The Role of the ADF Gene Family in Maize Response to Abiotic Stresses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Download and Arrangement of ADF Gene Family in Maize
2.2. Clustering, Collinearity, Gene Structure and Promoter Analysis of ADF Family
2.3. Drought Stress Treatment in Seedling Stage of Maize
2.4. Real-Time Fluorescence Quantitative PCR Detection
2.5. Analysis of Transcription Level of Maize ADF Family under Abiotic Stresses
2.6. Candidate-Gene Association Analysis in ZmADF5
3. Results
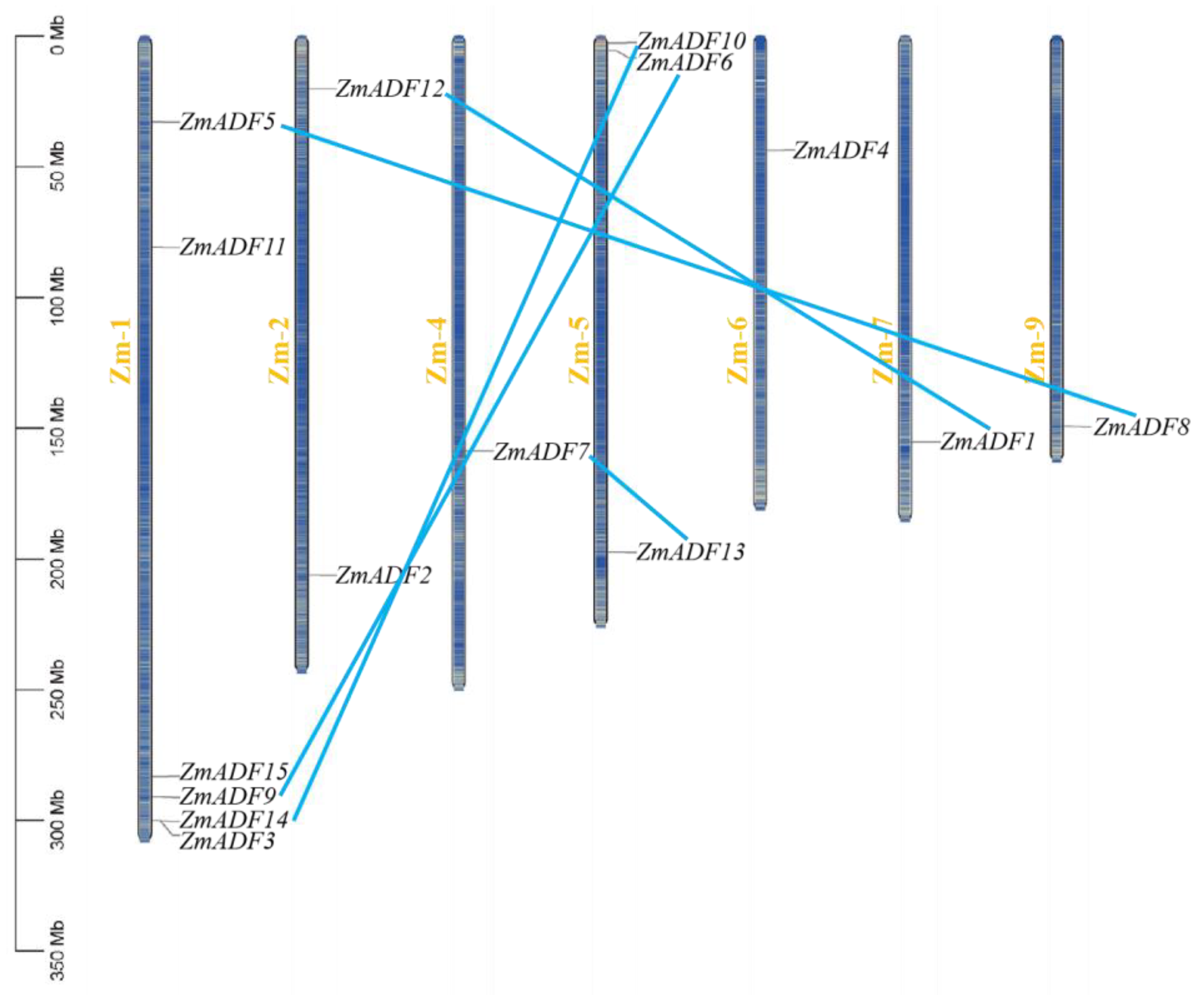
3.1. Chromosome Location and Physicochemical Properties of ADF Family in Maize
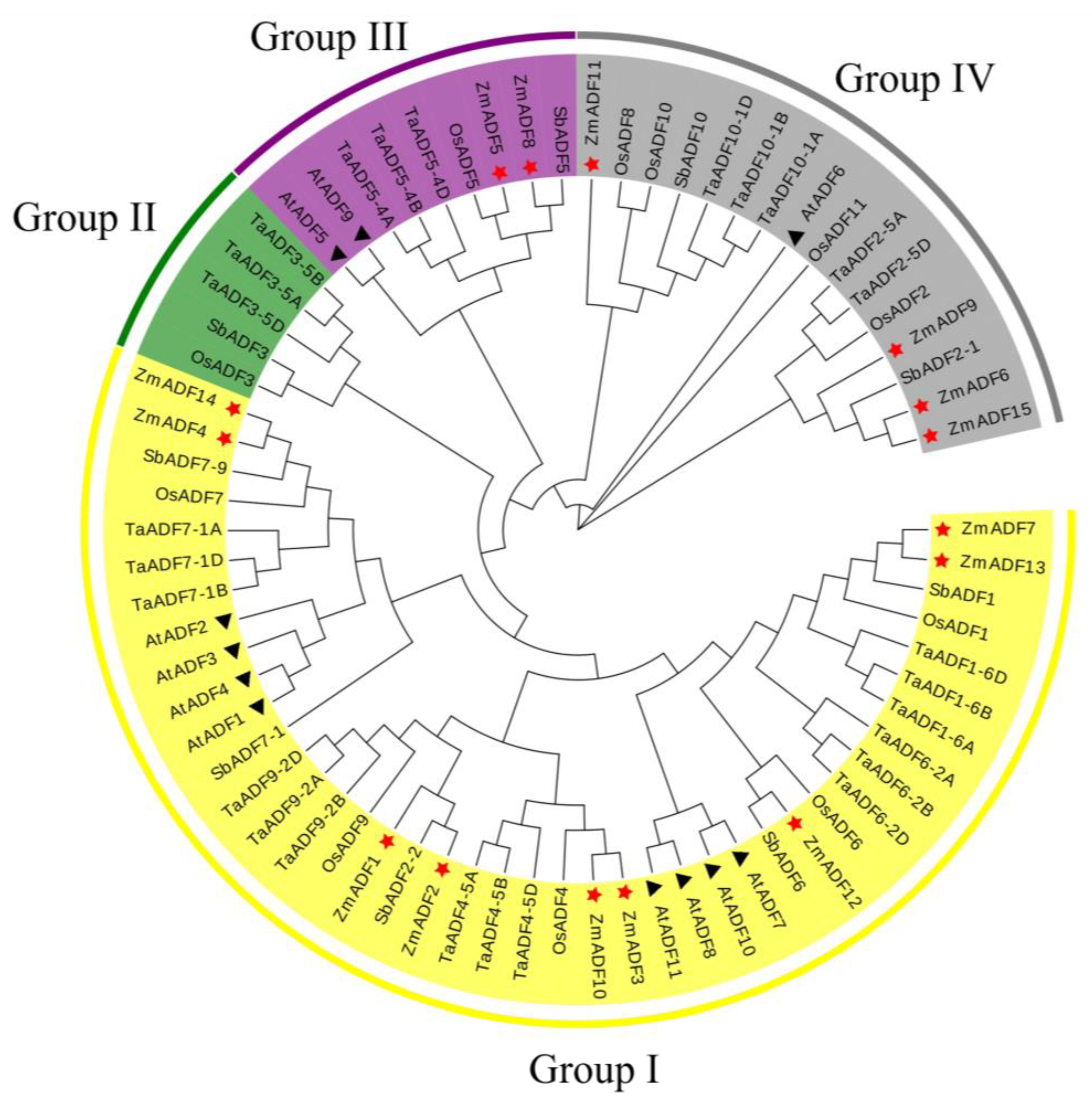
3.2. Phylogenetic Analysis of Maize ADF Family with Arabidopsis, Wheat, Rice and Sorghum
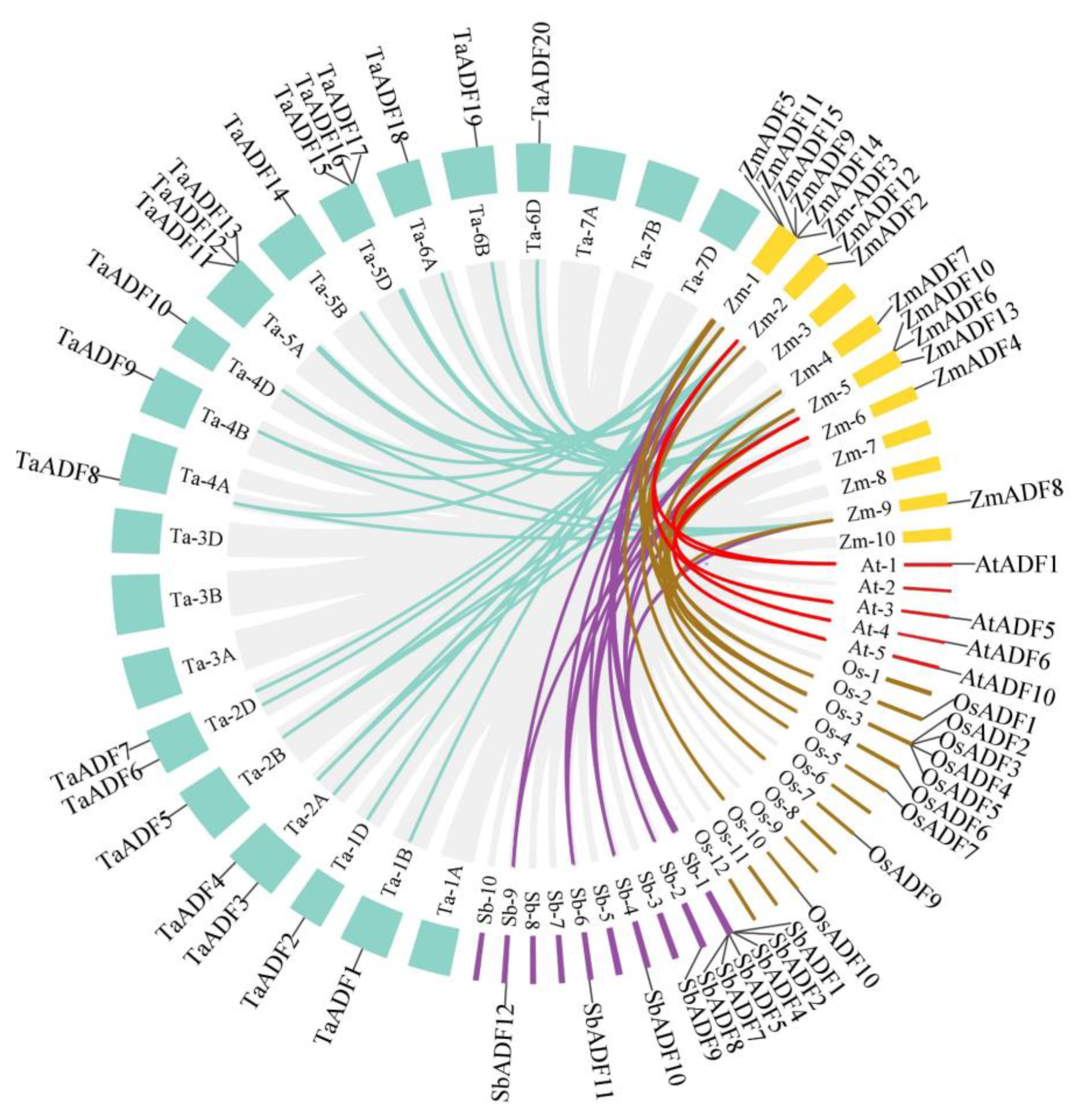
3.3. Collinearity Analysis of Maize ADF Gene Family with Arabidopsis, Wheat, Rice and Sorghum
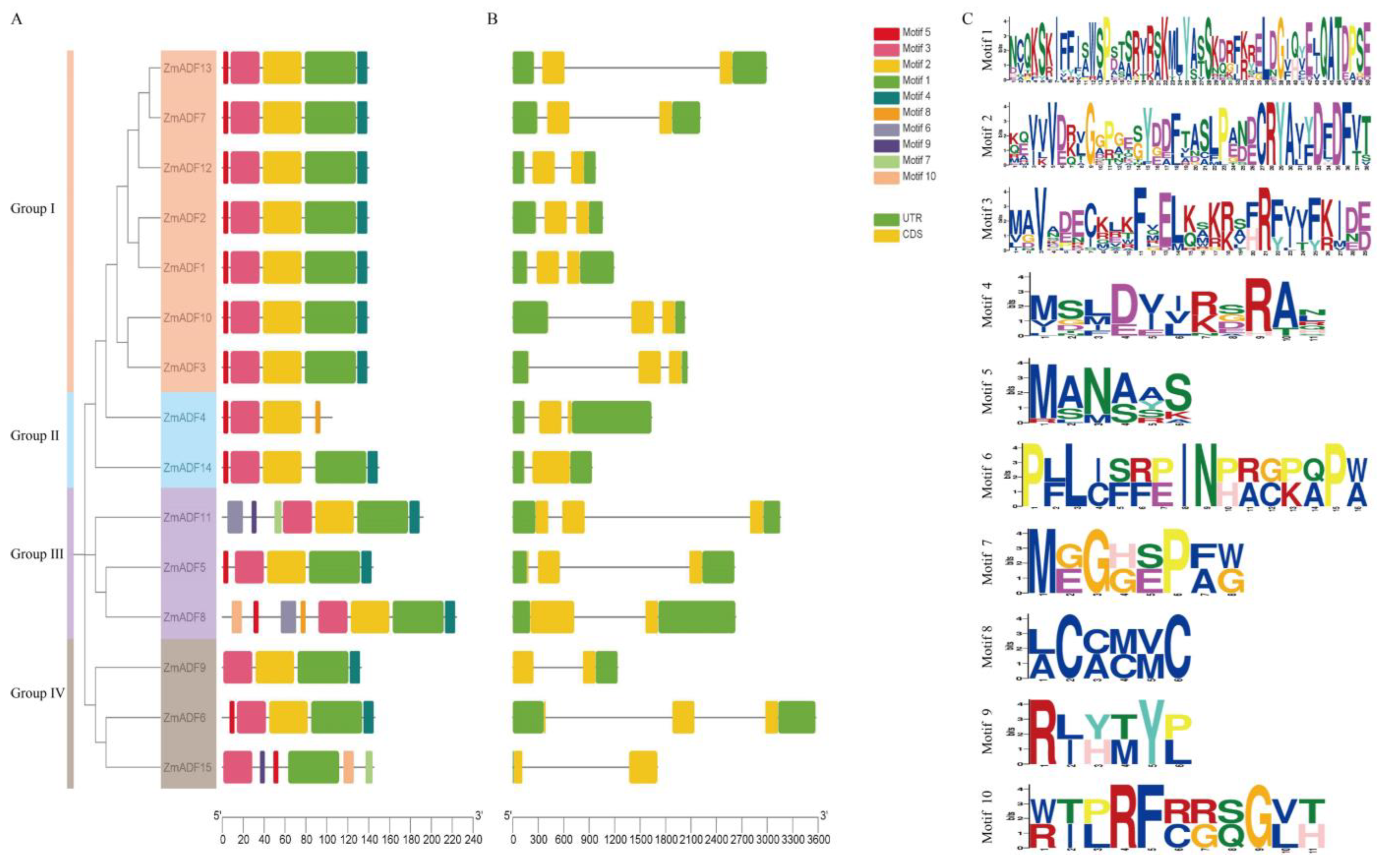
3.4. Gene Structure and Conserved Sequence of ADF Family in Maize
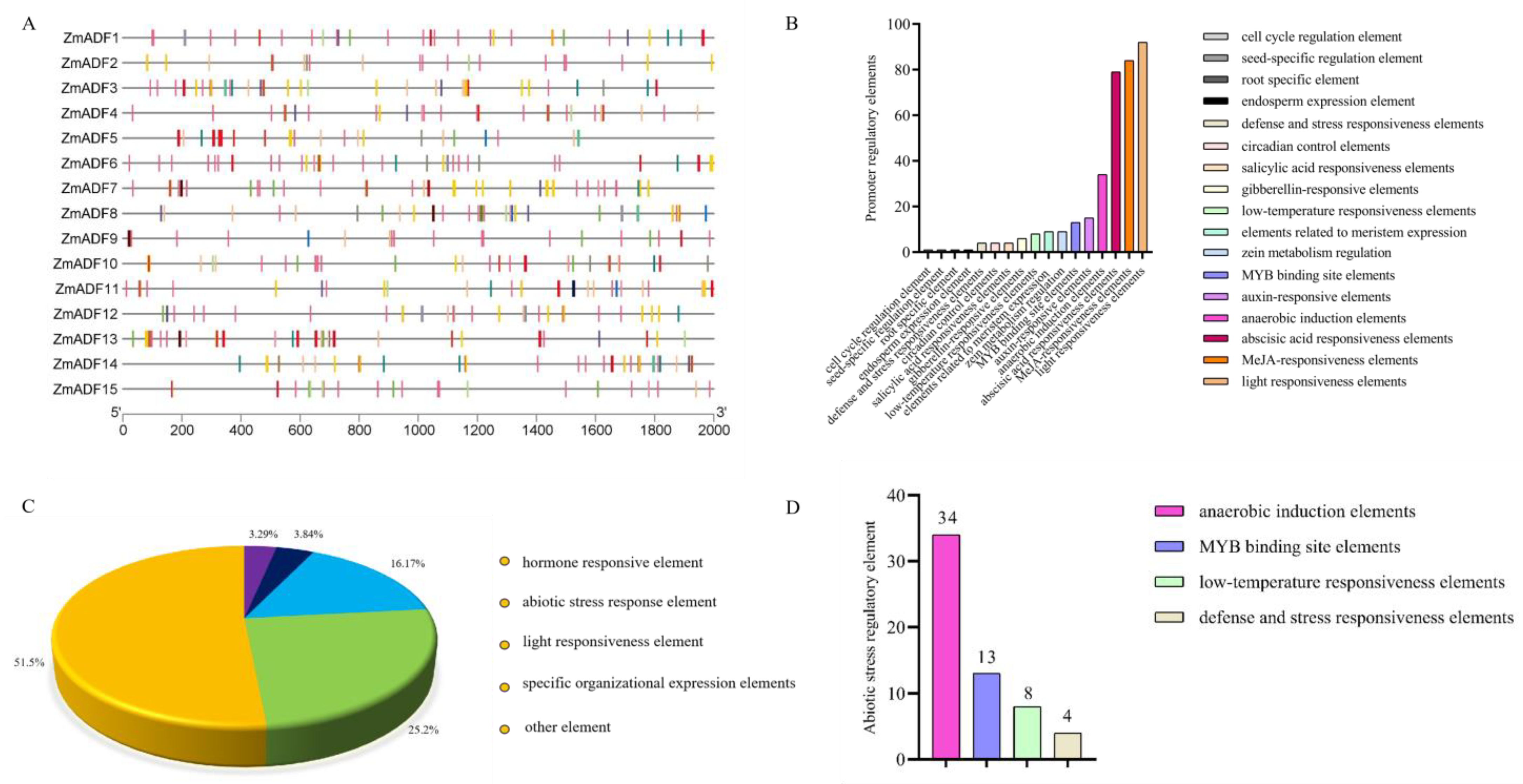
3.5. Analysis of Cis-Acting Elements in the Promoter Regions of Maize ADF Family Genes
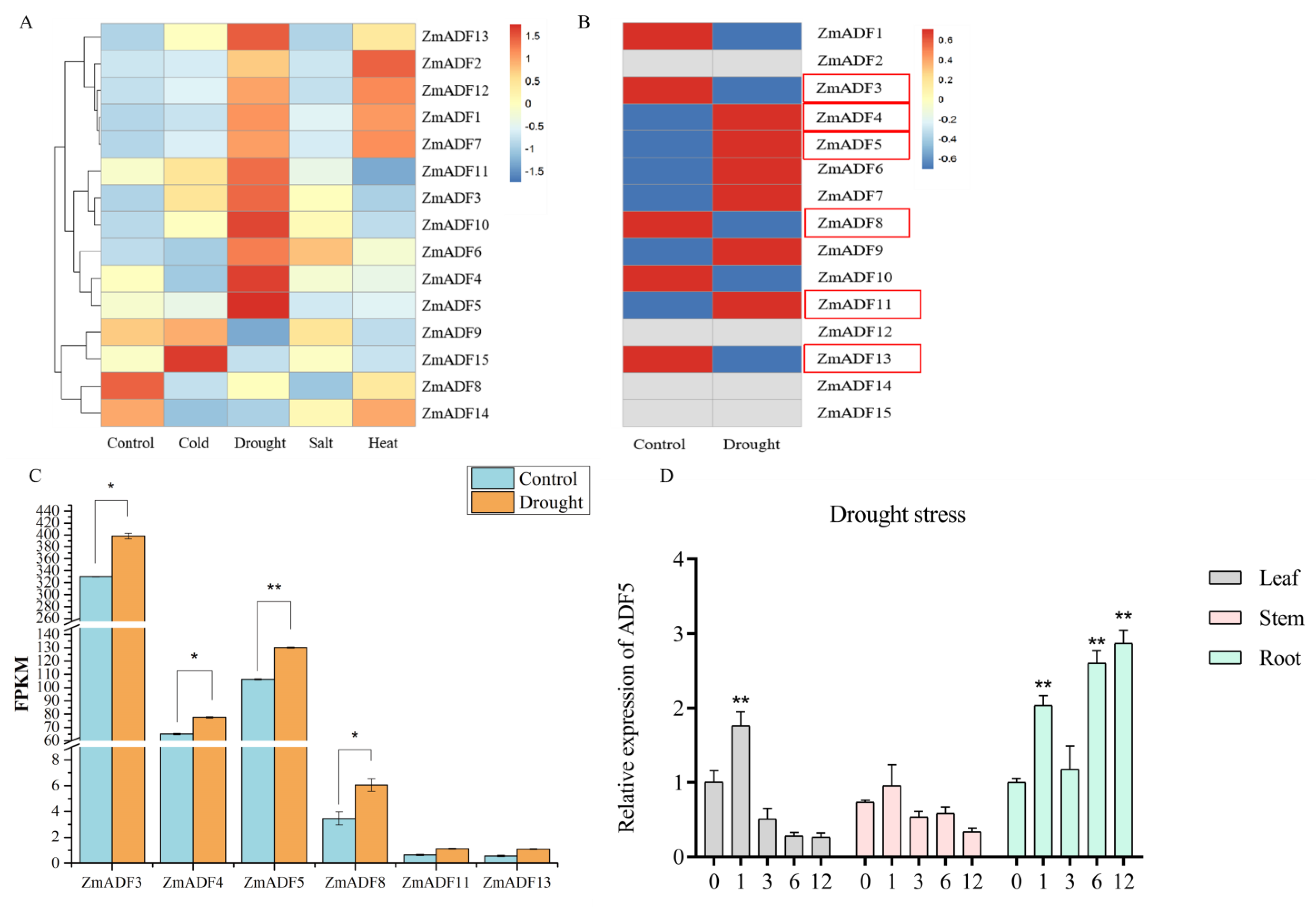
3.6. Transcriptome Analysis of Maize ADF Gene Family under Abiotic Stresses
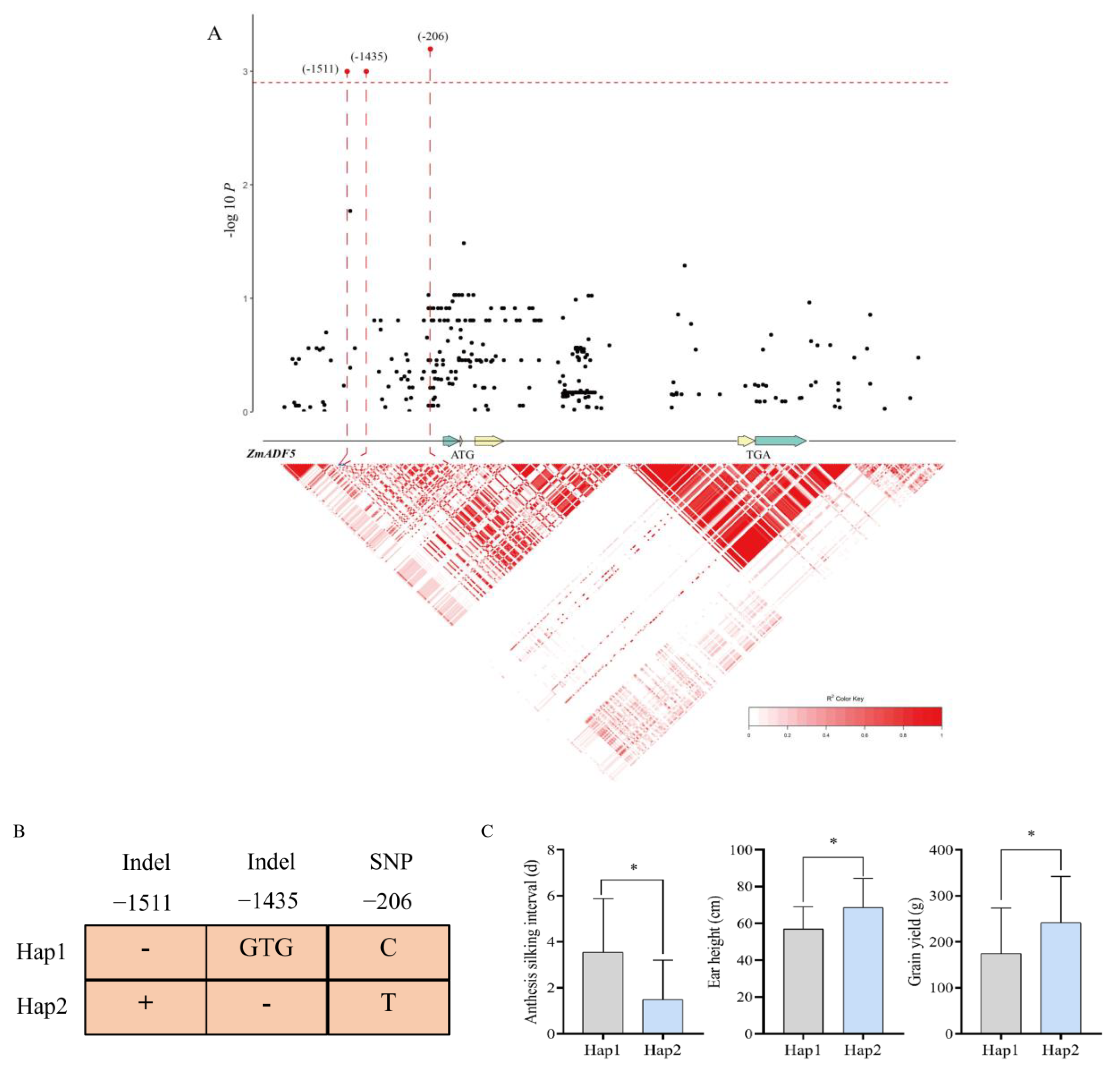
3.7. Association Analysis of ZmADF5 as a Candidate Gene for Drought Tolerance
4. Discussion
4.1. Gene Duplication in ADF Gene Family and Its Possible Function in the Evolutionary Process of Genes
4.2. The Characterization of ADF Gene Family and Its Regulation in Response to Abiotic Stresses
4.3. Excellent Allelic Variations Associated with Drought Tolerance in ZmADF5
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tigchelaar, M.; Battisti, D.S.; Naylor, R.L.; Ray, D.K. Future warming increases probability of globally synchronized maize production shocks. Proc. Natl. Acad. Sci. USA 2018, 115, 6644–6649. [Google Scholar] [CrossRef]
