Changes in the Rhizosphere Biome Depending on the Variety of Wheat, Timing of Its Growing Season, and Agrochemical Components in the Soils of Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Experiment and Soil Description
2.2. Experimental Design
2.3. Soil Sampling Experimental Set-Up
2.4. Soil Enzyme Activities and Microbiological Analyses
2.5. Statistical Analyses
3. Results and Discussion
3.1. How the Period Affects the Soil Microbiological Properties
3.2. Soil Organic Matter and Microbiological Parameters as a Function of the Treatment along the Period
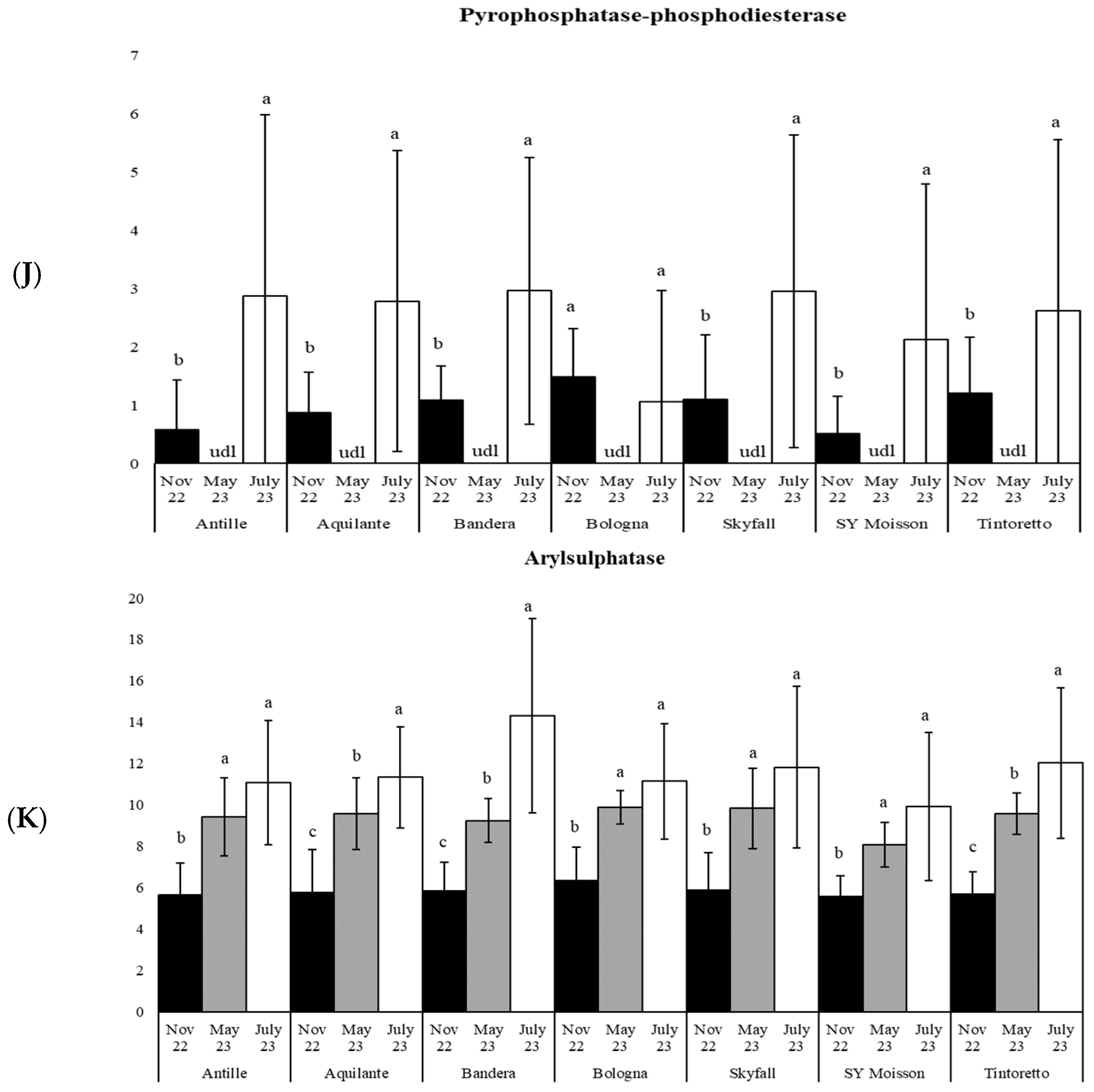
3.3. How the Soil Organic Matter and Microbiological Parameters Change along the Period in the Commercial Bread Wheat Varieties within the Untreated Field Plots
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hiltner, L. Über neuere Erfahrungen und Probleme auf dem Gebiet der Bodenbakteriologie und unter besonderer Berücksichtigung der Gründüng und Brache. Arb. DLG 1904, 98, 59–78. [Google Scholar]
- Foster, R.C.; Rovira, A.D.; Cock, T.W. Ultrastructure of the Root-Soil Interface; American Phytopathological Society: Paul, MN, USA, 1983; p. 157. [Google Scholar]
- Hartmann, A.; Rothballer, M.; Schmid, M. Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 2008, 312, 7–14. [Google Scholar] [CrossRef]
- Ober, E.S.; Alahmad, S.; Cockram, J.; Forestan, C.; Hickey, L.T.; Kant, J.; Maccaferri, M.; Marr, E.; Milner, M.; Pinto, F.; et al. Wheat root systems as a breeding target for climate resilience. Theor. Appl. Genet. 2021, 134, 1645–1662. [Google Scholar] [CrossRef]
- Bardgett, R.D.; Mommer, L.; De Vries, F.T. Going underground: Root traits as drivers of ecosystem processes. Trends. Ecol. Evol. 2014, 29, 692–699. [Google Scholar] [CrossRef]
- Herms, C.H.; Hennessy, R.C.; Bak, F.; Dresbøll, D.B.; Nicolaisen, M.H. Back to our roots: Exploring the role of root morphology as a mediator of beneficial plant-microbe interactions. Environ. Microbiol. 2022, 24, 3264–3272. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P.A.H.M. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Castrillo, G.; Pereira Lima Teixera, P.J.; Paredes, S.H.; Law, F.T.; de Lorenzo, L.; Feltcher, M.E.; Finkel, O.M.; Breakfield, N.W.; Mieczkowski, P.; Jones, D.C.; et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature 2017, 543, 513–518. [Google Scholar] [CrossRef]
- Stringlis, I.A.; Zhang, H.; Pieterse, C.M.J.; Bolton, M.D.; de Jonge, R. Microbial small molecules—Weapons of plant subversion. Nat. Prod. Rep. 2018, 5, 410–433. [Google Scholar] [CrossRef] [PubMed]
- DeAngelis, K.; Ji, P.; Firestone, M.K.; Lindow, S.E. Two novel bacterial biosensors for detection of nitrate availability in the rhizosphere. Appl. Environ. Microbiol. 2005, 71, 8537–8547. [Google Scholar] [CrossRef]
