A Valuable Source of Promising Extremophiles in Microbial Plastic Degradation
Abstract
1. Introduction
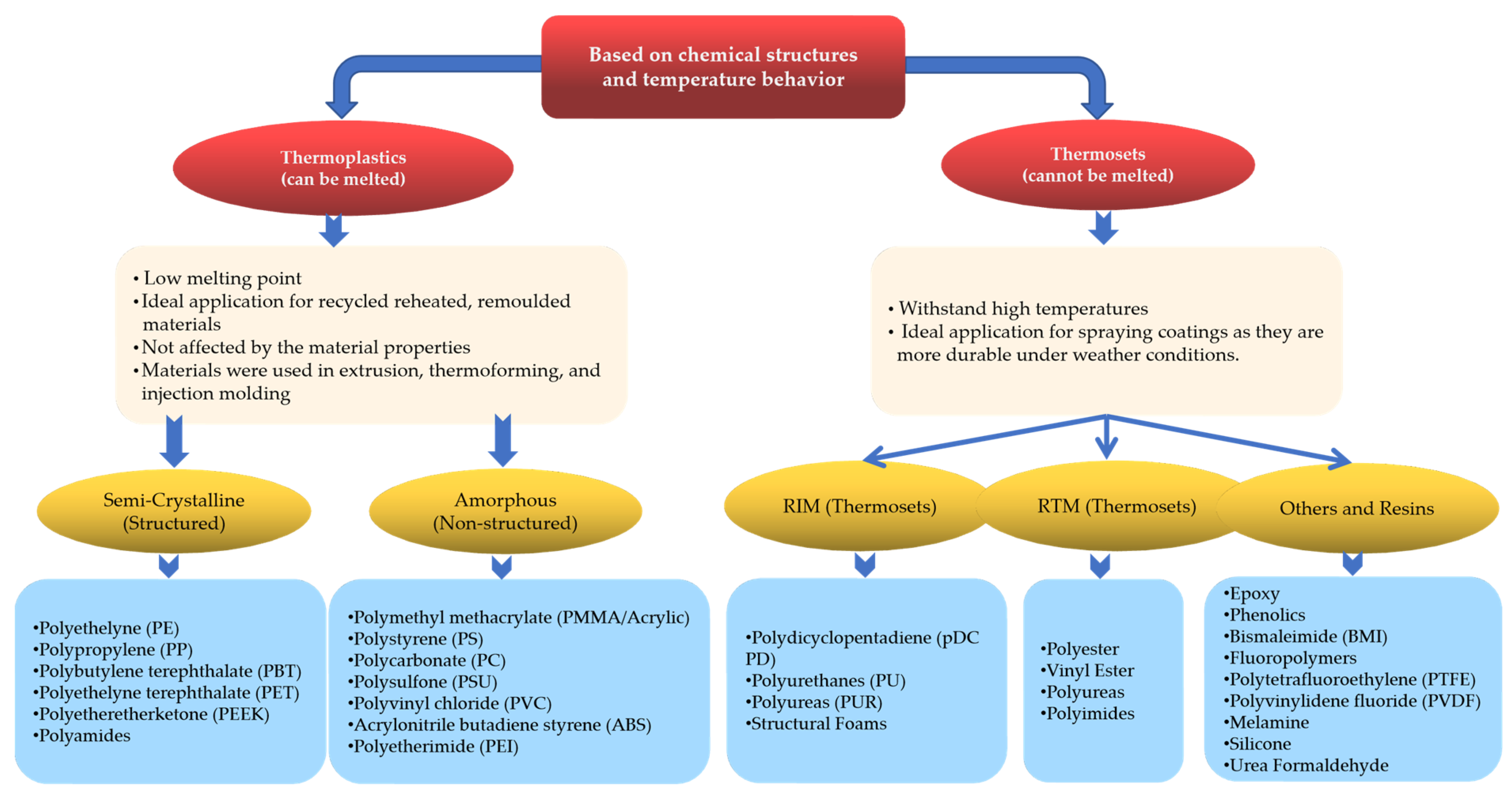
2. Classification and Identification of Plastics
2.1. Classification Based on Chemical Structures and Temperatures
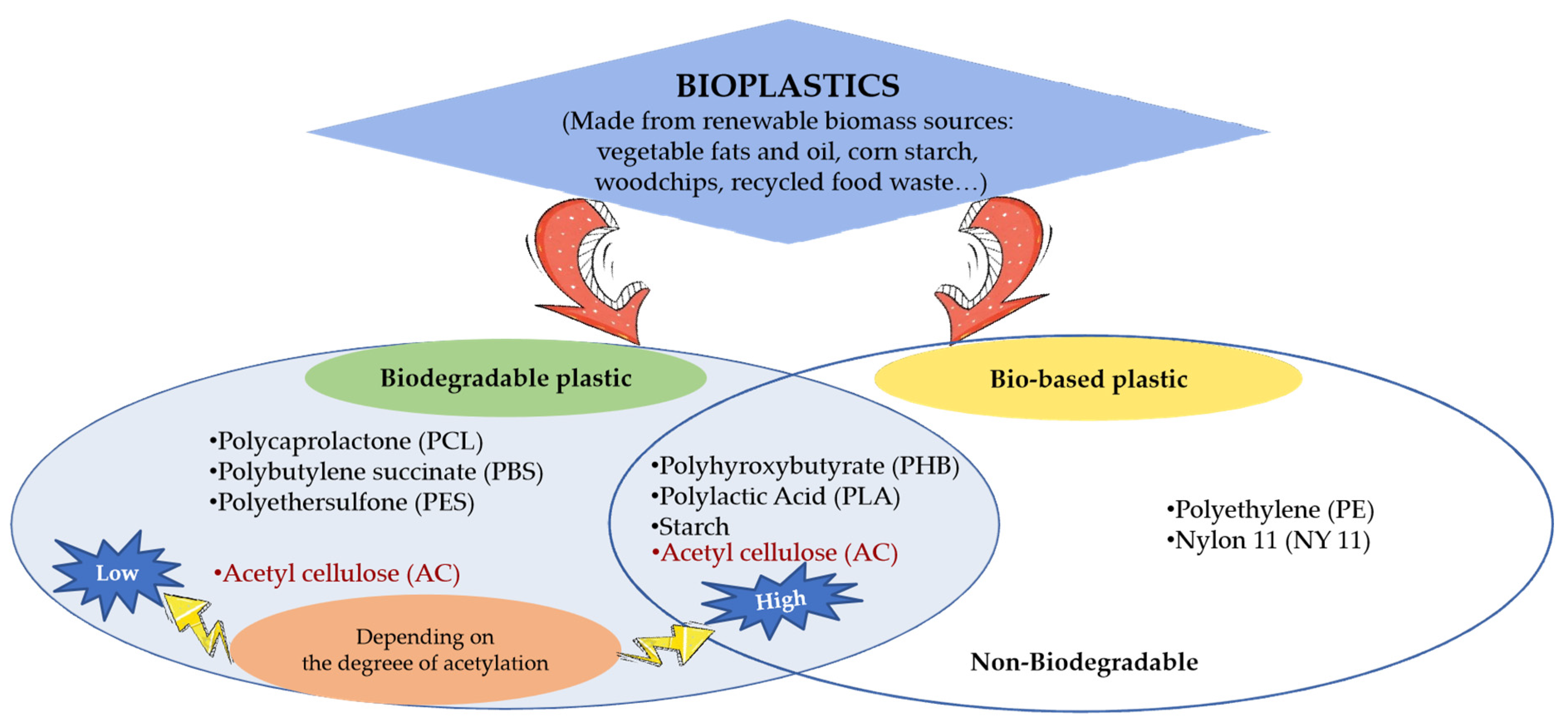
2.2. Classification Based on Physiochemical Properties and Biodegradability
3. Biodegradation of Plastics Using Capable Bacteria
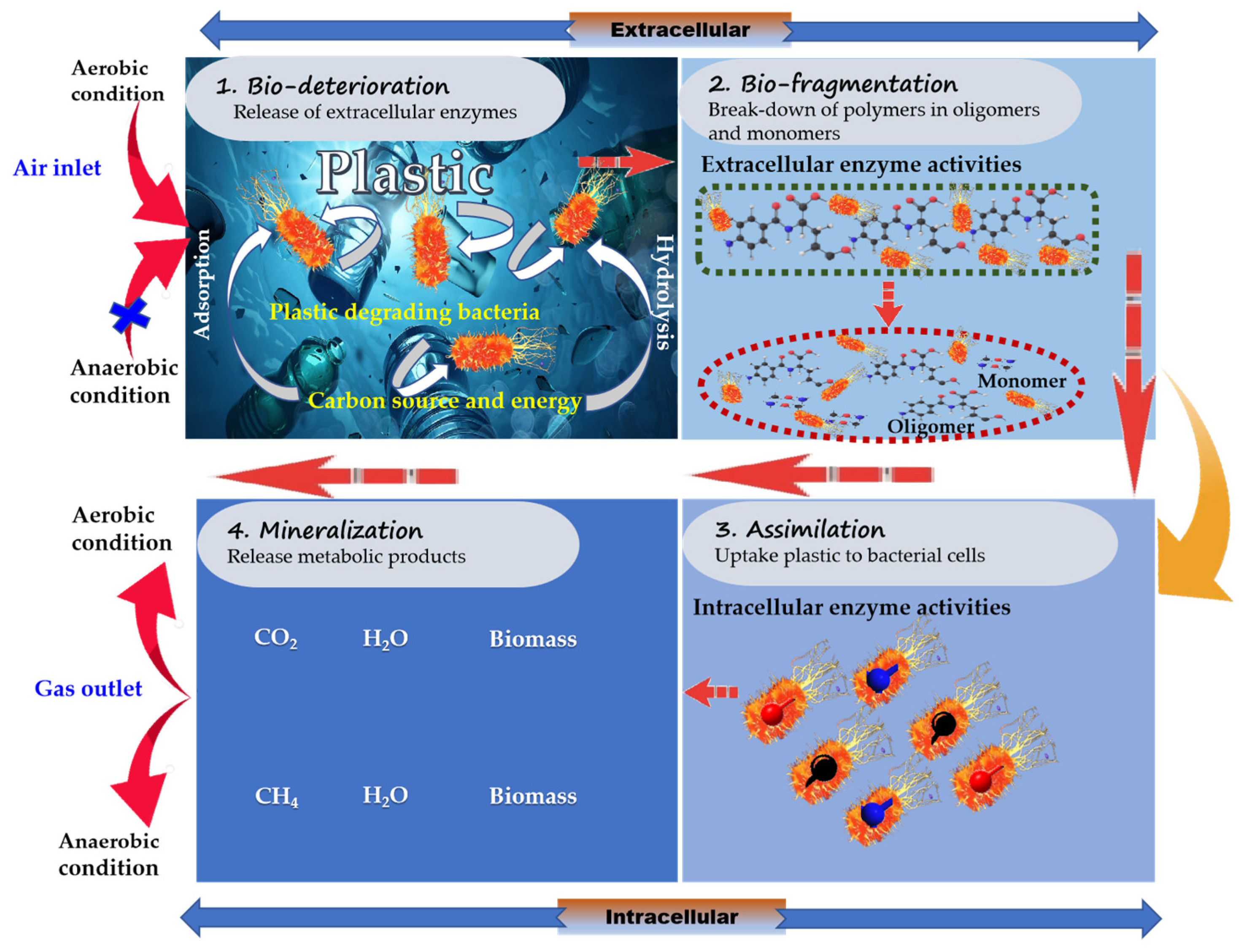
3.1. Investigations of Plastic-Degrading Bacteria and Their Degradation Mechanisms
3.2. Classification of Plastic-Degrading Bacterial Groups Based on Biodegradability
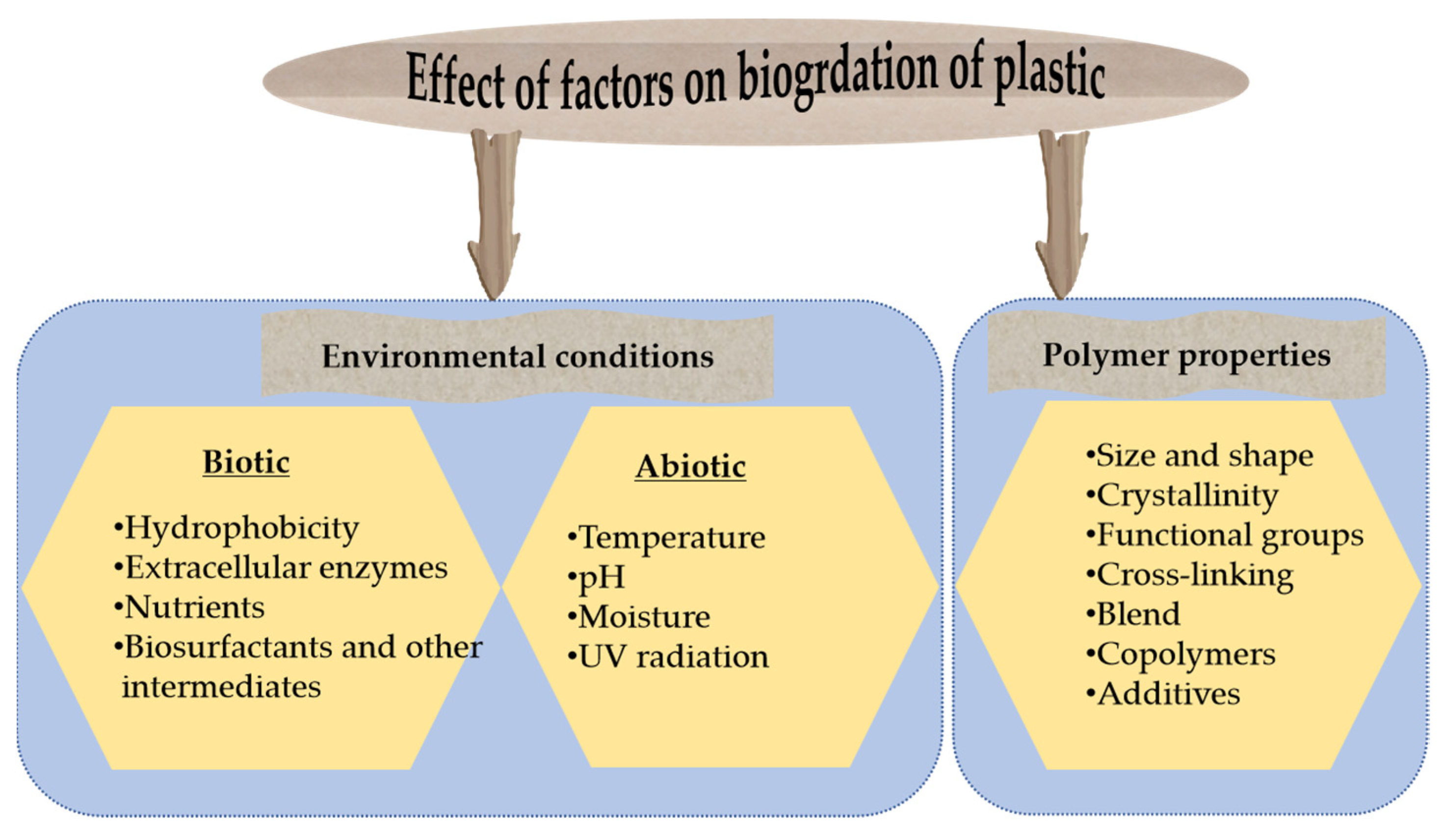
4. Factors Affecting Plastic Degradation Efficiency
5. The Special Characteristics of Extremophiles Reveal Interesting Mechanisms in Plastic Degradation
5.1. How Do Extremophiles Make a “Plasticsphere”?
5.2. From Which Places Should Plastic-Degrading Bacteria Be Taken for Investigation?
5.3. Typical Extremophiles Participating in Plastic Degradation
6. Drawbacks and Future Prospects
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Huerta, E.; Thapa, B.; Yang, X.; Gertsen, H.; Salánki, T.; Geissen, V. Decay of low-density polyethylene by bacteria extracted from earthworm’s guts: A potential for soil restoration. Sci. Total Environ. 2018, 624, 753–757. [Google Scholar] [CrossRef] [PubMed]
- Raddadi, N.; Fava, F. Biodegradation of oil-based plastics in the environment: Existing knowledge and needs of research and innovation. Sci. Total Environ. 2019, 679, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.M.; Robertson, M.L. The future of plastics recycling. Science 2017, 358, 870–872. [Google Scholar] [CrossRef]
- Geyer, R.; Jambeck, J.; Law, K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef]
- Ragaert, K.; Delva, L.; Van Geem, K. Mechanical and chemical recycling of solid plastic waste. Waste Manag. 2017, 69, 24–58. [Google Scholar] [CrossRef]
- Scalenghe, R. Resource or waste? A perspective of plastics degradation in soil with a focus on end-of-life options. Heliyon 2018, 4, e00941. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.; Belchior, A.; da Silva Verônica, D.; Rotter, A.; Petrovski, Z.; Almeida Pedro, L.; Lourenço Nídia, D.; Gaudêncio Susana, P. Marine Environmental Plastic Pollution: Mitigation by Microorganism Degradation and Recycling Valorization. Front. Mar. Sci. 2020, 7, 567126. [Google Scholar] [CrossRef]
- Sharma, M.D.; Elanjickal, A.I.; Mankar, J.S.; Krupadam, R.J. Assessment of cancer risk of microplastics enriched with polycyclic aromatic hydrocarbons. J. Hazard. Mater. 2020, 398, 122994. [Google Scholar] [CrossRef]
- Rahimi, A.; García, J.M. Chemical recycling of waste plastics for new materials production. Nat. Rev. Chem. 2017, 1, 0046. [Google Scholar] [CrossRef]
- Álvarez-Hernández, C.; Cairós, C.; López-Darias, J.; Mazzetti, E.; Hernández-Sánchez, C.; González-Sálamo, J. Microplastic debris in beaches of Tenerife (Canary Islands, Spain). Mar. Pollut. Bull. 2019, 146, 26–32. [Google Scholar] [CrossRef]
- Bergmann, M.; Mützel, S.; Primpke, S.; Tekman, M.B.; Trachsel, J.; Gerdts, G. White and wonderful? Microplastics prevail in snow from the Alps to the Arctic. Sci. Adv. 2019, 5, eaax1157. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Kong, T.; Li, Y.; Li, Q.; Li, Z.; Zhang, J. Biodegradation of Microplastic Derived from Poly(ethylene terephthalate) with Bacterial Whole-Cell Biocatalysts. Polymers 2018, 10, 1326. [Google Scholar] [CrossRef] [PubMed]
- Serra, S.; Simeis, D.D. Use of Lactobacillus rhamnosus (ATCC 53103) as Whole-Cell Biocatalyst for the Regio- and Stereoselective Hydration of Oleic, Linoleic, and Linolenic Acid. Catalysts 2018, 8, 109. [Google Scholar] [CrossRef]
- Pham, V.H.T.; Kim, J.; Chang, S.; Chung, W. Investigation of lipolytic-secreting bacteria from an artificially polluted soil using a modified culture method and optimization of their lipase production. Microorganisms 2021, 9, 2590. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Wei, R.; Cui, Q.; Bornscheuer, U.T.; Liu, Y. Thermophilic whole-cell degradation of polyethylene terephthalate using engineered Clostridium thermocellum. Microb. Biotechnol. 2021, 14, 374–385. [Google Scholar] [CrossRef]
