2025. "Gene Expression Profiling and Phenotypic Characterization of Circulating Tumor Cells Derived from a Murine Osteosarcoma Model" Cancers 17, no. 7: 1210. https://doi.org/10.3390/cancers17071210
APA StyleBenje, M., Vitacchio, T., Fritsche, D., & Tinganelli, W. (2025). Gene Expression Profiling and Phenotypic Characterization of Circulating Tumor Cells Derived from a Murine Osteosarcoma Model. Cancers, 17(7), 1210. https://doi.org/10.3390/cancers17071210