- Carvalho-Estrada, P.A.; de Andrade, P.A.M.; Paziani, S.F.; Nussio, L.G.; Quecine, M.C. Rehydration of dry maize preserves the desirable bacterial community during ensiling. FEMS Microbiol. Lett. 2020, 367, 139. [Google Scholar] [CrossRef]
- Liu, S.; Meng, J.; Lan, Y.; Cheng, X.E.Y.; Liu, Z.; Chen, W. Effect of maize straw biochar on maize straw composting by affecting effective bacterial community. Prep. Biochem. Biotechnol. 2021, 51, 792–802. [Google Scholar] [CrossRef]
- Wang, M.; Qiao, J.; Sheng, Y.; Wei, J.; Cui, H.; Li, X.; Yue, G. Bioconversion of maize fiber to bioethanol: Status and perspectives. Waste Manag. 2023, 15, 256–268. [Google Scholar] [CrossRef]
- Bray, E.A. Plant responses to water deficit. Trends Plant Sci. 1997, 2, 48–54. [Google Scholar] [CrossRef]
- Wang, B.; Liu, C.; Zhang, D.; He, C.; Zhang, J.; Li, Z. Effects of maize organ-specific drought stress response on yields from transcriptome analysis. BMC Plant Biol. 2019, 19, 335. [Google Scholar] [CrossRef]
- Roy-Zokan, E.M.; Dyer, K.A.; Meagher, R.B. Phylogenetic patterns of codon evolution in the actin-depolymerizing factor/cofilin (ADF/CFL) gene family. PLoS ONE 2015, 10, 0145917. [Google Scholar] [CrossRef] [PubMed]
- Bamburg, J.R.; Harris, H.E.; Weeds, A.G. Partial purification and characterization of an actin depolymerizing factor from brain. FEBS Lett. 1980, 121, 178–182. [Google Scholar] [CrossRef]
- Nishida, E.; Maekawa, S.; Sakai, H. Cofilin, a protein in porcine brain that binds to actin filaments and inhibits their interactions with myosin and tropomyosin. Biochemistry 1984, 23, 5307–5313. [Google Scholar] [CrossRef]
