Excavation of Genes Response to Heat Resistance by Transcriptome Analysis in Bottle Gourd (Lagenaria siceraria (Mol.) Standl.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Heat Stress Treatment
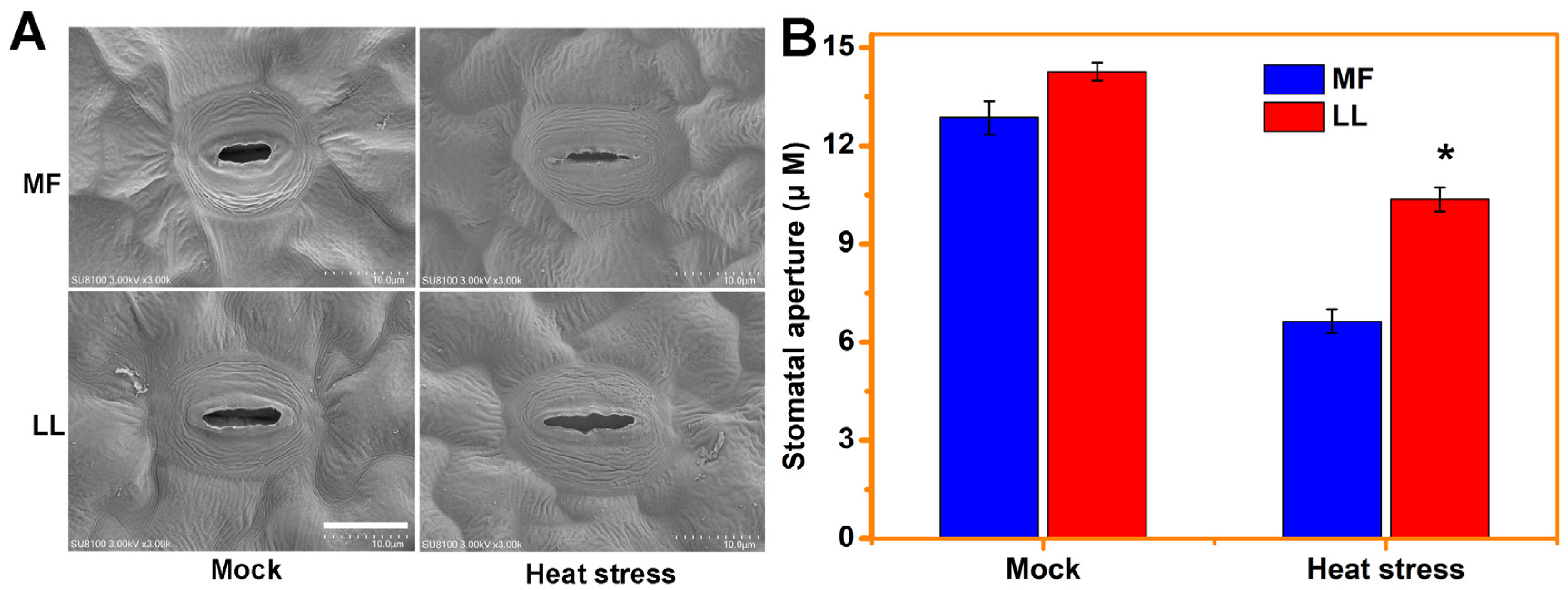
2.2. Stomatal Aperture Analyses
2.3. Transcriptome Sequencing
2.4. Bioinformatic Analysis of RNA-Seq Data
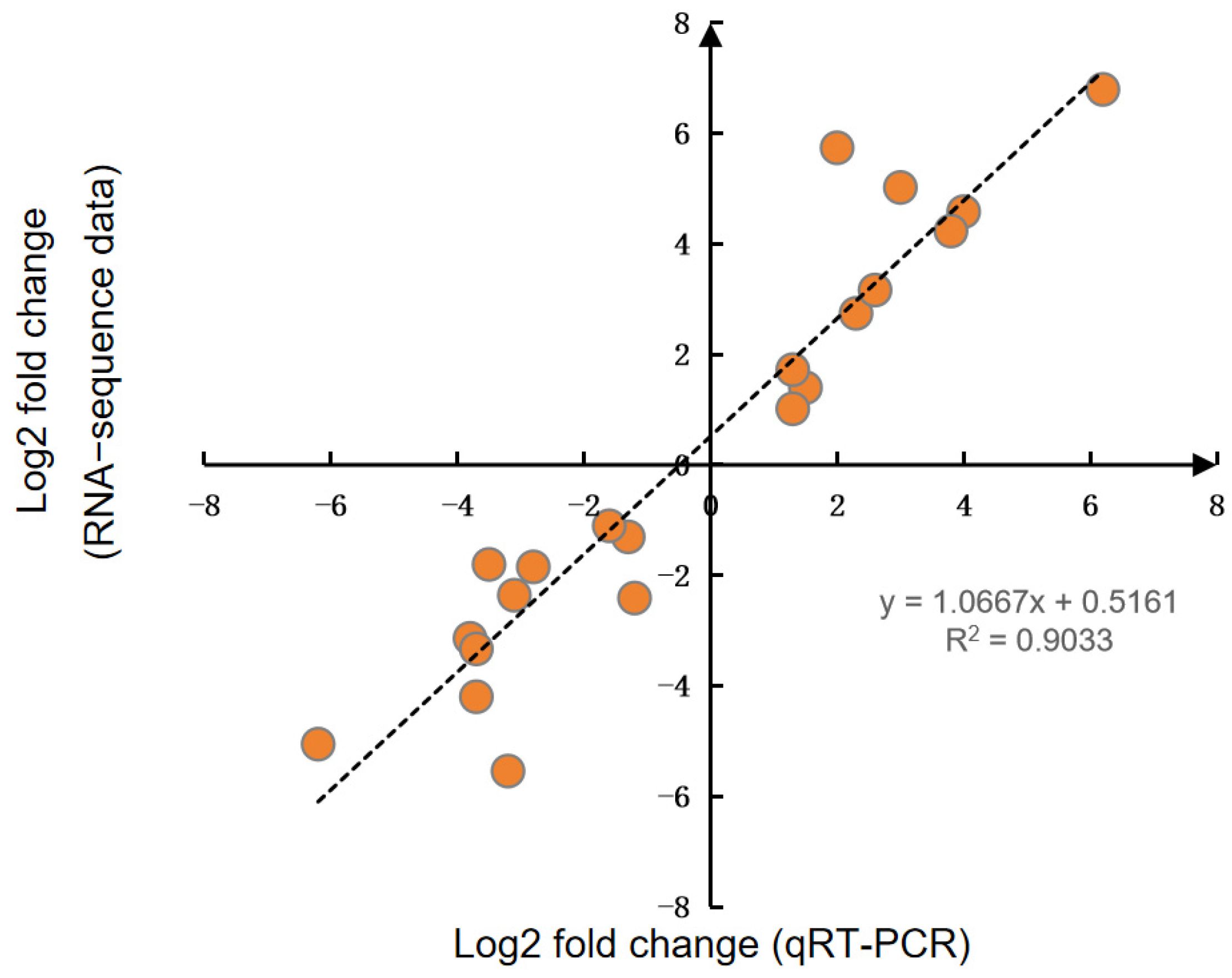
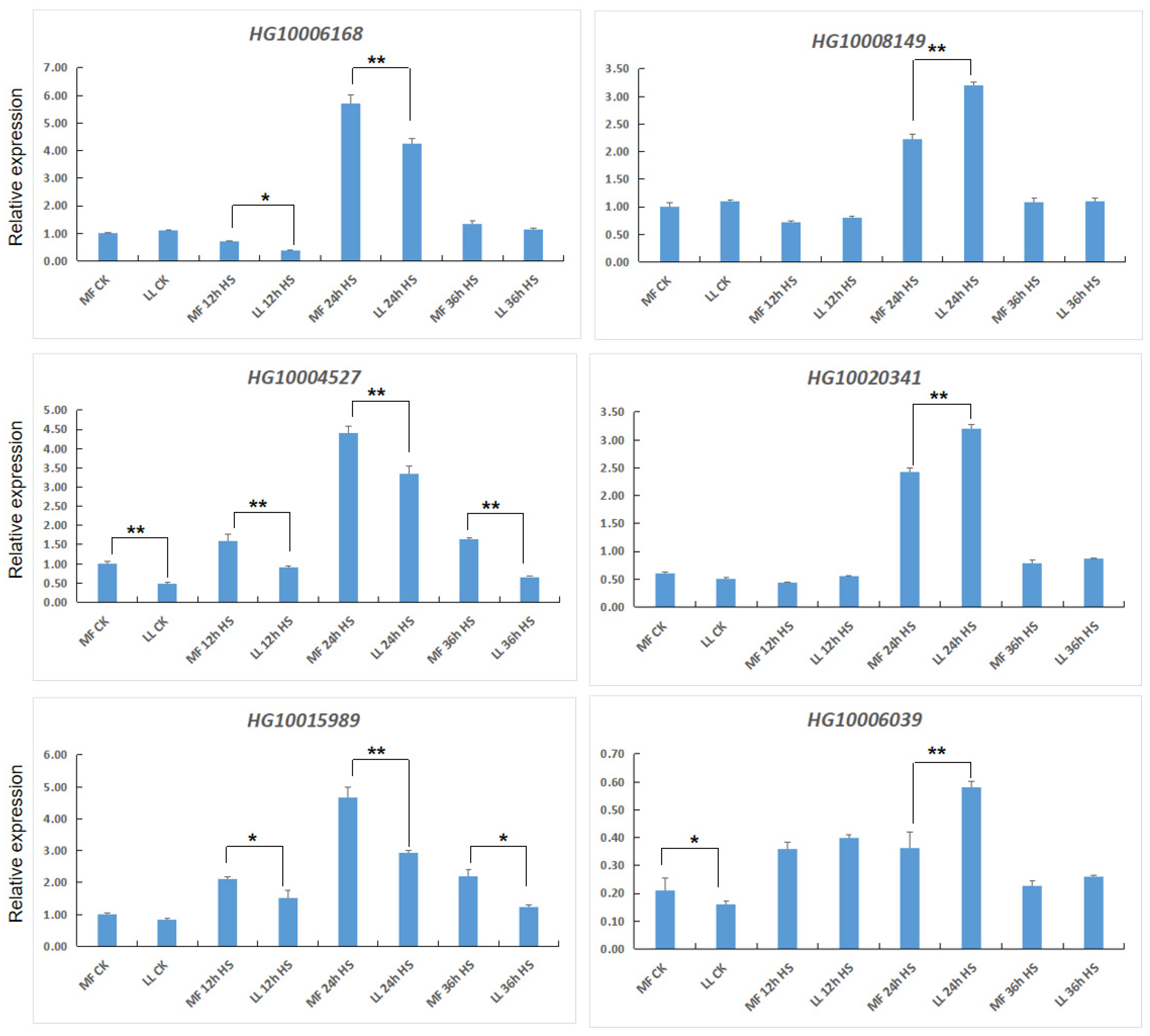
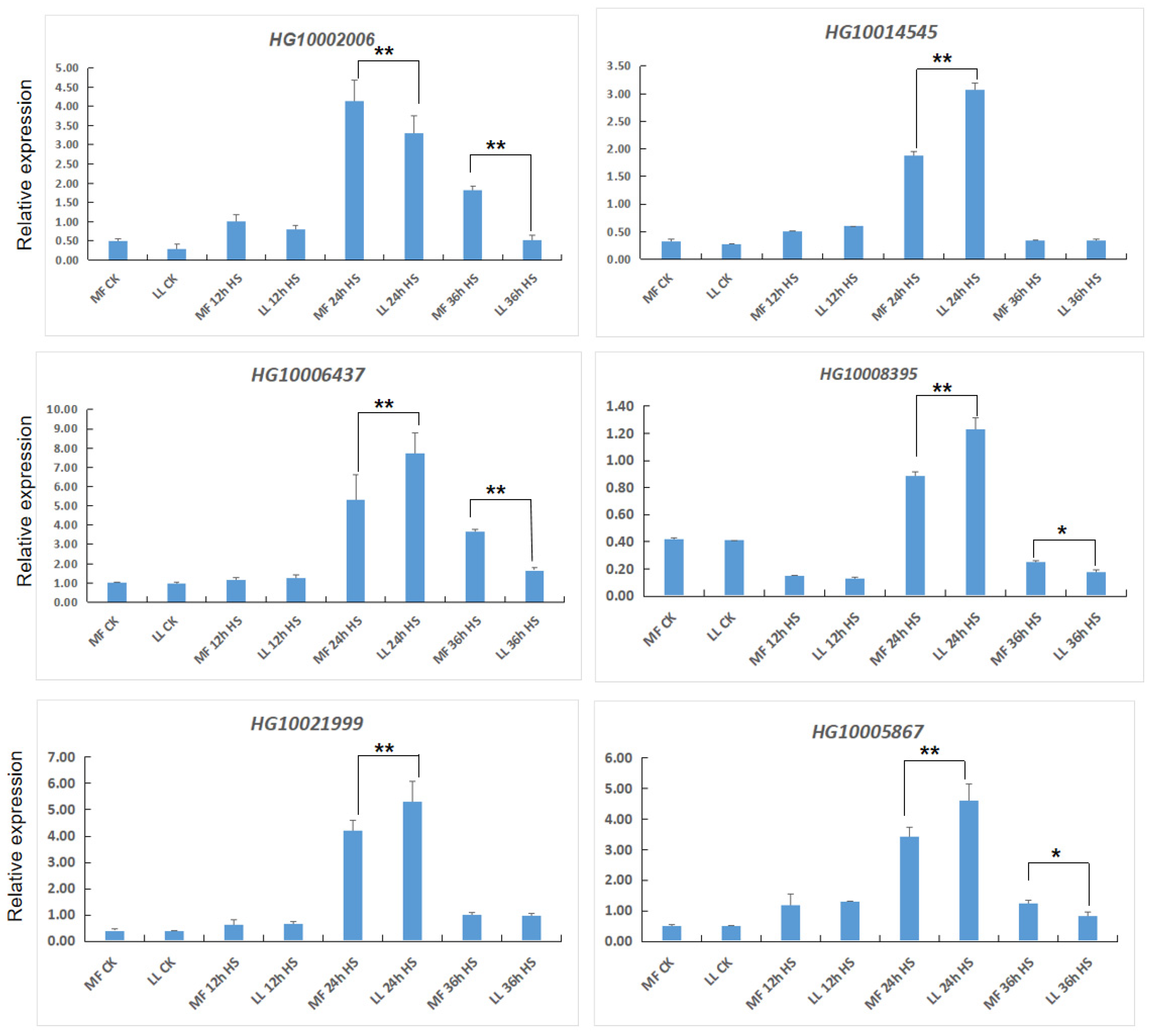
2.5. Quantitative Real-Time PCR (qRT-PCR)
3. Results
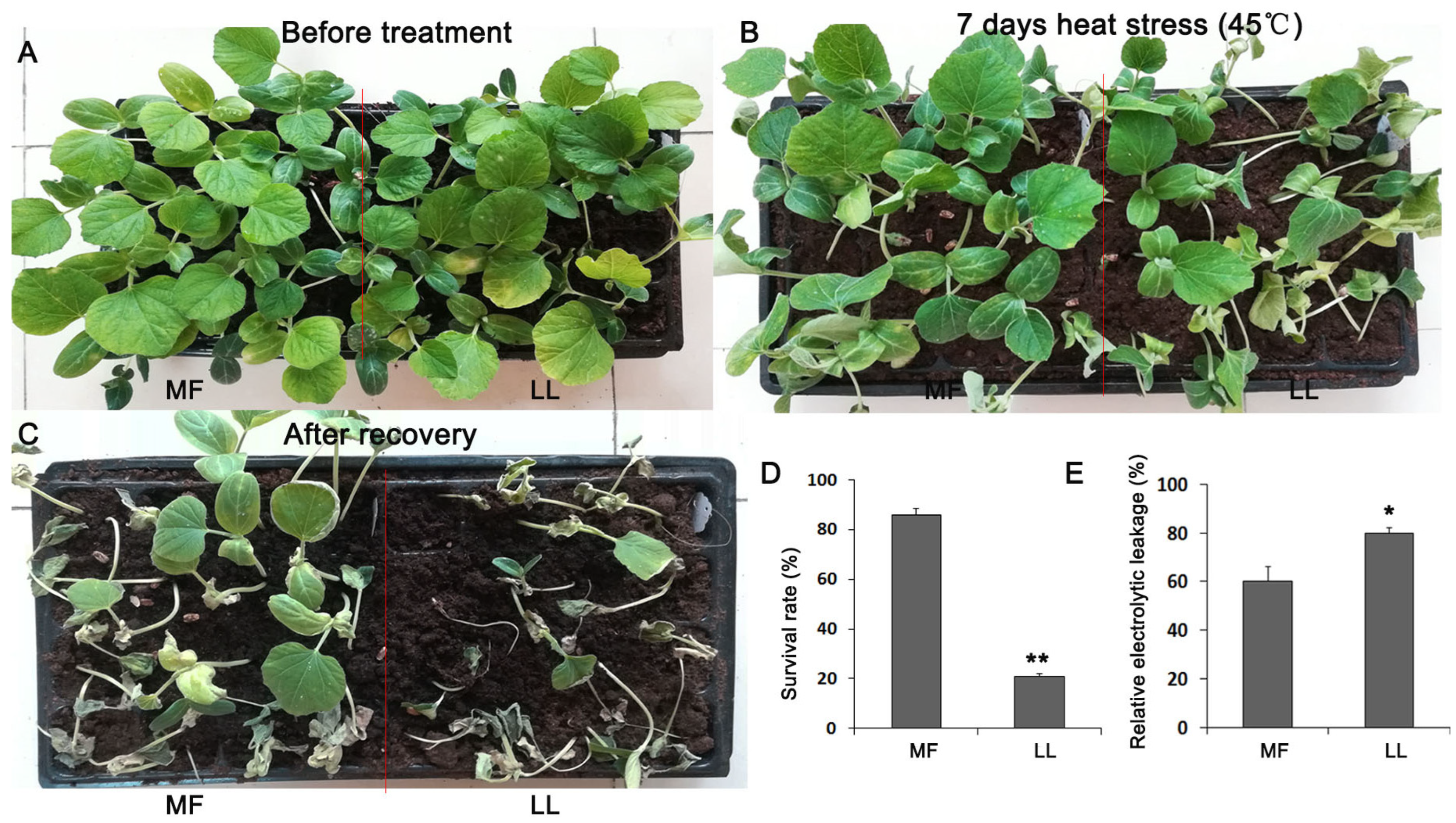
3.1. MF Is a Resistant Cultivar to Heat Stress
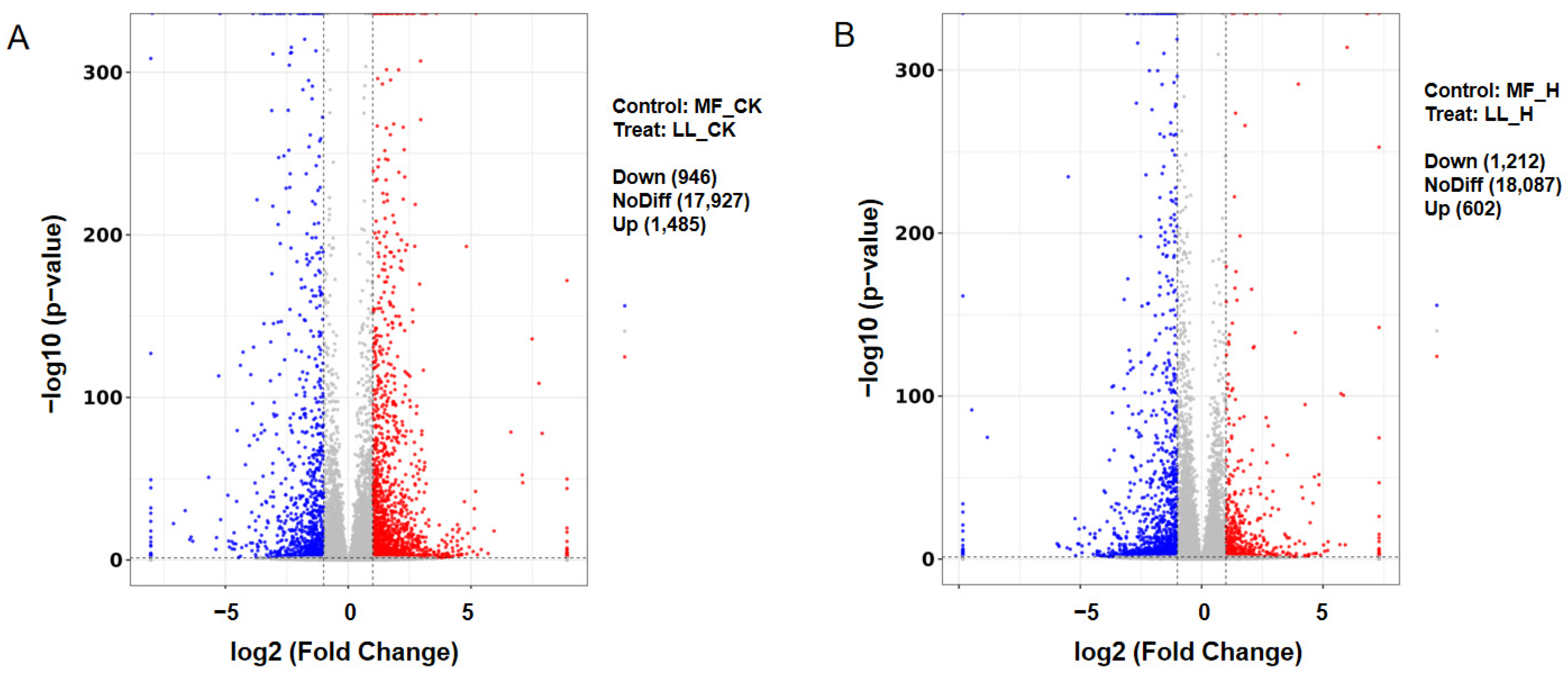
3.2. Heat Stress-Induced Transcriptional Changes
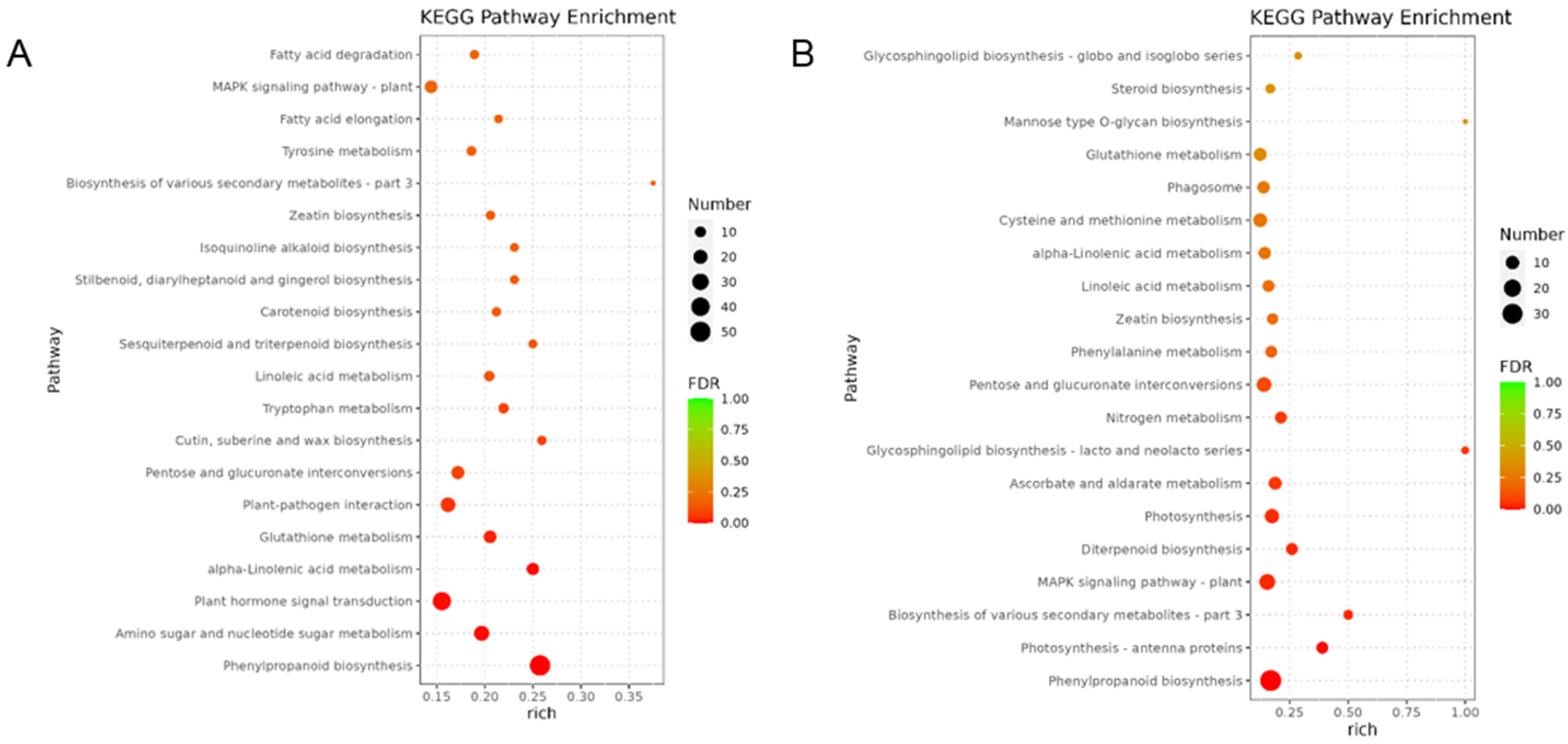
3.3. Enriched Gene Categories Related to Heat Stress
3.4. Expression of bHLH-Related Genes under Heat Stress
3.5. Expression of MAPK-Related Genes under Heat Stress
4. Discussion
4.1. Analysis of bHLHs under Heat Stress
4.2. Analysis of the MAPK Signal Pathway under Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Wang, D.; Qin, B.X.; Li, X.; Tang, D.; Zhang, Y.; Cheng, Z.K.; Xue, Y.B. Nucleolar DEAD-Box RNA helicase TOGR1 regulates thermotolerant growth as a Pre-rRNA chaperone in rice. PLoS Genet. 2016, 12, e1005844. [Google Scholar] [CrossRef]
- McClung, C.R.; Davis, S.J. Ambient thermometers in plants: From physiological outputs towards mechanisms of thermal sensing. Curr. Biol. 2010, 20, 1086–1092. [Google Scholar] [CrossRef]
- Mittler, R.; Finka, A.; Goloubinoff, P. How do plants feel the heat? Trends Biochem. Sci. 2012, 37, 118–125. [Google Scholar] [CrossRef]
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Damián, B.; Vicent, A.; Aurelio, G.C.; Inupakutika, M.A.; Ron, M. ABA is required for the accumulation of APX1 and MBF1c during a combination of water deficit and heat stress. J. Exp. Bot. 2016, 67, 5381–5390. [Google Scholar] [CrossRef]