- Pometto, A.L.; Lee, B.T.; Johnson, K.E. Production of an extracellular polyethylene-degrading enzyme(s) by Streptomyces species. Appl. Environ. Microbiol. 1992, 58, 731–733. [Google Scholar] [CrossRef]
- Austin, H.P.; Allen, M.D.; Donohoe, B.S.; Rorrer, N.A.; Kearns, F.L.; Silveira, R.L.; Pollard, B.C.; Domnick, G.; Duman, R. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. USA 2018, 115, 4350–4357. [Google Scholar] [CrossRef]
- de Castro, A.M.; Carniel, A.; Nicomedes Junior, J.; da Conceição Gomes, A.; Valoni, É. Screening of commercial enzymes for poly(ethylene terephthalate) (PET) hydrolysis and synergy studies on different substrate sources. J. Ind. Microbiol. Biotechnol. 2017, 44, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Zimmermann, W. Biocatalysis as a green route for recycling the recalcitrant plastic polyethylene terephthalate. Microb. Biotechnol. 2017, 10, 1302–1307. [Google Scholar]
- MacElroy, R.D. Some comments on the evolution of extremophiles. Biosystems 1974, 6, 74–75. [Google Scholar] [CrossRef]
- European Bioplastics. Bioplastics market development update 2020. In Proceedings of the European Bioplastics Conference, Berlin, Germany, 30 November 2020; p. 10117. [Google Scholar]
- Rahman, M.H.; Bhoi, P.R. An overview of non-biodegradable bioplastics. J. Clean. Prod. 2021, 294, 126218. [Google Scholar] [CrossRef]
- Montazer, Z.; Habibi Najafi, M.B.; Levin, D.B. Microbial degradation of low-density polyethylene and synthesis of polyhydroxyalkanoate polymers. Can. J. Microbiol. 2019, 65, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Montazer, Z.; Habibi Najafi, M.B.; Levin, D.B. In vitro degradation of low-density polyethylene by new bacteria from larvae of the Greater Wax Moth, Galleria melonella. Can. J. Microbiol. 2021, 67, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Chandra, P.; Enespa, P.; Singh, D.P. Microplastic degradation by bacteria in aquatic ecosystem. In Microorganisms for Sustainable Environment and Health; Chowdhary, P., Raj, A., Verma, D., Akhter, Y., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 431–467. [Google Scholar]
- Lacombe-Harvey, M.È.; Brzezinski, R.; Beaulieu, C. Chitinolytic functions in Actinobacteria: Ecology, enzymes, and evolution. Appl. Microbiol. Biotechnol. 2018, 102, 7219–7230. [Google Scholar] [CrossRef]
- Pathak, V.M.; Navneet. Review on the current status of polymer degradation: A microbial approach. Bioresour. Bioprocess. 2017, 4, 15. [Google Scholar] [CrossRef]
- Rodríguez-Fonseca, M.F.; Sánchez-Suárez, J.; Valero, M.F.; Ruiz-Balaguera, S.; Díaz, L.E. Streptomyces as Potential Synthetic Polymer Degraders: A Systematic Review. Bioengineering 2021, 8, 154. [Google Scholar] [CrossRef] [PubMed]
- Pham, V.H.T.; Kim, J.; Chang, S.; Chung, W. Biodegradation of Methylene Blue Using a Novel Lignin Peroxidase Enzyme Producing Bacteria, Named Bacillus sp. React3, as a Promising Candidate for Dye-Contaminated Wastewater Treatment. Fermentation 2022, 8, 190. [Google Scholar] [CrossRef]
- Pham, V.H.T.; Kim, J.; Chang, S.; Chung, W. Purification and characterization of strong simultaneous enzyme production of protease and α-Amylase from an extremophile-Bacillus sp. FW2 and its possibility in food waste degradation. Fermentation 2022, 8, 12. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef]
- Han, X.; Liu, W.; Huang, J.W.; Ma, J.; Zheng, Y.; Ko, T.P. Structural insight into catalytic mechanism of PET hydrolase. Nat. Commun. 2017, 8, 2106. [Google Scholar] [CrossRef]
- Palm, G.J.; Reisky, L.; Böttcher, D.; Müller, H.; Michels, E.A.; Walczak, M.C.; Berndt, L.; Weiss, M.S.; Bornscheuer, U.T.; Weber, G. Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate. Nat. Commun. 2019, 10, 1717. [Google Scholar] [CrossRef] [PubMed]
- Bollinger, A.; Thies, S.; Grünhagen, E.K.; Gertzen, C.; Kobus, S.; Höppner, A.; Ferrer, M.; Gohlke, H.; Smits, S.H.J.; Jaeger, K.E. A novel polyester hydrolase from the marine bacterium Pseudomonas aestusnigri–structural and functional insights. Front. Microbiol. 2020, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- Skariyachan, S.; Manjunatha, V.; Sultana, S.; Jois, C.; Bai, V.; Vasist, K.S. Novel bacterial consortia isolated from plastic garbage processing areas demonstrated enhanced degradation for low density polyethylene. Environ. Sci. Pollut. Res. 2016, 23, 18307–18319. [Google Scholar] [CrossRef]