- Kim, S.R.; Kim, Y.; An, G. Molecular cloning and characterization of anther-preferential cDNA encoding a putative actin-depolymerizing factor. Plant Mol. Biol. 1993, 21, 39–45. [Google Scholar] [CrossRef]
- Jiang, C.J.; Weeds, A.G.; Hussey, P.J. The maize actin-depolymerizing factor, ZmADF3, redistributes to the growing tip of elongating root hairs and can be induced to translocate into the nucleus with actin. Plant J. 1997, 12, 1035–1043. [Google Scholar] [CrossRef]
- Nishida, E.; Iida, K.; Yonezawa, N.; Koyasu, S.; Yahara, I.; Sakai, H. Cofilin is a component of intranuclear and cytoplasmic actin rods induced in cultured cells. Proc. Natl. Acad. Sci. USA 1987, 84, 5262–5266. [Google Scholar] [CrossRef]
- Xu, K.; Zhao, Y.; Zhao, S.H.; Liu, H.D.; Wang, W.W.; Zhang, S.H.; Yang, X.J. Genome-wide identification and low temperature responsive pattern of actin depolymerizing factor (ADF) gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 2021, 12, 618984. [Google Scholar] [CrossRef]
- Menand, B.; Calder, G.; Dolan, L. Both chloronemal and caulonemal cells expand by tip growth in the moss physcomitrella patens. J. Exp. Bot. 2007, 58, 1843–1849. [Google Scholar] [CrossRef]
- Bou Daher, F.; van Oostende, C.; Geitmann, A. Spatial and temporal expression of actin depolymerizing factors ADF7 and ADF10 during male gametophyte development in Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 1177–1192. [Google Scholar] [CrossRef]
- Niu, Y.; Qian, D.; Liu, B.; Ma, J.; Wan, D.; Wang, X.; He, W.; Xiang, Y. ALA6, a P4-type ATPase is involved in heat stress responses in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 01732. [Google Scholar] [CrossRef]
- Sengupta, S.; Mangu, V.; Sanchez, L.; Bedre, R.; Joshi, R.; Rajasekaran, K.; Baisakh, N. An actin-depolymerizing factor from the halophyte smooth cordgrass, Spartina alterniflora (SaADF2), is superior to its rice homolog (OsADF2) in conferring drought and salt tolerance when constitutively overexpressed in rice. Plant Biotechnol. J. 2019, 17, 188–205. [Google Scholar] [CrossRef]