- Larkindale, J.; Hall, J.D.; Knight, M.R.; Vierling, E. Heat Stress Phenotypes of Arabidopsis Mutants Implicate Multiple Signaling Pathways in the Acquisition of Thermotolerance. Plant Physiol. 2005, 138, 882–897. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Liu, J.P.; Zhang, C.C.; Wei, C.C.; Liu, X.; Wang, M.G.; Yu, F.F.; Xie, Q.; Tu, J.M. The RING finger ubiquitin E3 ligase OsHTAS enhances heat tolerance by promoting H2O2-induced stomatal closure in rice. Plant Physiol. 2015, 170, 429–443. [Google Scholar] [CrossRef]
- Takabatake, R.; Karita, E.; Seo, S.; Mitsuhara, I.; Kuchitsu, K.; Ohashi, Y. Pathogen-induced calmodulin isoforms in basal resistance against bacterial and fungal pathogens in tobacco. Plant Cell Physiol. 2007, 48, 414–423. [Google Scholar] [CrossRef]
- Yang, X.H.; Liang, Z.; Lu, C.M. Genetic engineering of the biosynthesis of glycinebetaine enhances photosynthesis against high temperature stress in transgenic tobacco plants. Plant Physiol. 2005, 138, 2299–2309. [Google Scholar] [CrossRef]
- Zhu, W.; Lu, M.H.; Gong, Z.H.; Chen, R.G. Cloning and expression of a small heat shock protein gene CaHSP24 from pepper under abiotic stress. Afr. J. Biotechnol. 2011, 10, 4968–4976. [Google Scholar]
- Li, H.; Liu, S.S.; Yi, C.Y.; Wang, F.; Zhou, J.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Yu, J.Q. Hydrogen peroxide mediates abscisic acid-induced HSP70 accumulation and heat tolerance in grafted cucumber plants. Plant Cell Environ. 2015, 37, 2768–2780. [Google Scholar] [CrossRef]
- Yu, B.W.; Yan, S.S.; Zhou, H.Y.; Dong, R.; Lei, J.J.; Chen, C.M.; Cao, B.H. Overexpression of CsCaM3 Improves High Temperature Tolerance in Cucumber. Front. Plant Sci. 2018, 9, 797. [Google Scholar] [CrossRef]
- Erickson, D.L.; Smith, B.D.; Clarke, A.C.; Sandweiss, D.H.; Tuross, N. An Asian origin for a 10,000-year-old domesticated plant in the Americas. Proc. Natl. Acad. Sci. USA 2005, 102, 18315–18320. [Google Scholar] [CrossRef]
