Differentially Expressed Transcription Factors during Male and Female Cone Development in Pinus halepensis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Histological Analysis
2.3. Sample Selection for RNA Sequencing
2.4. RNA Isolation, Library Preparation and Sequencing
2.5. Transcriptome Analysis: Assembly and Differential Expression Analysis
2.6. Sequence Similarity and Functional Annotation
2.7. Phylogenetic Analysis
2.8. Quantitative Polymerase Chain Reaction (qPCR) Analysis
3. Results
3.1. Phenotypic Characterization of the Developmental Stages
3.2. Histological Analysis
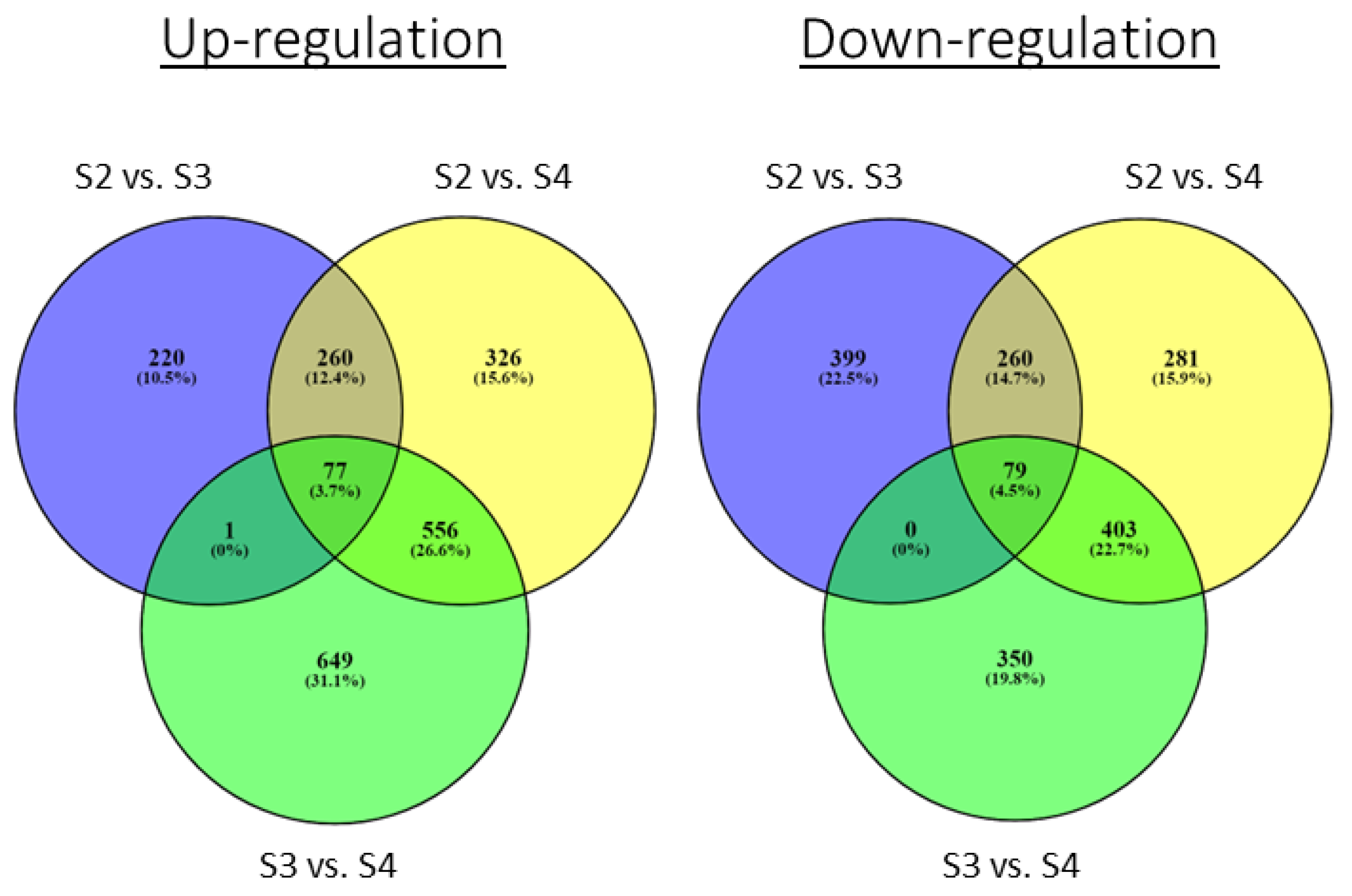
3.3. Transcriptome Analysis of the Three Bud Types at Early and Late Differentiation Stages
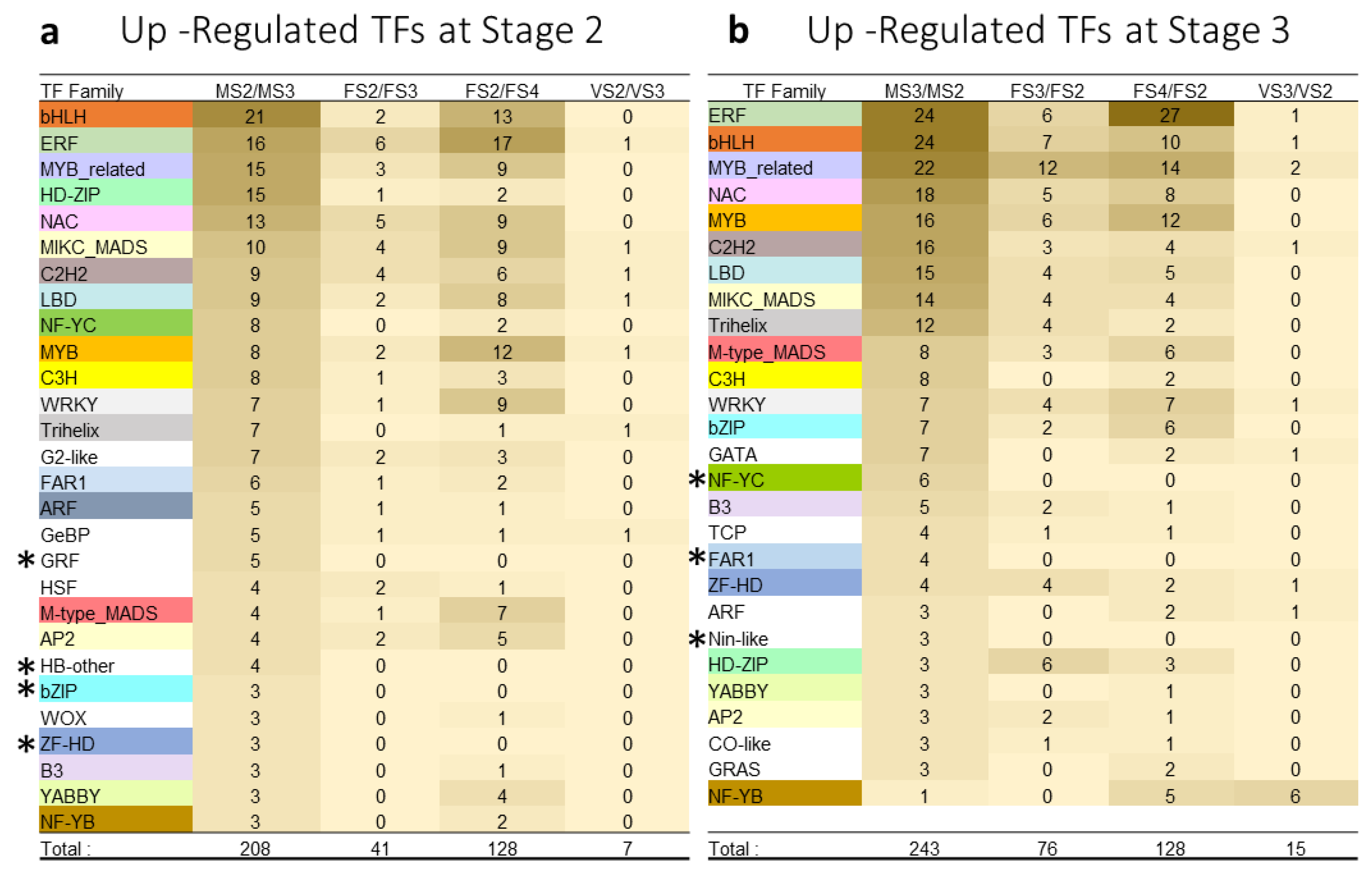
3.4. Transcription Factors Families Involved in Early and Late Stages of Cone Development
3.5. Differentially Expressed TFs Involved in Flowering Induction
3.6. Differentially Expressed TF Involved in Female Cone Differentiation
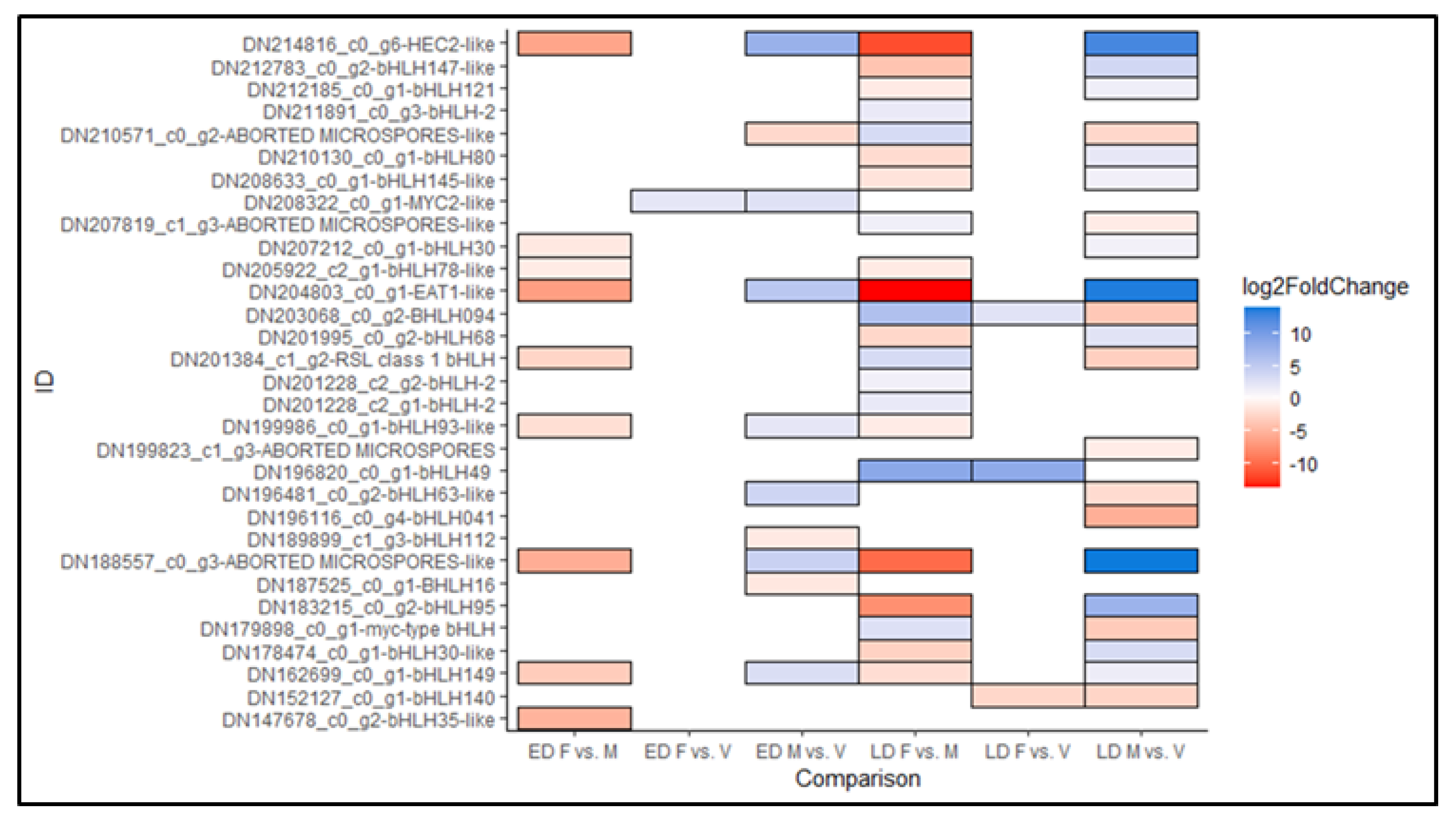
3.7. Differentially Expressed TF Involved in Male Cone Differentiation
3.8. Phylogenetic Analysis
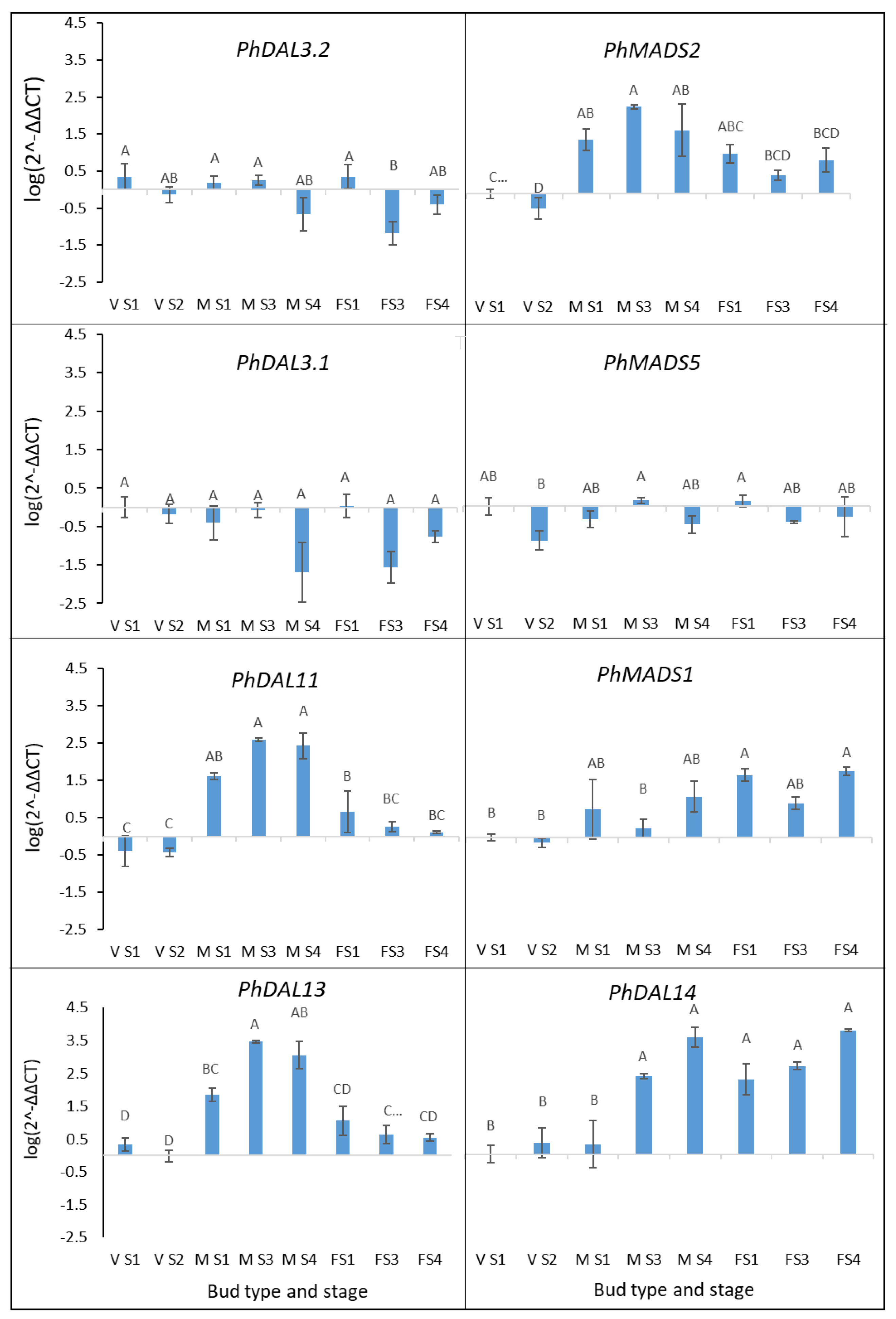
3.9. Differentially Expressed Gene Validation
4. Discussion
4.1. Reproductive Development Is Accompanied by Enriched TF Expression
4.2. Floral Induction Homologous Genes
4.3. Genes Involved in Male and Female Cone Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferrandiz, C.; Gu, Q.; Martienssen, R.; Yanofsky, M.F. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 2000, 127, 725–734. [Google Scholar] [CrossRef]
- Lee, J.; Lee, I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010, 61, 2247–2254. [Google Scholar] [CrossRef]
- Ito, S.; Song, Y.H.; Josephson-Day, A.R.; Miller, R.J.; Breton, G.; Olmstead, R.G.; Imaizumi, T. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 3582–3587. [Google Scholar] [CrossRef]