- Inada, N. Plant actin depolymerizing factor: Actin microfilament disassembly and more. J. Plant Res. 2017, 130, 227–238. [Google Scholar] [CrossRef]
- Huang, Y.C.; Huang, W.L.; Hong, C.Y.; Lur, H.S.; Chang, M.C. Comprehensive analysis of differentially expressed rice actin depolymerizing factor gene family and heterologous overexpression of OsADF3 confers Arabidopsis thaliana drought tolerance. Rice 2012, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Xie, Y.; Jiang, Y.; Qu, X.; Huang, S. Arabidopsis actin-depolymerizing factor7 severs actin filaments and regulates actin cable turnover to promote normal pollen tube growth. Plant Cell 2013, 25, 3405–3423. [Google Scholar] [CrossRef] [PubMed]
- Allard, A.; Bouzid, M.; Betz, T.; Simon, C.; Abou-Ghali, M.; Lemière, J.; Valentino, F.; Manzi, J.; Brochard-Wyart, F.; Guevorkian, K.; et al. Actin modulates shape and mechanics of tubular membranes. Sci. Adv. 2020, 22, 3050. [Google Scholar] [CrossRef]
- Zhu, J.; Nan, Q.; Qin, T.; Qian, D.; Mao, T.L.; Yuan, S.J.; Wu, X.R.; Niu, Y.; Bai, Q.F.; An, L.Z.; et al. Higher-ordered actin structures remodeled by Arabidopsis actin-depolymerizing factor 5 are important for pollen germination and pollen tube growth. Mol. Plant 2017, 10, 1065–1081. [Google Scholar] [CrossRef]
- Huang, J.; Sun, W.; Ren, J.; Yang, R.; Fan, J.; Li, Y.; Wang, X.; Joseph, S.; Deng, W.; Zhai, L. Genome-Wide Identification and Characterization of Actin-Depolymerizing Factor (ADF) Family Genes and Expression Analysis of Responses to Various Stresses in Zea mays L. Int. J. Mol. Sci. 2020, 21, 1751. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, N.N.; Song, W.L.; Yin, G.J.; Qin, Y.J.; Yan, Y.M.; Hu, Y.K. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆ Ct Method. Methods. 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.Y.; Dillies, M.A. SARTools: A DESeq2-and edgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Wang, N.; Wang, Z.P.; Liang, X.I.; Weng, J.F.; Lv, X.L.; Zhang, D.G.; Yang, J.; Yong, H.J.; Li, M.S.; Li, F.h.; et al. Identification of loci contributing to maize drought tolerance in a genome-wide association study. Euphytica 2016, 210, 165–179. [Google Scholar] [CrossRef]
- Ruzicka, D.R.; Kandasamy, M.K.; McKinney, E.C.; Burgos-Rivera, B.; Meagher, R.B. The ancient subclasses of Arabidopsis actin depolymerizing factor genes exhibit novel and differential expression. Plant J. 2007, 52, 460–472. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Shi, M.; Wang, D.; Gong, Y.; Sha, Q.; Lv, P.; Yang, J.; Chu, P.; Guo, S. Research progress on the roles of actin-depolymerizing factor in plant stress responses. Front. Plant Sci. 2023, 16, 1278311. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Ma, H.; DePamphilis, C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef]
- Jiang, Y.; Lu, Q.; Huang, S. Functional non-equivalence of pollen ADF isovariants in Arabidopsis. Plant J. 2022, 110, 1068–1081. [Google Scholar] [CrossRef]
- Lv, G.H.; Li, Y.F.; Wu, Z.X.; Zhang, Y.H.; Li, X.N.; Wang, T.Z.; Ren, W.C.; Liu, L.; Chen, J.J. Maize actin depolymerizing factor 1 (ZmADF1) negatively regulates pollen development. Biochem. Biophys. Res. Commun. 2024, 703, 149637. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Qian, D.; Luo, C.X.; Niu, Y.Z.; Li, T.; Li, C.Y.; Xiang, Y.; Wang, X.Y.; Niu, Y. Arabidopsis ADF5 acts as a downstream target gene of CBFs in response to low-temperature stress. Front. Cell Dev. Biol. 2021, 9, 635533. [Google Scholar] [CrossRef] [PubMed]