- Mashilo, J.; Odindo, A.O.; Shimelis, H.A.; Musenge, P.; Tesfay, S.Z.; Magwaza, L.S. Photosynthetic response of bottle gourd [Lagenaria siceraria (Molina) Standl.] to drought stress: Relationship between cucurbitacins accumulation and drought tolerance. Sci. Hortic. 2018, 231, 133–143. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, L.; Wang, L.; Guo, S. Bottle gourd rootstock-grafting promotes photosynthesis by regulating the stomata and non-stomata performances in leaves of watermelon seedlings under NaCl stress. J. Plant Physiol. 2015, 186, 50–58. [Google Scholar] [CrossRef]
- Singh, A.P.; Singh, T.; Rickard, C.L. Visualising impregnated chitosan in Pinus radiata early wood cells using light and scanning electron microscopy. Micron 2010, 41, 263–267. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.K.; Zhang, Y.; Chen, J.; Zhang, Z.J.; Wang, J.; Li, S.T.; Li, R.Q.; Bolund, L.; et al. WEGO, A web tool for plotting GO annotations. Nocleic Acids Res. 2006, 34, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, 277–280. [Google Scholar] [CrossRef]
- Li, Y.W.; Wang, Y.; Wu, X.Y.; Wang, J.; Wu, X.H.; Wang, B.G.; Lu, Z.F.; Li, G.J. Novel genomic regions of fusarium wilt resistance in bottle gourd [Lagenaria siceraria (Mol.) Standl.] Discovered in Genome-Wide Association Study. Front. Plant Sci. 2021, 12, 650157. [Google Scholar] [CrossRef]
- Xu, P.; Wang, Y.; Sun, F.S.; Wu, R.L.; Du, H.L.; Wang, Y.H.; Jiang, L.B.; Wu, X.H.; Wu, X.Y.; Yang, L.M.; et al. Long-read genome assembly and genetic architecture of fruit shape in the bottle gourd. Plant J. 2021, 107, 956–968. [Google Scholar] [CrossRef]
- Duek, P.D.; Fankhauser, C. bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 2005, 10, 51–54. [Google Scholar] [CrossRef]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef]