- Silva, C.S.; Puranik, S.; Round, A.; Brennich, M.; Jourdain, A.; Parcy, F.; Hugouvieux, V.; Zubieta, C. Evolution of the Plant Reproduction Master Regulators LFY and the MADS Transcription Factors: The Role of Protein Structure in the Evolutionary Development of the Flower. Front. Plant Sci. 2016, 6, 1193. [Google Scholar] [CrossRef]
- Theißen, G.; Melzer, R.; Rümpler, F. MADS-domain transcription factors and the floral quartet model of flower development: Linking plant development and evolution. Development 2016, 143, 3259–3271. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Kaya, H.; Goto, K.; Iwabuchi, M.; Araki, T. A Pair of Related Genes with Antagonistic Roles in Mediating Flowering Signals. Science 1999, 286, 1960–1962. [Google Scholar] [CrossRef]
- Wickland, D.P.; Hanzawa, Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 Gene Family: Functional Evolution and Molecular Mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar] [CrossRef]
- Moraes, T.S.; Dornelas, M.C.; Martinelli, A.P. FT/TFL1: Calibrating Plant Architecture. Front. Plant Sci. 2019, 10, 97. [Google Scholar] [CrossRef]
- Liljegren, S.J.; Gustafson-Brown, C.; Pinyopich, A.; Ditta, G.S.; Yanofsky, M.F. Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 1999, 11, 1007–1018. [Google Scholar] [CrossRef]
- Samach, A.; Onouchi, H.; Gold, S.E.; Ditta, G.S.; Schwarz-Sommer, Z.; Yanofsky, M.F.; Coupland, G. Distinct Roles of CONSTANS Target Genes in Reproductive Development of Arabidopsis. Science 2000, 288, 1613–1616. [Google Scholar] [CrossRef]
- Borner, R.; Kampmann, G.; Chandler, J.; Gleißner, R.; Wisman, E.; Apel, K.; Melzer, S. A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 2000, 24, 591–599. [Google Scholar] [CrossRef]
- De La Torre, A.R.; Piot, A.; Liu, B.; Wilhite, B.; Weiss, M.; Porth, I. Functional and morphological evolution in gymnosperms: A portrait of implicated gene families. Evol. Appl. 2020, 13, 210–227. [Google Scholar] [CrossRef]