- Massalha, H.; Korenblum, E.; Tholl, D.; Aharoni, A. Small molecules below-ground: The role of specialized metabolites in the rhizosphere. Plant J. 2017, 90, 788–807. [Google Scholar] [CrossRef]
- Mommer, L.; Kirkegaard, J.; van Ruijven, J. Root-root interactions: Towards a rhizosphere framework. Trends. Plant Sci. 2016, 21, 209–217. [Google Scholar] [CrossRef]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends. Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef]
- Kawasaki, A.; Dennis, P.G.; Forstner, C.; Raghavendra, A.K.H.; Mathesius, U.; Richardson, A.E.; Delhaize, E.; Gilliham, M.; Watt, M.; Ryan, P.R. Manipulating exudate composition from root apices shapes the microbiome throughout the root system. Plant Physiol. 2021, 187, 2279–2295. [Google Scholar] [CrossRef]
- Malusà, E.; Pinzari, F.; Canfora, L. Efficacy of biofertilizers: Challenges to improve crop production. In Microbial Inoculants in Sustainable Agricultural Productivity; Springer: New Delhi, India, 2016; pp. 17–40. [Google Scholar]
- Jamil, F.; Mukhtar, H.; Fouillaud, M.; Dufossé, L. Rhizosphere signalling: Insights into plant-rhizomicrobiome interactions for sustainable agronomy. Microorganisms 2022, 10, 899. [Google Scholar] [CrossRef] [PubMed]
- Vives-Peris, V.; de Ollas, C.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Root exudates: From plant to rhizosphere and beyond. Plant Cell Rep. 2020, 39, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Santoyo, G. How plants recruit their microbiome? New insights into beneficial interactions. J. Adv. Res. 2022, 40, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.N.; Schachtman, D.P. Root exudates impact plant performance under abiotic stress. Trends. Plant Sci. 2022, 27, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Stringlis, I.A.; Proietti, S.; Kickman, R.; Van Verk, M.C.; Zamioudis, C.; Pieterse, C.M.J. Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant J. 2018, 93, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, J.; Lu, X.; Bai, Y.; Wang, G. Two diversities meet in the rhizosphere: Root specialized metabolites and microbiome. J. Genet. Genom. 2023, 23, 1–12. [Google Scholar] [CrossRef]
- He, D.; Singh, S.K.; Li, P.; Kaushal, R.; Vílchez, J.I.; Chuyang, S.; Wu, X.; Zhang, H. Flavonoid-attracted Aeromonas sp. from the Arabidopsis root microbiome enhances plant dehydration resistance. ISME J. 2022, 16, 2622–2632. [Google Scholar] [CrossRef]