- Cao, Z.B.; Tang, H.W.; Cai, Y.H.; Zeng, B.H.; Zhao, J.L.; Tang, X.Y.; Lu, M.; Wang, H.M.; Zhu, X.J.; Wu, X.F.; et al. Natural variation of HTH5 from wild rice, Oryza rufipogon Griff., is involved in conferring high-temperature tolerance at the heading stage. Plant Biotechnol. 2022, 20, 1591–1605. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.E.; Meacham-Hensold, K.; Lemonnier, P.; Slattery, R.A.; Benjamin, C.; Bernacchi, C.J.; Lawson, T.; Cavanagh, A.P. The effect of increasing temperature on crop photosynthesis: From enzymes to ecosystems. J. Exp. Bot. 2021, 72, 2822–2844. [Google Scholar] [CrossRef]
- Tian, Y.L.; Chen, J.; Chen, C.Q.; Deng, A.X.; Song, Z.W.; Zheng, C.Y.; Willem, H.M.; Zhang, W.J. Warming impacts on winter wheat phenophase and grain yield under field conditions in Yangtze delta plain, China. Field Crops Res. 2012, 134, 193–199. [Google Scholar] [CrossRef]
- Khan, A.H.; Ma, Y.Z.; Wu, Y.L.; Akbar, A.D.; Shaban, M.H.; Ullah, A.B.; Deng, J.W.; Khan, A.S.; Chi, H.B.; Zhu, L.F.; et al. High-temperature stress suppresses allene oxide cyclase 2 and causes male sterility in cotton by disrupting jasmonic acid signaling. Crop J. 2023, 11, 33–45. [Google Scholar] [CrossRef]
- Abbas, S. Climate change and major crop production: Evidence from Pakistan. Environ. Sci. Pollut. Res. 2022, 29, 5406–5414. [Google Scholar] [CrossRef]
- Yokotani, N.; Sato, Y.; Tanabe, S.; Chujo, T.; Shimizu, T.; Okada, K.; Yamane, H.; Shimono, M.; Sugano, S.; Takatsuji, H.; et al. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 2013, 64, 5085–5097. [Google Scholar] [CrossRef]
- Chen, N.; Chi, X.Y.; Pan, L.J.; Yu, S.L. Advances in MYB transcription factors during salt-stress regulation in plants. Plant Physiol. J. 2015, 51, 1395–1399. [Google Scholar]
- Ma, H.Z.; Liu, C.; Li, Z.X.; Ran, Q.J.; Xie, G.N.; Wang, B.M.; Fang, S.; Chu, J.F.; Zhang, J.R. ZmbZIP4 contributes to stress resistance in maize by regulating ABA synthesis and root development. Plant Physiol. 2018, 178, 753–770. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef]