- Mouradov, A.; Glassick, T.; Hamdorf, B.; Teasdale, R.D. Molecular control of early cone development inPinus radiata. Protoplasma 1999, 208, 3–12. [Google Scholar] [CrossRef]
- Mellerowicz, E.J.; Horgan, K.; Walden, A.; Coker, A.; Walter, C. PRFLL—A Pinus radiata homologue of FLORICAULA and LEAFY is expressed in buds containing vegetative shoot and undifferentiated male cone primordia. Planta 1998, 206, 619–629. [Google Scholar] [CrossRef]
- Maizel, A.; Busch, M.A.; Tanahashi, T.; Perkovic, J.; Kato, M.; Hasebe, M.; Weigel, D. The Floral Regulator LEAFY Evolves by Substitutions in the DNA Binding Domain. Science 2005, 308, 260–263. [Google Scholar] [CrossRef]
- Carlsbecker, A.; Tandre, K.; Johanson, U.; Englund, M.; Engström, P. The MADS-box gene DAL1 is a potential mediator of the juvenile-to-adult transition in Norway spruce (Picea abies). Plant J. 2004, 40, 546–557. [Google Scholar] [CrossRef]
- Mouradov, A.; Glassick, T.; Hamdorf, B.; Murphy, L.; Fowler, B.; Marla, S.; Teasdale, R.D. NEEDLY, a Pinus radiata ortholog of FLORICAULA/LEAFY genes, expressed in both reproductive and vegetative meristems. Proc. Natl. Acad. Sci. USA 1998, 95, 6537–6542. [Google Scholar] [CrossRef]
- Uddenberg, D.; Reimegård, J.; Clapham, D.; Almqvist, C.; von Arnold, S.; Emanuelsson, O.; Sundström, J.F. Early Cone Setting in Picea abies acrocona Is Associated with Increased Transcriptional Activity of a MADS Box Transcription Factor. Plant Physiol. 2012, 161, 813–823. [Google Scholar] [CrossRef]
- Dornelas, M.C.; Rodriguez, A.P.M. A Floricaula/Leafy gene homolog is preferentially expressed in developing female cones of the tropical pine Pinus caribaea var. caribaea. Genet. Mol. Biol. 2005, 28, 299–307. [Google Scholar] [CrossRef]