- Yan, D.; Tajima, H.; Cline, L.C.; Fong, R.Y.; Ottaviani, J.I.; Shapiro, H.Y.; Blumwald, E. Genetic modification of flavone biosynthesis in rice enhances biofilm formation of soil diazotrophic bacteria and biological nitrogen fixation. Plant Biotechnol. J. 2022, 20, 2135–2148. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; He, X.; Baer, M. Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation. Nat. Plants 2021, 7, 481–499. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, H.; Nakayasu, M.; Aoki, Y.; Yamazaki, S.; Nagano, A.J.; Yazaki, K.; Sugiyama, A. Diurnal metabolic regulation of isoflavones and soyasaponins in soybean roots. Plant Direct 2020, 18, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, J.; Khashi, U.; Rahman, M.; Gao, D.; Wei, Z.; Wu, F.; Dini-Andreote, F. Interspecific plant interaction via root exudates structures the disease suppressiveness of rhizosphere microbiomes. Mol. Plant 2023, 16, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.R.; Dai, L.; Xu, G.F.; Wang, H.S. A strain of Phoma species improves drought tolerance of Pinus tabulaeformis. Sci. Rep. 2021, 11, 7637. [Google Scholar] [CrossRef] [PubMed]
- Koroney, A.S.; Plasson, C.; Pawlak, B.; Sidikou, R.; Driouich, A.; Menu-Bouaouiche, L.; Vicré-Gibouin, M. Root exudate of Solanum tuberosum is enriched in galactose-containing molecules and impacts the growth of Pectobacterium atrosepticum. Ann. Bot. 2016, 118, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Cannesan, M.G.; Gangneux, C.; Lanoue, A.; Giron, D.; Laval, K.; Hawes, M.; Driouch, A.; Vicré-Gibon, M. Association between border cell responses and localized root infection by pathogenic Aphanomyces euteiches. Ann. Bot. 2011, 108, 459–469. [Google Scholar] [CrossRef]
- Kim, J.; Felton, G.W. Priming of antiherbivore defensive responses in plants. Insect Sci. 2013, 20, 273–285. [Google Scholar] [CrossRef]
- Ameye, M.; Audenaert, K.; De Zutter, N.; Steppe, K.; Van Meulebroek, L.; Vanhaeck, L.; De Vleesschauwer, D.; Haesaert, G.; Smagghe, G. Priming of wheat with the green leaf volatile Z-3-hexenyl acetate enhances defense against Fusarium graminearum but boosts deoxynivalenol production. Plant Physiol. 2015, 167, 1671–1684. [Google Scholar] [CrossRef]
- Cofer, T.M.; Seidl-Adams, I.; Tumlinson, J.H. From Acetoin to (Z)-3-Hexen-1-ol: The Diversity of Volatile Organic Compounds that Induce Plant Responses. Agric. Food Chem. 2018, 66, 11197–11208. [Google Scholar] [CrossRef]
- Kong, H.G.; Song, G.C.; Sim, H.J.; Ryu, C.M. Achieving similar root microbiota composition in neighbouring plants through airborne signalling. ISME J. 2021, 15, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Li, Z.; Kuipers, O.P. Plant-Microbe Interaction: Transcriptional Response of Bacillus Mycoides to Potato Root Exudates. J. Vis. Exp. 2018, 137, 57606. [Google Scholar] [CrossRef]