- Qian, D.; Zhang, Z.; He, J.X.; Zhang, P.; Ou, X.B.; Li, T.; Niu, L.P.; Nan, Q.; Niu, Y.; He, W.L.; et al. Arabidopsis ADF5 promotes stomatal closure by regulating actin cytoskeleton remodeling in response to ABA and drought stress. J. Exp. Bot. 2019, 70, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.J.; Wang, N.; Yang, R.S.; Wang, X.N.; Luo, P.; Chen, Y.; Wang, F.; Li, M.S.; Weng, J.F.; Zhang, D.G.; et al. ZmADF5, a maize actin-depolymerizing factor conferring enhanced drought tolerance in maize. Plants 2024, 13, 619. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cheng, J.; Bi, S.; Wang, J.; Cheng, X.; Liu, S.; Gao, Y.; Lan, Q.K.; Shi, X.W.; Wang, Y.; et al. Actin depolymerization factor ADF1 regulated by MYB30 plays an important role in plant thermal adaptation. Int. J. Mol. Sci. 2023, 24, 5675. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zou, M.; Pan, Q.; Li, J. Analysis of actin array rearrangement during the plant response to bacterial stimuli. Methods Mol. Biol. 2023, 2604, 263–270. [Google Scholar] [PubMed]
- Wang, L.; Qiu, T.Q.; Yue, J.R.; Guo, N.N.; He, Y.J.; Han, X.P.; Wang, Q.Y.; Jia, P.F.; Wang, H.D.; Li, M.Z.; et al. Arabidopsis ADF1 is regulated by MYB73 and is involved in response to salt stress affecting actin filament organization. Plant Cell Physiol. 2021, 62, 1387–1395. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Li, X.; Peng, L.; Hua, X.Y.; Zhang, Q.; Li, K.X.; Huang, Y.L.; Ji, H.; Wu, X.B.; Chen, Y.H.; et al. Binding of 14-3-3κ to ADF4 is involved in the regulation of hypocotyl growth and response to osmotic stress in Arabidopsis. Plant Sci. 2022, 320, 111261. [Google Scholar] [CrossRef] [PubMed]