- Klintenäs, M.; Pin, P.A.; Benlloch, R.; Ingvarsson, P.K.; Nilsson, O. Analysis of conifer FLOWERING LOCUS T/TERMINAL FLOWER1-like genes provides evidence for dramatic biochemical evolution in the angiosperm FT lineage. New Phytol. 2012, 196, 1260–1273. [Google Scholar] [CrossRef]
- Niu, S.; Li, J.; Bo, W.; Yang, W.; Zuccolo, A.; Giacomello, S.; Chen, X.; Han, F.; Yang, J.; Song, Y.; et al. The Chinese pine genome and methylome unveil key features of conifer evolution. Cell 2022, 185, 204–217.e14. [Google Scholar] [CrossRef]
- Karlgren, A.; Gyllenstrand, N.; Källman, T.; Sundström, J.F.; Moore, D.; Lascoux, M.; Lagercrantz, U. Evolution of the PEBP Gene Family in Plants: Functional Diversification in Seed Plant Evolution. Plant Physiol. 2011, 156, 1967–1977. [Google Scholar] [CrossRef]
- Theißen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef]
- Weigel, D.; Meyerowitz, E.M. Activation of Floral Homeotic Genes in Arabidopsis. Science 1993, 261, 1723–1726. [Google Scholar] [CrossRef]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genetic interactions among floral homeotic genes of Arabidopsis. Development 1991, 112, 1–20. [Google Scholar] [CrossRef]
- Mouradov, A.; Hamdorf, B.; Teasdale, R.D.; Kim, J.T.; Winter, K.-U.; Theißen, G. A DEF/GLO-like MADS-box gene from a gymnosperm: Pinus radiata contains an ortholog of angiosperm B class floral homeotic genes. Dev. Genet. 1999, 25, 245–252. [Google Scholar] [CrossRef]
- Sundström, J.; Engström, P. Conifer reproductive development involves B-type MADS-box genes with distinct and different activities in male organ primordia. Plant J. 2002, 31, 161–169. [Google Scholar] [CrossRef]
- Mouradov, A.; Glassick, T.V.; Hamdorf, B.A.; Murphy, L.C.; Marla, S.S.; Yang, Y.; Teasdale, R.D. Family of MADS-Box Genes Expressed Early in Male and Female Reproductive Structures of Monterey Pine. Plant Physiol. 1998, 117, 55–62. [Google Scholar] [CrossRef][Green Version]
- Tandre, K.; Albert, V.A.; Sundas, A.; Engstrom, P. Conifer homologues to genes that control floral development in angiosperms. Plant Mol. Biol. 1995, 27, 69–78. [Google Scholar] [CrossRef]