- Yao, P.F.; Sun, Z.X.; Li, C.L.; Zhao, X.R.; Li, M.F.; Deng, R.Y.; Huang, Y.J.; Zhao, H.X.; Chen, H.; Wu, Q. Overexpression of Fagopyrum tataricum FtbHLH2 enhances tolerance to cold stress in transgenic Arabidopsis. Plant Physiol. Biochem. 2018, 125, 85–94. [Google Scholar] [CrossRef]
- Li, F.; Guo, S.Y.; Zhao, Y.; Chen, D.Z.; Chong, K.; Xu, Y.Y. Overexpression of a homopeptide repeat-containing bHLH protein gene (OrbHLH001) from Dongxiang Wild Rice confers freezing and salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 2010, 29, 977–986. [Google Scholar] [CrossRef]
- Seo, J.S.; Joo, J.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; Song, S.I.; Cheong, J.J.; Lee, J.S.; Kim, J.K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef]
- Li, M.; He, P.; Zhao, Z.; Liu, J.; Liu, H.; Ma, S.; Shen, Y.; Li, B. Effect of temperature on betacyanins synthesis and the transcriptome of Suaeda salsa. Front. Plant Sci. 2023, 14, 1203089. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, A.; Wu, X.; Zhu, Z.; Yang, Z.; Zhu, Y.; Zha, D. Transcriptome analysis revealed expression of genes related to anthocyanin biosynthesis in eggplant (Solanum melongena L.) under high-temperature stress. BMC Plant Biol. 2019, 19, 387. [Google Scholar] [CrossRef]
- Jiang, L.; Tian, X.; Li, S.; Fu, Y.; Xu, J.; Wang, G. The AabHLH35 transcription factor identified from Anthurium and raeanum is involved in cold and drought tolerance. Plants 2019, 8, 216. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, S.; Tian, Y.; Wang, Q.; Chen, S.; Li, H.; Ma, C.; Li, H. Functional characterization of a sugar beet BvbHLH93 transcription factor in salt stress tolerance. Int. J. Mol. Sci. 2021, 22, 3669. [Google Scholar] [CrossRef]