- Skariyachan, S.; Patil, A.A.; Shankar, A.; Manjunath, M.; Bachappanavar, N.; Kiran, S. Enhanced polymer degradation of polyethylene and polypropylene by novel thermophilic consortia of Brevibacillus sps. and Aneurinibacillus sp. screened from waste management landfills and sewage treatment plants. Polym. Degrad. Stab. 2018, 149, 52–68. [Google Scholar] [CrossRef]
- Matsumiya, Y.; Murata, N.; Tanabe, E.; Kubota, K.; Kubo, M. Isolation and characterization of an ether-type polyurethane-degrading micro-organism and analysis of degradation mechanism by Alternaria sp. J. Appl. Microbiol. 2010, 10, 1946–1953. [Google Scholar]
- Bahl, S.; Dolma, J.; Jyot Singh, J.; Sehgal, S. Biodegradation of plastics: A state of the art review. Mater. Today Proc. 2021, 39, 31–34. [Google Scholar] [CrossRef]
- Kliem, S.; Kreutzbruck, M.; Bonten, C. Review on the biological degradation of polymers in various environments. Materials 2020, 13, 4586. [Google Scholar] [CrossRef] [PubMed]
- Ruggero, F.; Gori, R.; Lubello, C. Methodologies to Assess Biodegradation of Bioplastics during Aerobic Composting and Anaerobic Digestion: A Review. Waste Manag. Res. 2019, 37, 959–975. [Google Scholar] [CrossRef]
- Andreeßen, C.; Steinbüchel, A. Recent developments in non-biodegradable biopolymers: Precursors, production processes, and future perspectives. Appl. Microbiol. Biotechnol. 2019, 103, 143–157. [Google Scholar] [CrossRef]
- Muenmee, S.; Chiemchaisri, W.; Chiemchaisri, C. Microbial consortium involving biological methane oxidation in relation to the biodegradation of waste plastics in a solid waste disposal open dump site. Int. Biodeterior. Biodegrad. 2015, 102, 172–181. [Google Scholar] [CrossRef]
- Ambika, D.K.; Lakshmi, B.K.M.; Hemalatha, K.P.J. Degradadtion of the low-density polythene by Achromobacter denitrificans strain s1, a novel marine isolate. Int. J. Rec. Sci. Res. 2015, 6, 5454–5464. [Google Scholar]
- Kowalczyk, A.; Chyc, M.; Ryszka, P. Achromobacter xylosoxidans as a new microorganism strain colonizing high-density polyethylene as a key step to its biodegradation. Environ. Sci. Pollut. Res. 2016, 23, 11349–11356. [Google Scholar] [CrossRef]
- Muhonja, C.N.; Makonde, H.; Magoma, G.; Imbug, M. Biodegradability of polyethylene by bacteria and fungi from Dandora dumpsite Nairobi-Kenya. PLoS ONE 2018, 13, e0198446. [Google Scholar] [CrossRef] [PubMed]
- Novotný, C.; Malachová, K.; Adamus, G.; Kwiecień, M.; Lotti, N.; Soccio, M.; Verney, V.; Fava, F. Deterioration of irradiation/high-temperature pretreated, linear low-density polyethylene (LLDPE) by Bacillus amyloliquefaciens. Int. Biodeterior. Biodegrad. 2018, 132, 259–267. [Google Scholar] [CrossRef]
- Dela Torre, D.Y.Z.; Delos Santos, L.A.; Reyes, M.L.C.; Baculi, R.Q. Biodegradation of low-density polyethylene by bacteria isolated from serpentinization-driven alkaline spring. Philipp. Sci. Lett. 2018, 11, 1–12. [Google Scholar]
- Mallisetty, R.; Veluru, S.; Hamzah, H.T.; Hamem, M.N.K.; Tukarambai, M.; Poiba, V.R.; Srikanth, R.; Mahdi, H.S. Biodegradation of low density polyethylene (LDPE) by Paenibacillus sp. and Serratia sp. isolated from marine soil sample. In Materials Today: Proceedings; Elsevier: Amsterdam, The Netherlands, 2023. [Google Scholar]
- Rajandas, H.; Parimannan, S.; Sathasivam, K.; Ravichandran, M.; Yin, L.S. A novel FTIR-ATR spectroscopy based technique for the estimation of low-density polyethylene biodegradation. Polym. Test. 2012, 31, 1094–1099. [Google Scholar] [CrossRef]
- Dey, A.S.; Bose, H.; Mohapatra, B.; Sar, P. Biodegradation of Unpretreated Low-Density Polyethylene (LDPE) by Stenotrophomonas sp. and Achromobacter sp., isolated from waste dumpsite and drilling fluid. Front. Microbiol. 2020, 11, 603210. [Google Scholar] [CrossRef] [PubMed]
- Sekhar, V.C.; Nampoothiri, K.M.; Mohan, A.J.; Nair, N.R.; Bhaskar, T.; Pandey, A. Microbial degradation of high impact polystyrene (HIPS), an e-plastic with decabromodiphenyl oxide and antimony trioxide. J. Hazard. Mater. 2016, 318, 347–354. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, R.; Lv, S.; Zhang, B.; Wang, J.; Shao, Z. Polystyrene-degrading bacteria in the gut microbiome of marine benthic polychaetes support enhanced digestion of plastic fragments. Commun. Earth Environ. 2024, 5, 162. [Google Scholar] [CrossRef]