- Bi, S.T.; Li, M.Y.; Liu, C.Y.; Liu, X.Y.; Cheng, J.N.; Wang, L.; Wang, J.S.; Lv, Y.L.; He, M.; Cheng, X.; et al. Actin depolymerizing factor ADF7 inhibits actin bundling protein VILLIN1 to regulate root hair formation in response to osmotic stress in Arabidopsis. PloS Genet. 2022, 18, e1010338. [Google Scholar] [CrossRef]
- Yang, Y.L.; Li, H.G.; Wang, J.; Wang, H.L.; He, F.; Su, Y.Y.; Zhang, Y.; Feng, C.H.; Niu, M.X.; Li, Z.H.; et al. ABF3 enhances drought tolerance via promoting ABA-induced stomatal closure by directly regulating ADF5 in Populus euphratica. J. Exp. Bot. 2020, 71, 7270–7285. [Google Scholar] [CrossRef]
| Name | Registration Number |
|---|---|
| ZmUBI-Forward primer | TGGTTGTGGCTTCGTTGGTT |
| ZmUBI-Reverse primer | GCTGCAGAAGAGTTTTGGGTACA |
| ZmADF5-Forward primer | CAGGGCCAAGATCCTGTACG |
| ZmADF5-Reverse primer | ATGACGTCGAAGCCCATCTC |
| Name | Registration Number | Molecular Weight (Da) | Chromosome | Gene Location | Number of Amino Acids (aa) | Isoelectric Point | GRAVY | Unstable Parameter | Aliphatic Amino Acid Index | Subcellular Localization | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ZmADF1 | Zm00001eb321460_T001 | 16,554.63 | 7 | 155289254 | 155290450 | 144 | 6.32 | −0.586 | 55.68 | 64.38 | Cytoplasmic |
| ZmADF2 | Zm00001eb105010_T001 | 16,083.09 | 2 | 206083396 | 206084459 | 139 | 5.57 | −0.642 | 45.49 | 63.17 | Cytoplasmic |
| ZmADF3 | Zm00001eb062600_T001 | 15,899.93 | 1 | 300195447 | 300197511 | 139 | 5.46 | −0.480 | 49.66 | 74.39 | Cytoplasmic |
| ZmADF4 | Zm00001eb266570_T001 | 15,855.13 | 6 | 43778235 | 43779873 | 139 | 7.66 | −0.369 | 51.09 | 77.88 | Cytoplasmic |
| ZmADF5 | Zm00001eb010370_T001 | 16,413.96 | 1 | 32840219 | 32842834 | 143 | 8.41 | −0.287 | 47.01 | 73.64 | Cytoplasmic |
| ZmADF6 | Zm00001eb213630_T001 | 16,833.10 | 5 | 5554015 | 5557587 | 145 | 6.15 | −0.471 | 53.27 | 70.62 | Cytoplasmic |
| ZmADF7 | Zm00001eb186790_T001 | 15,855.09 | 4 | 158650414 | 158652626 | 139 | 6.31 | −0.316 | 49.98 | 67.34 | Cytoplasmic |