- Ross-Elliot, T.J.; Jensen, K.H.; Haaning, K.S.; Wager, B.M.; Knoblauch, J.; Howell, A.H.; Mullendore, D.L.; Monteith, A.G.; Paultre, D.; Yan, D.; et al. Phloem unloading in Arabidopsis roots is convective and regulated by the phloem-pole pericycle. ELife 2017, 6, e24125. [Google Scholar] [CrossRef] [PubMed]
- Badri, D.V.; Weir, T.L.; van der Lelie, D.; Vivanco, J.M. Rhizosphere chemical dialogues: Plant-microbe interactions. Curr. Opin. Biotechnol. 2009, 20, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Vidal, A.; Hirte, J.; Bender, S.F.; Mayer, J.; Gattinger, A.; Höschen, C.; Schädler, S.; Iqbal, T.M.; Mueller, C.W. Linking 3D soil structure and plant-microbe-soil carbon transfer in the rhizosphere. Front. Environ. Sci. 2018, 6, 9. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [PubMed]
- Hassani, M.A.; Özkurt, E.; Seybold, H.; Dagan, T.; Stukenbrock, E.H. Interactions and Codaptation in Plant Metaorganisms. Annu. Rev. Phytopathol. 2019, 57, 483–503. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Raaijmakers, J.M. Saving seed microbiomes. ISME J. 2018, 12, 1167–1170. [Google Scholar] [CrossRef] [PubMed]
- Checcucci, A.; Marchetti, M. The Rhizosphere Talk Show: The rhizobia on Stage. Front. Agron. 2020, 2, 591494. [Google Scholar] [CrossRef]
- Elhady, A.; Abbasi, S.; Safaie, N.; Heuer, H. Responsiveness of Elite Cultivars vs. Ancestral Genotypes of Barley to Beneficial Rhizosphere Microbiome, Supporting Plant Defense Against Root-Lesion Nematodes. Front. Plant Sci. 2021, 12, 721016. [Google Scholar] [CrossRef]
- Lyu, D.; Zajonc, J.; Pagé, A.; Tanney, C.A.S.; Shah, A.; Monjezi, N.; Msimbira, L.A.; Antar, M.; Nazari, M.; Backer, R.; et al. Plant Holobiont Theory: The Phytomicrobiome Plays a Central Role in Evolution and Success. Microorganisms 2021, 24, 675. [Google Scholar] [CrossRef] [PubMed]
- Al-Busaidi, W.; Janke, R.; Blackburn, D.M.; Khan, M.M. Impact of long-term agricultural farming on soil and water chemical properties: A case study from Al-Batinah regions (Oman). J. Saudi Soc. Agric. Sci. 2022, 21, 397–403. [Google Scholar] [CrossRef]
- Roman, D.L.; Voiculescu, D.I.; Filip, M.; Ostafe, V.; Isvoran, A. Effects of Triazole Fungicides on Soil Microbiota and on the Activities of Enzymes Found in Soil: A Review. Agriculture 2021, 11, 893. [Google Scholar] [CrossRef]
- Zhang, L.; Yan, C.; Guo, Q.; Zhang, J.; Ruiz, J. The impact of agricultural chemical inputs on environment: Global evidence from informetrics analysis and visualization. Int. J. Low Carbon Technol. 2018, 13, 338–352. [Google Scholar] [CrossRef]
- Baćmaga, M.; Wyszkowska, J.; Kucharski, J. Response of soil microorganisms and enzymes to the foliar application of Helicur 250 EW fungicide on Horderum vulgare L. Chemosphere 2020, 242, 125163. [Google Scholar] [CrossRef]