- El-Kurdi, N.; El-Shatoury, S.; ElBaghdady, K.; Hammad, S.; Ghazy, M. Biodegradation of polystyrene nanoplastics by Achromobacter xylosoxidans M9 offers a mealworm gut-derived solution for plastic pollution. Arch. Microbiol. 2024, 206, 238. [Google Scholar] [CrossRef]
- Jiang, S.; Su, T.; Zhao, J.; Wang, Z. Isolation, Identification, and characterization of Polystyrene-degrading bacteria from the gut of Galleria Mellonella (Lepidoptera: Pyralidae) Larvae. Front. Bioeng. Biotechnol. 2021, 9, 736062. [Google Scholar] [CrossRef]
- Kim, H.W.; Jo, J.H.; Kim, Y.B.; Le, T.K.; Cho, C.W.; Yun, C.H.; Chi, W.S.; Yeom, S.J. Biodegradation of polystyrene by bacteria from the soil in common environments. J. Hazard. Mater. 2021, 416, 126239. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, J.; Wu, W.M.; Zhao, J.; Song, Y.; Gao, L. Biodegradation and mineralization of polystyrene by plastic-eating mealworms: Part 2. Role of gut microorganisms. Environ. Sci. Technol. 2015, 49, 12087–12093. [Google Scholar] [CrossRef] [PubMed]
- Ganesh Kumar, A.; Hinduja, M.; Sujitha, K.; Nivedha Rajan, N.; Dharani, G. Biodegradation of polystyrene by deep-sea Bacillus paralicheniformis G1 and genome analysis. Sci. Total Environ. 2021, 774, 145002. [Google Scholar] [CrossRef] [PubMed]
- Auta, H.S.; Abioye, O.P.; Aransiola, S.A.; Bala, J.D.; Chukwuemeka, V.I.; Hassan Aziz, A.; Fauziah, S.H. Enhanced microbial degradation of PET and PS microplastics under natural conditions in mangrove environment. J. Environ. Manag. 2022, 304, 114273. [Google Scholar] [CrossRef]
- Taniguchi, I.; Yoshida, S.; Hiraga, K.; Miyamoto, K.; Kimura, Y.; Oda, K. Biodegradation of PET: Current status and application aspects. ACS Catal. 2019, 9, 4089–4105. [Google Scholar] [CrossRef]
- Dąbrowska, G.B.; Janczak, K.; Richert, A. Combined use of Bacillus strains and Miscanthus for accelerating biodegradation of poly (lactic acid) and poly (ethylene terephthalate). PeerJ 2021, 9, e10957. [Google Scholar] [CrossRef]
- Dhaka, V.; Singh, S.; Ramamurthy, P.C.; Samuel, J.; Swamy Sunil Kumar Naik, T.; Khasnabis, S.; Prasad, R.; Singh, J. Biological degradation of polyethylene terephthalate by rhizobacteria. Environ. Sci. Pollut. Res. 2023, 30, 116488–116497. [Google Scholar] [CrossRef]
- Giacomucci, L.; Raddadi, N.; Soccio, M.; Lotti, N.; Fava, F. Polyvinyl chloride biodegradation by Pseudomonas citronellolis and Bacillus flexus. New Biotechnol. 2019, 52, 35–41. [Google Scholar] [CrossRef]
- Zhe, Z.; Peng, H.; Yang, D.; Zhang, G.; Zhang, J.; Ju, F. Polyvinyl Chloride Degradation by Intestinal Klebsiella of Pest larvae. BioRxiv 2021, 462898. [Google Scholar] [CrossRef]
- Yadav, M.; Manderia, S.; Singh, S.; Deva, M.A. Isolation and Characterization of Polyvinyl Chloride (PVC) Degrading Bacteria from Polluted Sites of Gwalior City, M.P., India. Nat. Environ. Pollut. Technol. 2022, 21, 201–207. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, T.; Zhang, W.; Lin, J.; Wang, Z.; Lyu, S.; Tong, H. Biodegradation of polylactic acid by a mesophilic bacteria Bacillus safensis. Chemosphere 2023, 318, 137991. [Google Scholar] [CrossRef] [PubMed]
- Jeon, A.M.; Park, S.J.; Choi, T.R.; Park, J.H.; Yang, Y.H.; Yoon, J.J. Biodegradation of polyethylene and polypropylene by Lysinibacillus species JJY0216 isolated from soil grove. Polym. Degrad. Stab. 2021, 191, 109662. [Google Scholar] [CrossRef]
- Aravinthan, A.; Arkatkar, A.; Juwarkar, A.A.; Doble, M. Synergistic growth of Bacillus and Pseudomonas and its degradation potential on pretreated polypropylene. Prep. Biochem. Biotechnol. 2016, 46, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Auta, H.S.; Emenike, C.U.; Fauziah, S.H. Screening for Polypropylene degradation potential of bacteria isolated from mangrove ecosystems in Penninsular Malaysia. Int. J. Biosci. Biochem. Bioinform. 2017, 7, 245–251. [Google Scholar]
- Jain, K.; Bhunia, H.; Sudhakara Reddy, M. Degradation of polypropylene-poly L lactide blend by bacteria isolated from compost. Bioremed J. 2018, 22, 3–4. [Google Scholar] [CrossRef]
- Nanthini Devi, K.; Raju, P.; Santhanam, P. Biodegradation of low-density polyethylene and polypropylene by microbes isolated from Vaigai River, Madurai, India. Arch. Microbiol. 2021, 203, 6253–6265. [Google Scholar] [CrossRef]