- Samarina, L.S.; Bobrovskikh, A.V.; Doroshkov, A.V.; Malyukova, L.S.; Matskiv, A.O.; Rakhmangulov, R.S.; Koninskaya, N.G.; Malyarovskaya, V.I.; Tong, W.; Xia, E. Comparative expression analysis of stress-inducible candidate genes in response to cold and drought in tea plant [Camellia sinensis (L.) Kuntze]. Front. Genet. 2020, 11, 611283. [Google Scholar] [CrossRef]
- Sharma, N.; Xin, R.; Kim, D.-H.; Sung, S.; Lange, T.; Huq, E. No flowering in short day (NFL) is a bHLH transcription factor that promotes flowering specifically under short-day conditions in Arabidopsis. Development 2016, 143, 682–690. [Google Scholar]
- Nadal, E.D.; Alepuz, P.M.; Posas, F. Dealing with osmostress through MAP kinase activation. Emro Rep. 2002, 3, 735–740. [Google Scholar] [CrossRef]
- Lin, L.; Wu, J.; Jiang, M.; Wang, Y. Plant mitogen-activated protein kinase cascades in environmental stresses. Int. J. Mol. Sci. 2021, 22, 1543. [Google Scholar] [CrossRef]
- Cargnello, M.; Roux, P.P. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. 2011, 75, 50–83. [Google Scholar] [CrossRef]
- Kyriakis, J.M.; Avruch, J. Mammalian MAPK signal transduction pathways activated by stress and inflammation: A 10-year update. Physiol. Rev. 2012, 92, 689–737. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, L.; An, H.L.; Wu, C.G.; Guo, X.Q. GhMPK16, a novel stress-responsive group D MAPK gene from cotton, is involved in disease resistance and drought sensitivity. BMC Mol. Biol. 2011, 12, 22. [Google Scholar] [CrossRef]
- Sadau, S.B.; Ahmad, A.; Tajo, S.M.; Ibrahim, S.; Kazeem, B.B.; Wei, H.; Yu, S. Overexpression of GhMPK3 from cotton enhances cold, drought, and salt stress in Arabidopsis. Agronomy 2021, 11, 1049. [Google Scholar] [CrossRef]
- Zhang, L.; Xi, D.M.; Li, S.W.; Gao, Z.; Zhao, S.L.; Shi, J.; Wu, C.G.; Guo, X.Q. A cotton group C MAP kinase gene, GhMPK2, positively regulates salt and drought tolerance in tobacco. Plant Mol. Biol. 2011, 77, 17–31. [Google Scholar] [CrossRef]
- Jammes, F.; Song, C.; Shin, D.; Munemasa, S.; Takeda, K.; Gu, D.; Cho, D.; Lee, S.; Giordo, R.; Sritubtim, S. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 20520–20525. [Google Scholar] [CrossRef]