- Koch, F.; Aisenberg, G.R.; Monteiro, M.A.; Pedó, T.; Zimmer, P.D.; Villela, F.A.; Aumonde, T.Z. Growth of wheat plants submitted to the application of the growth regulator trinexapac-ethyl and vigor of the produced seeds. Agrociencia 2017, 21, 24–32. [Google Scholar] [CrossRef][Green Version]
- Pricinotto, L.F.; Claudemir, Z.; Inês, C.; de Batista, F.; Alves de Oliveira, M.; Sampaio Ferreira, A.; Spolaor, T.L. Trinexapac-ethyl in the vegetative and reproductive performance of corn. Afr. J. Agric. Res. 2015, 10, 1735–1742. [Google Scholar] [CrossRef][Green Version]
- Gonçalves, I.C.R.; Araújo, A.S.F.; Carvalho, E.M.S.; Carneiro, R.F.V. Effect of paclobutrazol on microbial biomass, respiration and cellulose decomposition in soil. Eur. J. Soil Biol. 2009, 45, 235–238. [Google Scholar] [CrossRef]
- Fornasier, F.; Ascher, J.; Ceccherini, M.T.; Tomat, E.; Pietramellara, G. A simplified rapid, low-cost and versatile DNA-based assessment of soil microbial biomass. Ecol. Indic. 2014, 45, 75–82. [Google Scholar] [CrossRef]
- Kottek, M.; Grieser, J.; Beck, C.; Rudolf, B.; Rubel, F. World map of the Köppen-Geiger climate classification update. Meteorol. Z. 2006, 15, 259–263. [Google Scholar] [CrossRef]
- Beck, H.E.; Zimmermann, N.E.; McVicar, T.R.; Vergopolan, N.; Berg, A.; Wood, E.F. Present and future Köppen-Geiger climate classification maps at 1-km resolution. Sci. Data 2018, 5, 180214. [Google Scholar] [CrossRef] [PubMed]
- Van Reeuwijk, L.P. Procedures for Soil Analysis, 6th ed.; Technical Paper No. 9; FAO/ISRIC, International Soil Reference an Information Centre: Wageningen, The Netherlands, 2022; p. 120. [Google Scholar]
- Fornasier, F.; Margon, A. Bovine serum albumin and Triton X-100 greatly increase phosphomonoesterases and arylsulphatase extraction yield from the soil. Soil Biol. Biochem. 2007, 39, 2682–2684. [Google Scholar] [CrossRef]
- Bardelli, T.; Gomez-Brandon, M.; Ascher-Jenull, J.; Fornasier, F.; Arfaioli, P.; Francioli, D.; Egli, M.; Sartori, G.; Insam, H.; Pietramellara, G. Effect of slope exposure on soil physico-chemical and microbiological properties along an altitudinal climosequence in the Italian Alps. Sci. Total Environ. 2017, 575, 1041–1055. [Google Scholar] [CrossRef]
- Bragato, G.; Fornasier, F.; Brus, D.J. Characterization of soil fertility and soil biodiversity with dsDNA as a covariate in a regression estimator for mean microbial biomass C: Soil dsDNA as a covariate for microbial biomass C. Eur. J. Soil Sci. 2016, 67, 827–834. [Google Scholar] [CrossRef]