- Peng, Y.H.; Shih, Y.H.; Lai, Y.C.; Liu, Y.Z.; Liu, Y.T.; Lin, N.C. Degradation of polyurethane by bacterium isolated from soil and assessment of polyurethanolytic activity of a Pseudomonas putida strain. Environ. Sci. Pollut. Res. 2014, 21, 9529–9537. [Google Scholar] [CrossRef]
- Kim, J.H.; Choi, S.H.; and Park, M.G.; Park, D.H.; Son, K.H.; Park, H.Y. Polyurethane biodegradation by Serratia sp. HY-72 isolated from the intestine of the Asian mantis Hierodula patellifera. Front. Microbiol. 2022, 13, 1005415. [Google Scholar] [CrossRef]
- Ali, S.S.; Elsamahy, T.; Koutra, E.; Kornaros, M.; El-Sheekh, M.; Abdelkarim, E.A.; Zhu, D.; Sun, J. Degradation of conventional plastic wastes in the environment: A review on current status of knowledge and future perspectives of disposal. Sci. Total Environ. 2021, 771, 144719. [Google Scholar] [CrossRef]
- Banerjee, A.; Chatterjee, K.; Madras, G. Enzymatic degradation of polymers: A brief review. Mater. Sci. Technol. 2014, 30, 567–573. [Google Scholar] [CrossRef]
- Ahmed, T.; Shahid, M.; Azeem, F.; Rasul, I.; Shah, A.A.; Noman, M.; Hameed, A.; Manzoor, N.; Manzoor, I.; Muhammad, S. Biodegradation of plastics: Current scenario and future prospects for environmental safety. Environ. Sci. Pollut. Res. 2018, 25, 7287–7298. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Zimmermann, W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: How far are we? Microb. Biotechnol. 2017, 10, 1308–1322. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, Y.S.; Ueda, H.; Koitabashi, M.; Kitamoto, H. Pretreatment with an esterase from the yeast Pseudozyma antarctica accelerates biodegradation of plastic mulch film in soil under laboratory conditions. J. Biosci. Bioeng. 2019, 127, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.S.; Zingarelli, S.; Nadeau, L.J.; Biffinger, J.C.; Drake, C.A.; Crouch, A.L.; Barlow, D.E.; Russell, J.N. Carbon catabolite repression and impranil polyurethane degradation in Pseudomonas protegens strain Pf-5. Appl. Environ. Microbiol. 2016, 82, 6080–6090. [Google Scholar] [CrossRef]
- Bano, K.; Kuddus, M.; Zaheer, M.R.; Zaheer, R.E. A novel, thermotolerant, extracellular PHB depolymerase producer Paenibacillus alvei PHB28 for bioremediation of biodegradable plastics. Turk. J. Biochem. 2019, 44, 344–353. [Google Scholar] [CrossRef]
- Kawai, F.; Kawase, T.; Shiono, T.; Urakawa, H.; Sukigara, S.; Tu, C. Enzymatic hydrophilization of polyester fabrics using a recombinant cutinase Cut190 and their surface characterization. J. Fiber Sci. Technol. 2017, 73, 8–18. [Google Scholar] [CrossRef]
- Knott, B.C.; Erickson, E.; Allen, M.D.; Gado, J.E.; Graham, R.; Kearns, F.L.; Pardo, I.; Topuzlu, E.; Anderson, J.J.; Austin, H.P.; et al. Characterization and engineering of a two-enzyme system for plastics depolymerisation. Proc. Natl. Acad. Sci. USA 2020, 117, 25476–25485. [Google Scholar] [CrossRef]
- Joo, S.; Cho, I.J.; Seo, H.; Son, H.F.; Sagong, H.Y.; Shin, T.J. Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nat. Commun. 2018, 9, 382. [Google Scholar] [CrossRef]
- Santo, M.; Weitsman, R.; Sivan, A. The role of the copper-binding enzyme, laccase, in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. Int. Biodeterior. Biodegrad. 2013, 84, 204–210. [Google Scholar] [CrossRef]
- Shyam, K.P.; Rajkumar, P.; Ramya, V.; Monisha Miriam, L.R. Biorefining Polyvinyl Alcohol (PVA) by Enterobacter cloacae and its Polyhydroxy Butyrate (PHB) Production Ability. Ind. Biotechnol. 2021, 17, 92–99. [Google Scholar] [CrossRef]
- Hibi, M.; Fukuda, D.; Kenchu, C.; Nojiri, M.; Hara, R.; Takeuchi, M. A three-component monooxygenase from Rhodococcus wratislaviensis may expand industrial applications of bacterial enzymes. Commun. Biol. 2021, 4, 16. [Google Scholar] [CrossRef] [PubMed]
- Takei, D.; Washio, K.; Morikawa, M. Identification of alkane hydroxylase genes in Rhodococcus sp. strain TMP2 that degrades a branched alkane. Biotechnol. Lett. 2008, 30, 1447–1452. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, P.; Dineshraj, D.; Yoganathan, K. Production and screening of depolymerising enymes by potential bacteria and fungi isolated from plastic waste dump yard sites. Int. J. Appl. Res. 2017, 3, 693–695. [Google Scholar]