- Englund, M.; Carlsbecker, A.; Engström, P.; Vergara-Silva, F. Morphological “primary homology” and expression of AG-subfamily MADS-box genes in pines, podocarps, and yews. Evol. Dev. 2011, 13, 171–181. [Google Scholar] [CrossRef]
- Carlsbecker, A.; Sundström, J.; Tandre, K.; Englund, M.; Kvarnheden, A.; Johanson, U.; Engström, P. The DAL10 gene from Norway spruce (Picea abies) belongs to a potentially gymnosperm-specific subclass of MADS-box genes and is specifically active in seed cones and pollen cones. Evol. Dev. 2003, 5, 551–561. [Google Scholar] [CrossRef]
- Ne’eman, G.; Goubitz, S.; Werger, M.J.A.; Shmida, A. Relationships between tree size, crown shape, gender segregation and sex allocation in Pinus halepensis, a Mediterranean pine tree. Ann. Bot. 2011, 108, 197–206. [Google Scholar] [CrossRef]
- Climent, J.; Prada, M.A.; Calama, R.; Chambel, M.R.; de Ron, D.S.; Alía, R. To grow or to seed: Ecotypic variation in reproductive allocation and cone production by young female Aleppo pine (Pinus halepensis, Pinaceae). Am. J. Bot. 2008, 95, 833–842. [Google Scholar] [CrossRef]
- Weinstein, A. Geographic variation and phenology of Pinus halepensis, P. brutia and P. eldarica in Israel. For. Ecol. Manag. 1989, 27, 99–108. [Google Scholar] [CrossRef]
- Brandizzi, F. Ruzin SE. 1999. Plant microtechnique and microscopy. 322 pp. Oxford, New York: Oxford University Press. £32.50 (softback). Ann. Bot. 2000, 86, 708. [Google Scholar] [CrossRef]
- Asgari, F.; Irian, S.; Jonoubi, P.; Majd, A. Meristem Structure, Development of Cones and Microsporogenesis of Tehran Pine (Pinus Eldarica Medw.). J. Plant Dev. 2014, 21, 83–93. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Upton, G.J.G. Fisher’s exact test. J. R. Stat. Soc. Ser. A (Stat. Soc.) 1992, 155, 395–402. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hocberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bramlett, D.L. Recognizing Developmental Stages in Southern Pine Flowers: The Key to Controlled Pollination; Southeastern Forest Experiment Station: Asheville, NC, USA, 1980. [Google Scholar]