- Dragan, A.I.; Casas-Finet, J.R.; Bishop, E.S.; Strouse, R.I.; Schenerman, M.A.; Geddes, C.D. Characterization of PicoGreen interaction with dsDNA and the origin of its fluorescence enhancement upon bindings. Biophys. J. 2010, 99, 3010–3019. [Google Scholar] [CrossRef] [PubMed]
- Mijangos, I.; Becerril, J.M.; Albizu, I.; Epelde, L.; Garbisu, C. Effects of glyphosate on rhizosphere soil microbial communities under two different plant compositions by cultivation-dependent and independent methodologies. Soil Biol. Biochem. 2009, 41, 505–513. [Google Scholar] [CrossRef]
- Walder, F.; Schmid, M.W.; Riedo, J.; Valzano-Held, A.Y.; Banerjee, S.; Büchi, L.; Van Der Heijden, M.G. Soil microbiome signatures are associated with pesticide residues in arable landscapes. Soil Biol. Biochem. 2022, 174, 108830. [Google Scholar] [CrossRef]
- Han, L.; Kong, X.; Xu, M.; Nie, J. Repeated exposure to fungicide tebuconazole alters the degradation characteristics, soil microbial community and functional profiles. Environ. Pollut. 2021, 287, 117660. [Google Scholar] [CrossRef]
- Munoz-Leoz, B.; Ruiz-Romera, E.; Antigüedad, I.; Garbisu, C. Tebuconazole application decreases soil microbial biomass and activity. Soil Biol. Biochem. 2011, 43, 2176–2183. [Google Scholar] [CrossRef]
- Nguyen, C. Rhizodeposition of organic C by plant: Mechanisms and controls. In Sustainable Agriculture; Springer: Berlin/Heidelberg, Germany, 2009; pp. 97–123. [Google Scholar] [CrossRef]
- Van der Krift, T.A.; Kuikman, P.J.; Möller, F.; Berendse, F. Plant species and nutritional-mediated control over rhizodeposition and root decomposition. Plant Soil 2001, 228, 191–200. [Google Scholar] [CrossRef]
- Hassan, M.K.; McInroy, J.A.; Kloepper, J.W. The Interactions of Rhizodeposits with Plant Growth-Promoting Rhizobacteria in the Rhizosphere: A Review. Agriculture 2019, 9, 142. [Google Scholar] [CrossRef]
- García-Orenes, F.; Morugán-Coronado, A.; Zornoza, R.; Scow, K. Changes in Soil Microbial Community Structure Influenced by Agricultural Management Practices in a Mediterranean Agro-Ecosystem. PLoS ONE 2013, 8, 80522. [Google Scholar] [CrossRef]
- Anderson, T.H.; Paulsen, H.M. Response time of soil microbial biomass after conversion from conventional to several different organic farming systems. Landbauforsch. Appl. Agric. For. Res. 2016, 66, 258–271. [Google Scholar] [CrossRef]
- Kumar, B.; Dhar, S.; Paul, S.; Paramesh, V.; Dass, A.; Upadhyay, P.K.; Kumar, A.; Abdelmohsen, S.A.M.; Alkallas, F.H.; El-Abedin, T.K.Z.; et al. Microbial Biomass Carbon, Activity of Soil Enzymes, Nutrient Availability, Root Growth, and Total Biomass Production in Wheat Cultivars under Variable Irrigation and Nutrient Management. Agronomy 2021, 11, 669. [Google Scholar] [CrossRef]