| ZmADF8 | Zm00001eb398870_T001 | 20,038.07 | 9 | 149321956 | 149324583 | 172 | 9.51 | −0.505 | 64.63 | 70.45 | Cytoplasmic |
| ZmADF9 | Zm00001eb059710_T001 | 15,620.91 | 1 | 291011670 | 291012909 | 132 | 7.78 | −0.448 | 37.04 | 81.97 | Cytoplasmic |
| ZmADF10 | Zm00001eb211740_T001 | 15,913.96 | 5 | 2677695 | 2679728 | 139 | 5.47 | −0.485 | 48.80 | 74.39 | Cytoplasmic |
| ZmADF11 | Zm00001eb021290_T002 | 22,733.91 | 1 | 80880988 | 80884908 | 210 | 5.64 | −0.627 | 38.45 | 61.05 | Cytoplasmic |
| ZmADF12 | Zm00001eb074420_T001 | 15,981.05 | 2 | 20224012 | 20227127 | 139 | 5.27 | −0.570 | 51.55 | 62.45 | Cytoplasmic |
| ZmADF13 | Zm00001eb249860_T001 | 15,893.23 | 5 | 197451938 | 197454933 | 139 | 7.56 | −0.271 | 52.66 | 71.58 | Cytoplasmic |
| ZmADF14 | Zm00001eb062580_T001 | 15,698.58 | 1 | 300194049 | 300194984 | 149 | 4.81 | 0.019 | 38.49 | 84.56 | Cytoplasmic |
| ZmADF15 | Zm00001eb057310_T001 | 16,725.47 | 1 | 283186285 | 283187993 | 144 | 9.44 | −0.274 | 59.98 | 75.90 | Cytoplasmic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, R.; Wang, F.; Luo, P.; Xu, Z.; Wang, H.; Zhang, R.; Li, W.; Yang, K.; Hao, Z.; Gao, W. The Role of the ADF Gene Family in Maize Response to Abiotic Stresses. Agronomy 2024, 14, 717. https://doi.org/10.3390/agronomy14040717
Yang R, Wang F, Luo P, Xu Z, Wang H, Zhang R, Li W, Yang K, Hao Z, Gao W. The Role of the ADF Gene Family in Maize Response to Abiotic Stresses. Agronomy. 2024; 14(4):717. https://doi.org/10.3390/agronomy14040717
Chicago/Turabian StyleYang, Ruisi, Fei Wang, Ping Luo, Zhennan Xu, Houwen Wang, Runze Zhang, Wenzhe Li, Ke Yang, Zhuanfang Hao, and Wenwei Gao. 2024. "The Role of the ADF Gene Family in Maize Response to Abiotic Stresses" Agronomy 14, no. 4: 717. https://doi.org/10.3390/agronomy14040717