- Yamaoka, S.; Nishihama, R.; Yoshitake, Y.; Ishida, S.; Inoue, K.; Saito, M.; Okahashi, K.; Bao, H.; Nishida, H.; Yamaguchi, K.; et al. Generative Cell Specification Requires Transcription Factors Evolutionarily Conserved in Land Plants. Curr. Biol. 2018, 28, 479–486.e5. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.-J.; Chen, X.; Song, Y.-T.; Zhang, G.-F.; Zhou, X.-Q.; Que, S.-P.; Mao, F.; Pervaiz, T.; Lin, J.-X.; Li, Y.; et al. MADS-box transcription factors MADS11 and DAL1 interact to mediate the vegetative-to-reproductive transition in pine. Plant Physiol. 2021, 187, 247–262. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Hou, X.-L.; Xing, G.-M.; Liu, J.-X.; Duan, A.-Q.; Xu, Z.-S.; Li, M.-Y.; Zhuang, J.; Xiong, A.-S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Shigyo, M.; Ito, M. Analysis of gymnosperm two-AP2-domain-containing genes. Dev. Genes Evol. 2004, 214, 105–114. [Google Scholar] [CrossRef]
- Vahala, T.; Oxelman, B.; Arnold, S.v. Two APETALA2-like genes of Picea abies are differentially expressed during development. J. Exp. Bot. 2001, 52, 1111–1115. [Google Scholar] [CrossRef]
- Nilsson, L.; Carlsbecker, A.; Sundås-Larsson, A.; Vahala, T. APETALA2 like genes from Picea abies show functional similarities to their Arabidopsis homologues. Planta 2007, 225, 589–602. [Google Scholar] [CrossRef]
- Carlsbecker, A.; Sundström, J.F.; Englund, M.; Uddenberg, D.; Izquierdo, L.; Kvarnheden, A.; Vergara-Silva, F.; Engström, P. Molecular control of normal and acrocona mutant seed cone development in Norway spruce (Picea abies) and the evolution of conifer ovule-bearing organs. New Phytol. 2013, 200, 261–275. [Google Scholar] [CrossRef]