- Cheng, W.; Parton, W.J.; Gonzalez-Meler, M.A.; Asao, S.; Mcnickle, G.G.; Brzostek, E.; Jastrow, J.D. Synthesis and modeling perspectives of rhizosphere priming. New Phytol. 2014, 201, 31–44. [Google Scholar] [CrossRef]
- Yin, L.; Dijkstra, F.A.; Wang, P.; Zhu, B.; Cheng, W. Rhizosphere priming effects on soil carbon and nitrogen dynamics among tree species with and without intraspecific competition. New Phytol. 2018, 218, 1036–1048. [Google Scholar] [CrossRef]
- Boyrahmadi, M.; Raiesi, F. Plant roots and species moderate the salinity effect on microbial respiration, biomass, and enzyme activities in a sandy clay soil. Biol. Fertil. Soils 2018, 54, 509–521. [Google Scholar] [CrossRef]
- Lareen, A.; Burton, F.; Schäfer, P. Plant root-microbe communication in shaping root microbiomes. Plant Mol. Biol. 2016, 90, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, A.K.; Yin, C.; Hulbert, S.H. Community Structure, Species Variation, and Potential Functions of Rhizosphere-Associated Bacteria of Different Winter Wheat (Triticum aestivum) Cultivars. Front. Plant Sci. 2017, 13, 132. [Google Scholar] [CrossRef]
- Lakshmanan, V.; Castaneda, R.; Rudrappa, T.; Bais, H.P. Root transcriptome analysis of Arabidopsis thaliana exposed to beneficial Bacillus subtilis FB17 rhizobacteria revealed genes for bacterial recruitment and plant defense independent of malate efflux. Planta 2013, 238, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Corneo, P.E.; Suenaga, H.; Kertesz, M.A.; Dijkstra, F.A. Effect of twenty four wheat genotypes on soil biochemical and microbial properties. Plant Soil 2016, 404, 141–155. [Google Scholar] [CrossRef]
- Karthik, N.; Binod, P.; Pandey, A. Purification and characterisation of an acidic and antifungal chitinase produced by a Streptomyces sp. Bioresour. Technol. 2015, 188, 195–201. [Google Scholar] [CrossRef]
- Medina-de la Rosa, G.; López-Reyes, L.; Carcaño-Montiel, M.G.; López-Olguín, J.F.; Hernández-Espinosa, M.Á.; Rivera-Tapia, J.A. Rhizosphere bacteria of maize with chitinolytic activity and its potential in the control of phytopathogenic fungi. Arch. Phytopathol. 2016, 49, 310–321. [Google Scholar] [CrossRef]
- Vaghela, B.; Vashi, R.; Rajput, K.; Joshi, R. Plant chitinases and their role in plant defense: A comprehensive review. Enzyme Microb. Technol. 2022, 159, 110055. [Google Scholar] [CrossRef]
- Yin, C.; Casa Vargas, J.M.; Schlatter, D.C.; Hagerty, C.H.; Hulbert, S.H.; Paulitz, T.C. Rhizosphere community selection reveals bacteria associated with reduced root disease. Microbiome 2021, 9, 86. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, R.; Hashem, A.; Abd Allah, E.F. Bacillus: A Biological Tool for Crop Improvement through Bio-Molecular Changes in Adverse Environments. Front. Physiol. 2017, 8, 667. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.Q.; Wang, F.Z.; Li, J.F. Hide-and-Seek: Chitin-Triggered Plant Immunity and Fungal Counterstrategies. Trends. Plant Sci. 2020, 25, 805–816. [Google Scholar] [CrossRef]