- Khokon, M.A.R.; Salam, M.A.; Jammes, F.; Ye, W.; Hossain, M.A.; Okuma, E.; Nakamura, Y.; Mori, I.C.; Kwak, J.M.; Murata, Y. MPK9 and MPK12 function in SA-induced stomatal closure in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2017, 81, 1394–1400. [Google Scholar] [CrossRef]
- Jammes, F.; Yang, X.; Xiao, S.; Kwak, J.M. Two Arabidopsis guard cell-preferential MAPK genes, MPK9 and MPK12, function in biotic stress response. Plant Signal. Behav. 2011, 6, 1875–1877. [Google Scholar] [CrossRef]
- Savatin, D.; Gigli, B.; Marti, L.; Fabbri, C.; Cervone, F.; De Lorenzo, G. The Arabidopsis NPK1-related protein kinases ANPs are required for elicitor-induced oxidative burst and immunity. Plant Physiol. 2014, 165, 1188–1202. [Google Scholar] [CrossRef]
- Marti, L.; Savatin, D.V.; Gigli-Bisceglia, N.; De Turris, V.; Cervone, F.; De Lorenzo, G. The intracellular ROS accumulation in elicitor-induced immunity requires the multiple organelle-targeted Arabidopsis NPK1-related protein kinases. Plant Cell Environ. 2021, 44, 931–947. [Google Scholar] [CrossRef]
| Sample | Reads No. | Bases (bp) | Q30 (bp) | Clean Reads No. | Clean Data (bp) | Q30 (%) | Clean Reads % | Clean Data % |
|---|---|---|---|---|---|---|---|---|
| MF_CK_1 | 48713312 | 7,355,710,112 | 7,019,875,701 | 45173544 | 6,821,205,144 | 95.43 | 92.73 | 92.73 |
| MF_CK_2 | 45757598 | 6,909,397,298 | 6,587,698,893 | 42487732 | 6,415,647,532 | 95.34 | 92.85 | 92.85 |
| MF_CK_3 | 43690728 | 6,597,299,928 | 6,275,922,552 | 40610484 | 6,132,183,084 | 95.12 | 92.94 | 92.94 |
| LL_CK_1 | 43525386 | 6,572,333,286 | 6,259,514,252 | 40395314 | 6,099,692,414 | 95.24 | 92.8 | 92.8 |
| LL_CK_2 | 49867998 | 7,530,067,698 | 7,180,347,054 | 46316818 | 6,993,839,518 | 95.35 | 92.87 | 92.87 |
| LL_CK_3 | 46377310 | 7,002,973,810 | 6,575,537,624 | 43003502 | 6,493,528,802 | 93.89 | 92.72 | 92.72 |