- Klocko, A.L.; Lu, H.; Magnuson, A.; Brunner, A.M.; Ma, C.; Strauss, S.H. Phenotypic Expression and Stability in a Large-Scale Field Study of Genetically Engineered Poplars Containing Sexual Containment Transgenes. Front. Bioeng. Biotechnol. 2018, 6, 100. [Google Scholar] [CrossRef]
- Li, C.; Chen, C.; Gao, L.; Yang, S.; Nguyen, V.; Shi, X.; Siminovitch, K.; Kohalmi, S.E.; Huang, S.; Wu, K.; et al. The Arabidopsis SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene SVP. PLoS Genet. 2015, 11, e1004944. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, X.; Liu, X.; Zhang, L. Evolutionary Analysis of MIKCc-Type MADS-Box Genes in Gymnosperms and Angiosperms. Front. Plant Sci. 2017, 8, 895. [Google Scholar] [CrossRef]
- Niu, S.; Yuan, H.; Sun, X.; Porth, I.; Li, Y.; El-Kassaby, Y.A.; Li, W. A transcriptomics investigation into pine reproductive organ development. New Phytol. 2016, 209, 1278–1289. [Google Scholar] [CrossRef]
- Dreni, L.; Zhang, D. Flower development: The evolutionary history and functions of the AGL6 subfamily MADS-box genes. J. Exp. Bot. 2016, 67, 1625–1638. [Google Scholar] [CrossRef]
- Gramzow, L.; Weilandt, L.; Theißen, G. MADS goes genomic in conifers: Towards determining the ancestral set of MADS-box genes in seed plants. Ann. Bot. 2014, 114, 1407–1429. [Google Scholar] [CrossRef]



| Meristem | Stage | Description |
|---|---|---|
| Vegetative | 1 * | Undifferentiated lateral meristems. |
| 2 | Differentiated non-visible needles. | |
| 3 | Visible elongated needles. | |
| Male | 1 * | Male flowering bud clearly identified. |
| 2 | Differentiated microsporophyll. | |
| 3 | Developed pollen sacs with a tapetal layer on the edges, containing pollen mother cells. | |
| Female | 1 * | Female flowering bud (ovulate cone) clearly identified. |
| 2 | Bract differentiation, ovuliferous scales are not differentiated. | |