- Ghatge, S.; Yang, Y.; Ahn, J.H.; Hur, H.G. Biodegradation of polyethylene: A brief review. Appl. Biol. Chem. 2020, 63, 27. [Google Scholar] [CrossRef]
- Raghavan, D.; Torma, A.E. DSC and FTIR characterization of biodegradation of polyethylene. Polym. Eng. Sci. 1992, 32, 438–444. [Google Scholar] [CrossRef]
- Islam, S.; Apitius, L.; Jakob, F.; Schwaneberg, U. Targeting microplastic particles in the void of diluted suspensions. Environ. Int. 2019, 123, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Zumstein, M.T.; Rechsteiner, D.; Roduner, N.; Perz, V.; Ribitsch, D.; Guebitz, G.M. Enzymatic hydrolysis of polyester thin films at the nanoscale: Effects of polyester structure and enzyme active-site accessibility. Environ. Sci. Technol. 2017, 51, 7476–7485. [Google Scholar] [CrossRef]
- Aguado, R.; Olazar, M.; San José, M.J.; Gaisán, B.; Bilbao, J. Wax formation in the pyrolysis of polyolefins in a conical spouted bed reactor. Energy Fuel 2002, 16, 1429–1437. [Google Scholar] [CrossRef]
- Rose, R.S.; Richardson, K.H.; Latvanen, E.J.; Hanson, C.A.; Resmini, M.; Sanders, I.A. Microbial degradation of plastic in aqueous solutions demon strated by CO2 evolution and quantifcation. Int. J. Mol. Sci. 2020, 21, 1176. [Google Scholar] [CrossRef]
- Yang, H.; Chen, G.; Wang, J. Microplastics in the Marine Environment: Sources, Fates, Impacts and Microbial Degradation. Toxics 2021, 9, 41. [Google Scholar] [CrossRef] [PubMed]
- Shi, A.; Fan, F.; Broach, J.R. Microbial adaptive evolution. J. Ind. Microbiol. Biotechnol. 2022, 49, kuab076. [Google Scholar] [CrossRef]
- Abebe, G.M. The Role of Bacterial Biofilm in Antibiotic Resistance and Food Contamination. Int. J. Microbiol. 2020, 25, 1705814. [Google Scholar] [CrossRef]
- Penesyan, A.; Paulsen, I.T.; Kjelleberg, S.; Gillings, M.R. Three faces of biofilms: A microbial lifestyle, a nascent multicellular organism, and an incubator for diversity. NPJ Biofilms Microbiomes 2021, 7, 80. [Google Scholar] [CrossRef] [PubMed]
- Sudhakar, M.; Trishul, A.; Mukesh, D.; Suresh Kumar, K.; Syed Jahan, S.; Inbakandan, D.; Viduthalai, R.R.; Umadevi, V.R.; Sriyutha Murthy, P.; Venkatesan, R. Biofouling and biodegradation of polyolefins in ocean waters. Polym. Degrad. Stab. 2007, 92, 1743–1752. [Google Scholar] [CrossRef]
- Tribedi, P.; Sil, A.K. Cell surface hydrophobicity: A key component in the degradation of polyethylene succinate by Pseudomonas sp. AKS2. J. Appl. Microbiol. 2014, 116, 295–303. [Google Scholar] [CrossRef]
- Zeenat; Elahi, A.; Bukhari, D.A.; Shamim, S.; Rehman, A. Plastics degradation by microbes: A sustainable approach. J. King Saud. Univ. Sci. 2021, 33, 101538. [Google Scholar] [CrossRef]
- Mevarech, M.; Felix Frolow, F.; Gloss, L.M. Halophilic enzymes: Proteins with a grain of salt. Biophys. Chem. 2000, 86, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Mozaheb, N.; Mingeot-Leclercq, M.P. Membrane Vesicle Production as a Bacterial Defense Against Stress. Front. Microbiol. 2020, 11, 600221. [Google Scholar] [CrossRef]
- Baumgarten, T.; Sperling, S.; Seifert, J.; von Bergen, M.; Steiniger, F.; Wick, L.Y.; Heipieper, H.J. Membrane vesicle formation as a multiple-stress response mechanism enhances Pseudomonas putida DOT-T1E cell surface hydrophobicity and biofilm formation. Appl. Environ. Microbiol. 2012, 78, 6217–6224. [Google Scholar] [CrossRef]
- Shree, P.; Singh, C.K.; Sodhi, K.K.; Surya, J.N.; Singh, D.K. Biofilms: Understanding the structure and contribution towards bacterial resistance in antibiotics. Med. Microecol. 2023, 16, 100084. [Google Scholar] [CrossRef]
- Jiménez-Arroyo, C.; Tamargo, A.; Molinero, N.; Moreno-Arribas, M.V. The gut microbiota, a key to understanding the health implications of micro(nano)plastics and their biodegradation. Microb. Biotechnol. 2023, 16, 34–53. [Google Scholar] [CrossRef]
- Chen, C.C.; Dai, L.; Ma, L.; Guo, R.T. Enzymatic degradation of plant biomass and synthetic polymers. Nat. Rev. Chem. 2020, 4, 114–126. [Google Scholar] [CrossRef]
- Pham, V.H.T.; Kim, J.; Shim, J.; Chang, S.; Chung, W. Coconut Mesocarp-based lignocellulosic waste as a substrate for cellulase production from high promising multienzyme-producing Bacillus amyloliquefaciens FW2 without pretreatments. Microorganisms 2022, 10, 327. [Google Scholar] [CrossRef]