| MF_H_1 | 48711970 | 7,355,507,470 | 7,003,097,638 | 45215592 | 6,827,554,392 | 95.2 | 92.82 | 92.82 |
| MF_H_2 | 50998488 | 7,700,771,688 | 7,335,822,954 | 47313752 | 7,144,376,552 | 95.26 | 92.77 | 92.77 |
| MF_H_3 | 48670696 | 7,349,275,096 | 7,003,542,935 | 45160744 | 6,819,272,344 | 95.29 | 92.78 | 92.78 |
| LL_H_1 | 46504370 | 7,022,159,870 | 6,697,944,713 | 43154332 | 6,516,304,132 | 95.38 | 92.79 | 92.79 |
| LL_H_2 | 47952574 | 7,240,838,674 | 6,914,489,745 | 44463080 | 6,713,925,080 | 95.49 | 92.72 | 92.72 |
| LL_H_3 | 50213476 | 7,582,234,876 | 7,255,490,965 | 46615128 | 7,038,884,328 | 95.69 | 92.83 | 92.83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Liu, W.; Peng, Q.; Shi, S.; Wang, Y.; Cao, L.; Jiang, B.; Lin, Y.; Zhao, T.; Cui, X.; et al. Excavation of Genes Response to Heat Resistance by Transcriptome Analysis in Bottle Gourd (Lagenaria siceraria (Mol.) Standl.). Agronomy 2024, 14, 299. https://doi.org/10.3390/agronomy14020299
Wang M, Liu W, Peng Q, Shi S, Wang Y, Cao L, Jiang B, Lin Y, Zhao T, Cui X, et al. Excavation of Genes Response to Heat Resistance by Transcriptome Analysis in Bottle Gourd (Lagenaria siceraria (Mol.) Standl.). Agronomy. 2024; 14(2):299. https://doi.org/10.3390/agronomy14020299
Chicago/Turabian StyleWang, Min, Wenrui Liu, Qingwu Peng, Shaoqi Shi, Ying Wang, Liqin Cao, Biao Jiang, Yu’e Lin, Tianyue Zhao, Xiaojuan Cui, and et al. 2024. "Excavation of Genes Response to Heat Resistance by Transcriptome Analysis in Bottle Gourd (Lagenaria siceraria (Mol.) Standl.)" Agronomy 14, no. 2: 299. https://doi.org/10.3390/agronomy14020299
APA StyleWang, M., Liu, W., Peng, Q., Shi, S., Wang, Y., Cao, L., Jiang, B., Lin, Y., Zhao, T., Cui, X., & Yang, S. (2024). Excavation of Genes Response to Heat Resistance by Transcriptome Analysis in Bottle Gourd (Lagenaria siceraria (Mol.) Standl.). Agronomy, 14(2), 299. https://doi.org/10.3390/agronomy14020299