| 3 | Ovuliferous scales are at early differentiation. | |
| 4 | Ovuliferous scales are differentiated, ovules are visible. |
| Comparison | Up-Regulated | Down-Regulated | Total | |
|---|---|---|---|---|
| Early developmental Stage | FS2 vs. MS2 | 417 | 405 | 822 |
| FS2 vs. VS2 | 92 | 88 | 180 | |
| MS2 vs. VS2 | 519 | 689 | 1208 | |
| Late developmental Stage | FS3 vs. MS3 | 2382 | 2115 | 4497 |
| FS3 vs. VS3 | 1020 | 1300 | 2320 | |
| MS3 vs. VS3 | 2042 | 2521 | 5563 | |
| Vegetative bud development | VS2 vs. VS3 | 139 | 224 | 363 |
| Male cone development | MS2 vs. MS3 | 2200 | 2155 | 4355 |
| Female cone development | FS2 vs. FS3 | 558 | 738 | 1296 |
| FS2 vs. FS4 | 1219 | 1023 | 2242 | |
| FS3 vs. FS4 | 1283 | 832 | 2115 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reisfeld, G.; Faigenboim, A.; Fox, H.; Zemach, H.; Eshed Williams, L.; David-Schwartz, R. Differentially Expressed Transcription Factors during Male and Female Cone Development in Pinus halepensis. Agronomy 2022, 12, 1588. https://doi.org/10.3390/agronomy12071588
Reisfeld G, Faigenboim A, Fox H, Zemach H, Eshed Williams L, David-Schwartz R. Differentially Expressed Transcription Factors during Male and Female Cone Development in Pinus halepensis. Agronomy. 2022; 12(7):1588. https://doi.org/10.3390/agronomy12071588
Chicago/Turabian StyleReisfeld, Gilad, Adi Faigenboim, Hagar Fox, Hanita Zemach, Leor Eshed Williams, and Rakefet David-Schwartz. 2022. "Differentially Expressed Transcription Factors during Male and Female Cone Development in Pinus halepensis" Agronomy 12, no. 7: 1588. https://doi.org/10.3390/agronomy12071588
APA StyleReisfeld, G., Faigenboim, A., Fox, H., Zemach, H., Eshed Williams, L., & David-Schwartz, R. (2022). Differentially Expressed Transcription Factors during Male and Female Cone Development in Pinus halepensis. Agronomy, 12(7), 1588. https://doi.org/10.3390/agronomy12071588