- Pham, V.H.T.; Kim, J.; Chang, S.; Bang, D. Investigating Bio-Inspired Degradation of Toxic Dyes Using Potential Multi-Enzyme Producing Extremophiles. Microorganisms 2023, 11, 1273. [Google Scholar] [CrossRef]
- Pham, V.H.T.; Kim, J.; Chang, S.; Shim, J.; Chung, W.; Bang, D. Rice Husk—Cellulose-Based Agricultural Waste Enhances the Degradation of Synthetic Dyes Using Multiple Enzyme-Producing Extremophiles. Microorganisms 2023, 11, 1974. [Google Scholar] [CrossRef] [PubMed]
- Mahdi, M.S.; Ameen, R.S.; Ibrahim, H.K. Study on degradation of nylon 6 by thermophilic bacteria Anoxybacillus rupiensis Ir3 (JQ912241). Int. J. Adv. Res. Biol. Sci. 2016, 3, 200–209. [Google Scholar]
- Cada, E.J.G.; Muyot, M.L.C.; Sison, J.M.C.; Baculi, R.Q. Enhanced in vitro biodegradation of low-density polyethylene using alkaliphilic bacterial consortium supplemented with iron oxide nanoparticles. Philipp. Sci. Lett. 2019, 12, 55–69. [Google Scholar]
- Sekiguchi, T.; Sato, T.; Enoki, M.; Kanehiro, H.; Uematsu, K.; Kato, C. Isolation and characterization of biodegradable plastic degrading bacteria from deep-sea environments. JAMSTEC Rep. Res. Dev. 2011, 11, 33–41. [Google Scholar] [CrossRef]
- Dussud, C.; Meistertzheim, A.; Conan, P.; Pujo-Pay, M.; George, M.; Fabre, P.; Coudane, J.; Higgs, P.; Elineau, A.; Pedrotti, M. Evidence of niche partitioning among bacteria living on plastics, organic particles and surrounding seawaters. Environ. Pollut. 2018, 236, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, G.; Arndt, F.; Kahnt, J.; Heider, J. Adaptations to a Loss-of-Function Mutation in the Betaproteobacterium Aromatoleum aromaticum: Recruitment of alternative enzymes for anaerobic Phenylalanine degradation. J. Bacteriol. 2017, 199, e00383-17. [Google Scholar] [CrossRef] [PubMed]
- Heimowska, A.; Rutkowska, M. Thermoplastic Elastomers-Synthesis and Applications. In Environmental Degradability of Polyurethanes; IntechOpen: London, UK, 2015; pp. 75–94. [Google Scholar]
- Atanasova, N.; Stoitsova, S.; Paunova-Krasteva, T.; Kambourova, M. Plastic Degradation by Extremophilic Bacteria. Int. J. Mol. Sci. 2021, 11, 5610. [Google Scholar] [CrossRef] [PubMed]
- Devi, R.S.; Kannan, V.R.; Natarajan, K.; Nivas, D.; Kannan, K.; Chandru, S.; Antony, A.R. The role of microbes in plastic degradation. Environ. Waste Manag. 2015, 2015, 341–370. [Google Scholar]
- Rüthi, J.; Cerri, M.; Brunner, I.; Stierli, B.; Sander, M.; Frey, B. Discovery of plastic-degrading microbial strains isolated from the alpine and Arctic terrestrial plastisphere. Front. Microbiol. 2023, 14, 1178474. [Google Scholar] [CrossRef] [PubMed]
- Yoon, M.; Jeong Jeon, H.; Nam Kim, M. Biodegradation of polyethylene by a soil bacterium and AlkB cloned recombinant cell. J. Bioremed. Biodegrad. 2012, 3, 4. [Google Scholar]
- Shah, A.A.; Hasan, F.; Hameed, A.; Ahmed, S. Biological degradation of plastics: A comprehensive review. Biotechnol. Adv. 2008, 26, 246–265. [Google Scholar] [CrossRef]
- Moyer, C.L.; Morita, R.Y. Psychrophiles and Psychrotrophs; eLS; Wiley &Sons: Hoboken, NJ, USA, 2007; pp. 1–5. [Google Scholar]
- Salvador, M.; Abdulmutalib, U.; Gonzalez, J.; Kim, J.; Smith, A.; Faulon, J.L. Microbial genes for a circular and sustainable bio-PET economy. Genes 2019, 10, 373. [Google Scholar] [CrossRef]
- Blank, L.M.; Narancic, T.; Mampel, J.; Tiso, T.; O’Connor, K. Biotechnological upcycling of plastic waste and other non-conventional feedstocks in a circular economy. Curr. Opin. Biotechnol. 2020, 62, 212–219. [Google Scholar] [CrossRef]
| Thermoplastics | Thermosets | |
| Advantages |
|
|
| Disadvantages |
|
|
| Semi-Crystalline Thermoplastics | Amorphous Thermoplastics | |
| Advantages |
|
|
| Disadvantages |
|
|
| No. | Bacteria | Source of Isolation | Plastic Types | Efficiency of Degradation (%) | References |
|---|---|---|---|---|---|
| 1 | Achromobacter denitrificans | A coastal area | PE | 40% in 2 months | [43] |
| 2 | Achromobacter xylosoxidans | Soil | PE | 9.38% in 150 days | [44] |
| 3 | Bacillus cereus strain A5 | Surface water | PE | 35.72% in 16 weeks | [45] |
| 4 | Bacillus amyloliquefaciens | Composted plastic material | PE | 3.2% in 60 days | [46] |
| 5 | Bacillus krulwichiae | Rock crevices | PE | 9.9% in 90 days | [47] |
| 6 | Consortia of Brevibacillus sp. and Aneurinibacillus sp. | Sewage treatment plants and waste management landfills | PE | 58.21% in 140 days | [36] |
| 7 | Paenibacillus sp. | Soil | PE | 22.46% in 35 days | [48] |
| 8 | Microbacterium paraoxydans | A clinical sample | PE | 61% in 60 days | [49] |
| 9 | Micrococcus luteus IRN20 | Plastic dump landfill soil | PE | 18.9% in 21 days | [23] |
| 10 | Stenotrophomonas sp. | Landfill soil | PE | 8% in 100 days | [50] |
| 11 | Achromobacter sp. | Drilling fluid | PE | 8% in 100 days | [50] |
| 12 | Exiguobacterium sp. | Plastic waste | PS | 12.4% (w/w) in 30 days | [51] |
| 13 | Acinetobacter sp. | The gut microbiome marine benthic polychaetes support | PS | 12.14% in 60 days | [52] |
| 14 | Achromobacter xylosoxidans M9 | A mealworm gut-derived solution | PS | 7% in 30 days | [53] |
| 16 | Massilia sp. FS1903 | The gut of Galleria Mellonella (Lepidoptera: Pyralidae) larvae | PS | 12.97% in 30 days | [54] |
| 17 | Pseudomonas lini JNU01 | Soil | PS | 1.45% in 30 days | [55] |
| 18 | Exiguobacterium sp. | The gut of Tenebrio molitor larvae | PS | 7.4% in 60 days | [56] |
| 19 | Bacillus paralicheniformis G1 | Sediment samples from Arabian sea | PS | 34% PS in 60 days | [57] |
| 20 | Bacillus sp., Pseudomonas sp., Staphylococcus sp. | Mangrove environments | PS, PET | 18% in 90 days | [58] |
| 21 | Ideonella sakaiensis sp. | A recycling plant in Japan | PET | 75% in 70 days | [59] |
| 22 | Clostridium thermocellum | A plant compost metagenome | PET | 60% in 14 days | [15] |
| 23 | Bacillus cereus | Soil | PET | 70–55% in 180 days | [60] |
| 24 | Bacillus pseudomycoides | Soil | PET | >65% in 28 days | [61] |
| 25 | Pseudomonas citronellolis and Bacillus flexus | Soil under pine trees and compost | PVC | 19% in 30 days | [62] |
| 27 | Klebsiella sp. | Pest larvae | PVC | 7% in 10 days | [63] |
| 28 | Bacillus sp. and Micrococcus sp. | Soil | PVC | 87.3% and 91.6%, respectively in 6 months | [64] |
| 29 | Bacillus safensis PLA1006 | Landfill soil | PLA | 8% in 30 days | [65] |
| 30 | Bacillus sp. and Miscanthus sp. | A heavy-metal-contaminated environment | PLA | 70% in 6 months | [60] |
| 31 | Lysinibacillus JJY0216 | A soil grove | PP | 4% in 6 days | [66] |
| 32 | Bacillus flexus, Pseudomonas azotoformans, Bacillus subtilis | Soil | PP | 22.7% in one year | [67] |
| 33 | Bacillus cereus and Sporosarcina globispora | Mangrove sediments | PP | 12% and 11%, respectively in 40 days | [68] |
| 34 | Bacillus sp. | Municipal compost waste | PP | 12% in 15 days | [69] |
| 35 | Bacillus paramycoides | A river | PE, PP | 78.99% in 21 days | [70] |
| 36 | Pseudomonas putida | Farm soils | PU | 92% in 4 days | [71] |
| 37 | Serratia sp. HY-72 | The intestine of the Asian mantis | PU | 23.95% in 2 weeks | [72] |
| Enzyme Family | Enzyme Group | Enzyme-Producing Bacterial Source | Type of Plastics | References |
|---|---|---|---|---|
| Hydrolase | Esterases | Pseudomonas antarctica | PU | [77] |
| Lipases | Pseudomonas sp. | PU | [25] | |
| Lipases | Pseudomonas sp. | PES | [78] | |
| Depolymerase (extracellular enzymes) | Paenibacillus alvei | PHB | [79] | |
| Cutinase | Saccharomonospora viridis | PE | [31,80] | |
| PETase enzymes | Ideonella sakaiensis | PET | [32,81,82] | |
| Oxidase | Laccase | Rodococcus rubber | PE | [83] |
| PVA dehydrogenase | Enterobacter cloacae | PVA | [84] | |
| Monoxygenases | Rhodococcus wratislaviensis | PE | [85] | |
| Alkane hydroxylase | Rhodococcus sp. | PE | [86] | |
| Lignin peroxidase | Bacillus sp., Streptomyces sp. | PE | [76] | |
| Manganese peroxidase | Bacillus sp. | PE | [87] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pham, V.H.T.; Kim, J.; Chang, S. A Valuable Source of Promising Extremophiles in Microbial Plastic Degradation. Polymers 2024, 16, 2109. https://doi.org/10.3390/polym16152109
Pham VHT, Kim J, Chang S. A Valuable Source of Promising Extremophiles in Microbial Plastic Degradation. Polymers. 2024; 16(15):2109. https://doi.org/10.3390/polym16152109
Chicago/Turabian StylePham, Van Hong Thi, Jaisoo Kim, and Soonwoong Chang. 2024. "A Valuable Source of Promising Extremophiles in Microbial Plastic Degradation" Polymers 16, no. 15: 2109. https://doi.org/10.3390/polym16152109
APA StylePham, V. H. T., Kim, J., & Chang, S. (2024). A Valuable Source of Promising Extremophiles in Microbial Plastic Degradation. Polymers, 16(15), 2109. https://doi.org/10.3390/polym16152109