- Hui, C.; Jiang, H.; Liu, B.; Wei, R.; Zhang, Y.; Zhang, O.; Liang, Y.; Zhao, Y. Chitin degradation and the temporary response of bacterial chitinolytic communities to chitin amendment in soil under different fertilization regimes. Sci. Total Environ. 2020, 705, 136003. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Li, M.; Yang, H.; Liu, J. Dynamic changes of the retention capacity for phosphorus by emergent macrophytes in the Yeyahu Wetland. Acta Sci. Circumstantiae 2012, 32, 1874–1881. [Google Scholar]
- Khan, K.S.; Mack, R.; Castillo, X.; Kaiser, M.; Joergensen, R.G. Microbial biomass, fungal and bacterial residues, and their relationships to the soil organic matter C/N/P/S ratios. Geoderma 2016, 271, 115–123. [Google Scholar] [CrossRef]
- Mohammadi, K. Soil Microbial Activity and Biomass as Influenced by Tillage and Fertilization in Wheat Production. Am. Eurasian J. Agric. Environ. Sci. 2011, 10, 330–337. [Google Scholar]
- Nannipieri, P.; Giagnoni, L.; Renella, G.; Puglisi, E.; Ceccanti, B.; Masciandaro, G.; Fornasier, F.; Moscatelli, M.C.; Marinari, S. Soil enzymology: Classical and molecular approaches. Biol. Fertil. Soils. 2012, 48, 743–762. [Google Scholar] [CrossRef]
- Lemanowicz, J. Mineral fertilization as a factor determining selected sorption properties of soil against the activity of phosphatases. Plant Soil Environ. 2013, 59, 439–445. [Google Scholar] [CrossRef]
- Wesolowska, S.; Futa, B.; Myszura, M.; Kobyłka, A. Residual Effects of Different Cropping Systems on Physicochemical Properties and the Activity of Phosphatases of Soil. Agriculture 2022, 12, 693. [Google Scholar] [CrossRef]
- Maurya, S.; Abraham, J.S.; Somasundaram, S.; Toteja, S.R.; Gupta, R.; Makhija, S. Indicators for assessment of soil quality: A mini-review. Environ. Monit. Assess. 2020, 192, 604. [Google Scholar] [CrossRef]
- Bhattacharyya, S.S.; Furtak, K. Soil Plant Microbe Interactions Determine Soil Biological Fertility by Altering Rhizospheric Nutrient Cycling and Biocrust Formation. Sustainability 2023, 15, 625. [Google Scholar] [CrossRef]
- Sun, X.; Yuqian, Y.; Ma, Q.; Guan, Q.; Jones, D. Variation in enzyme activities involved in carbon and nitrogen cycling in rhizosphere and bulk soil after organic mulching. Rhizosphere 2021, 19, 100376. [Google Scholar] [CrossRef]


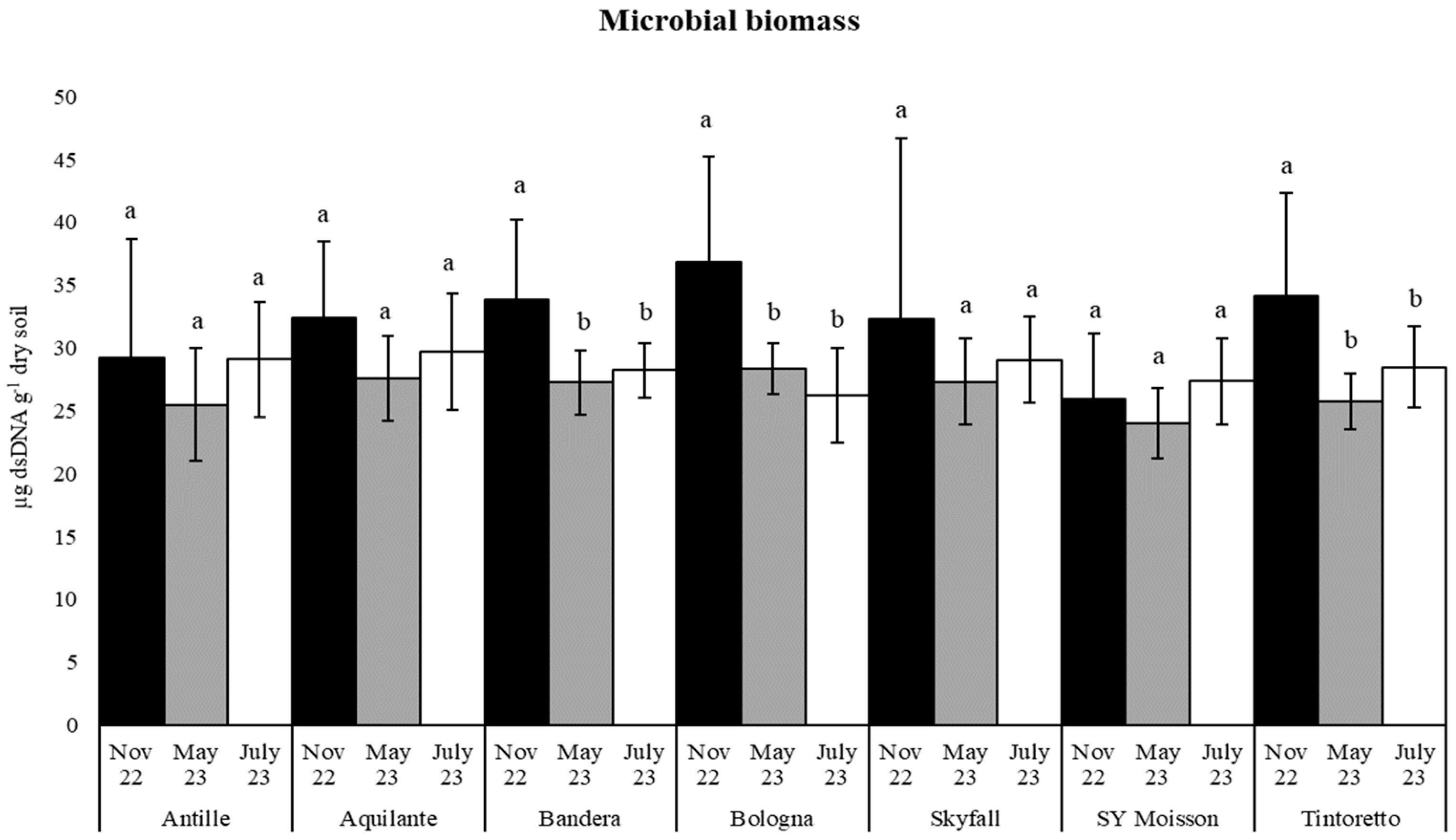
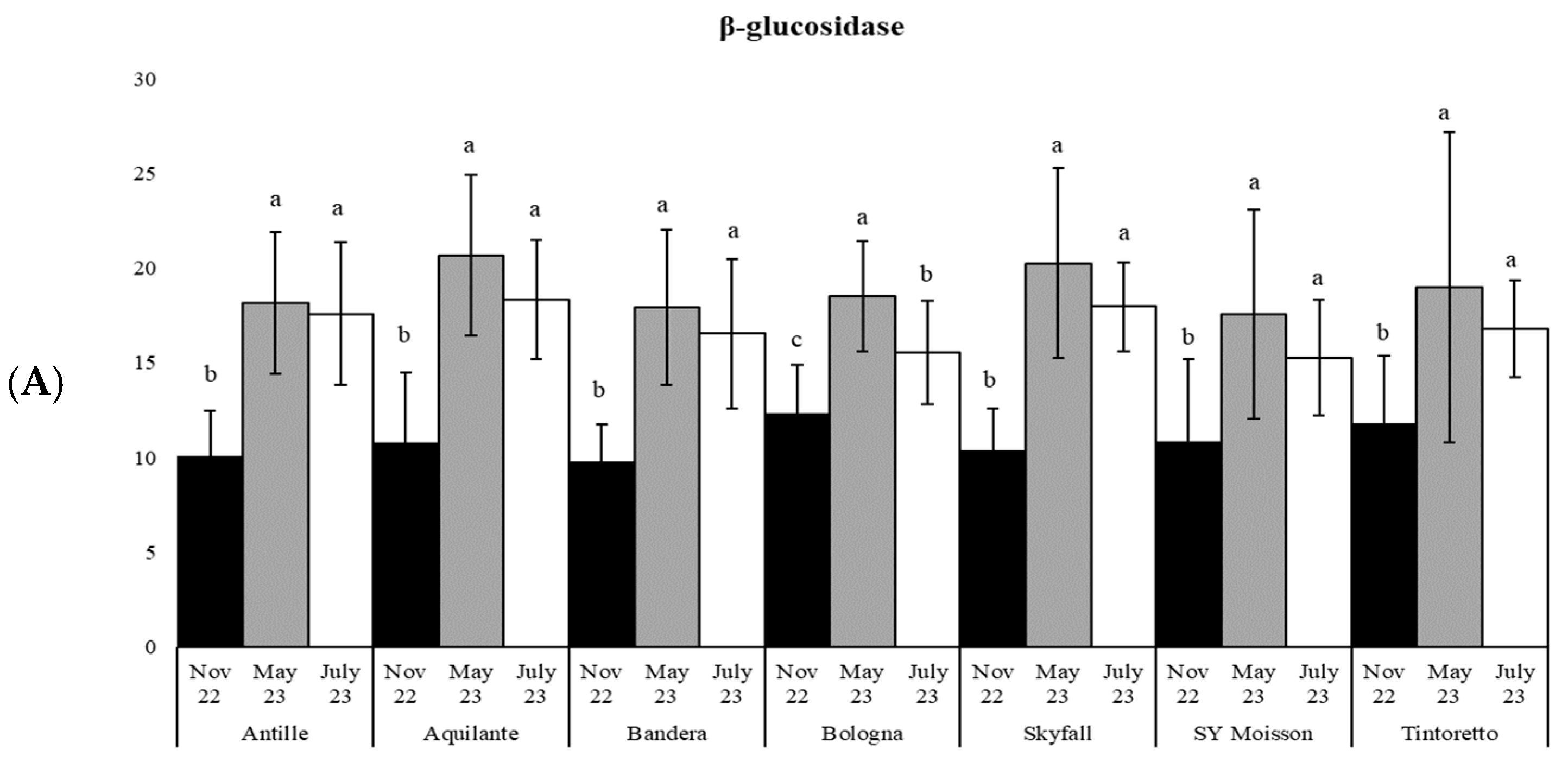
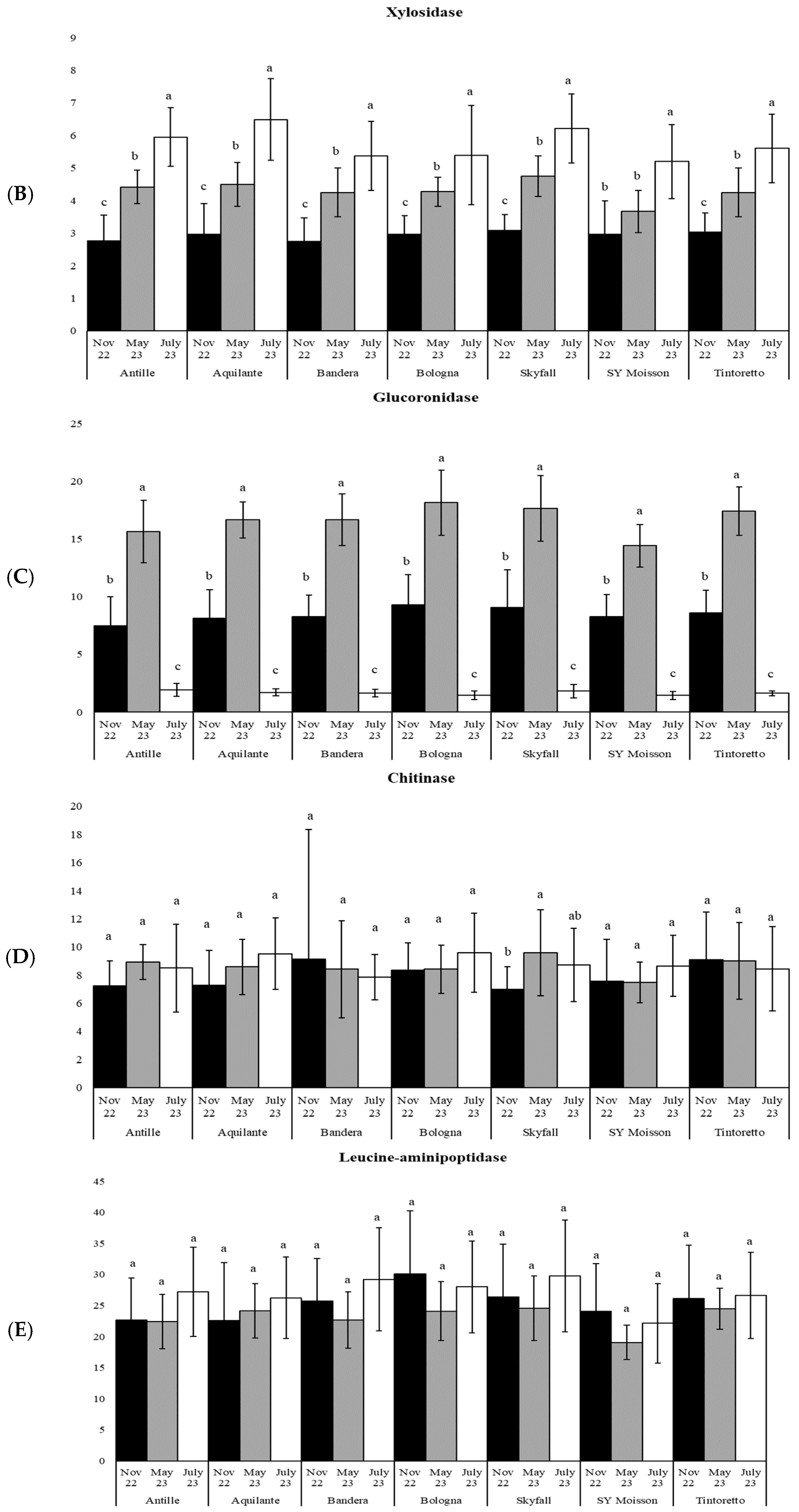
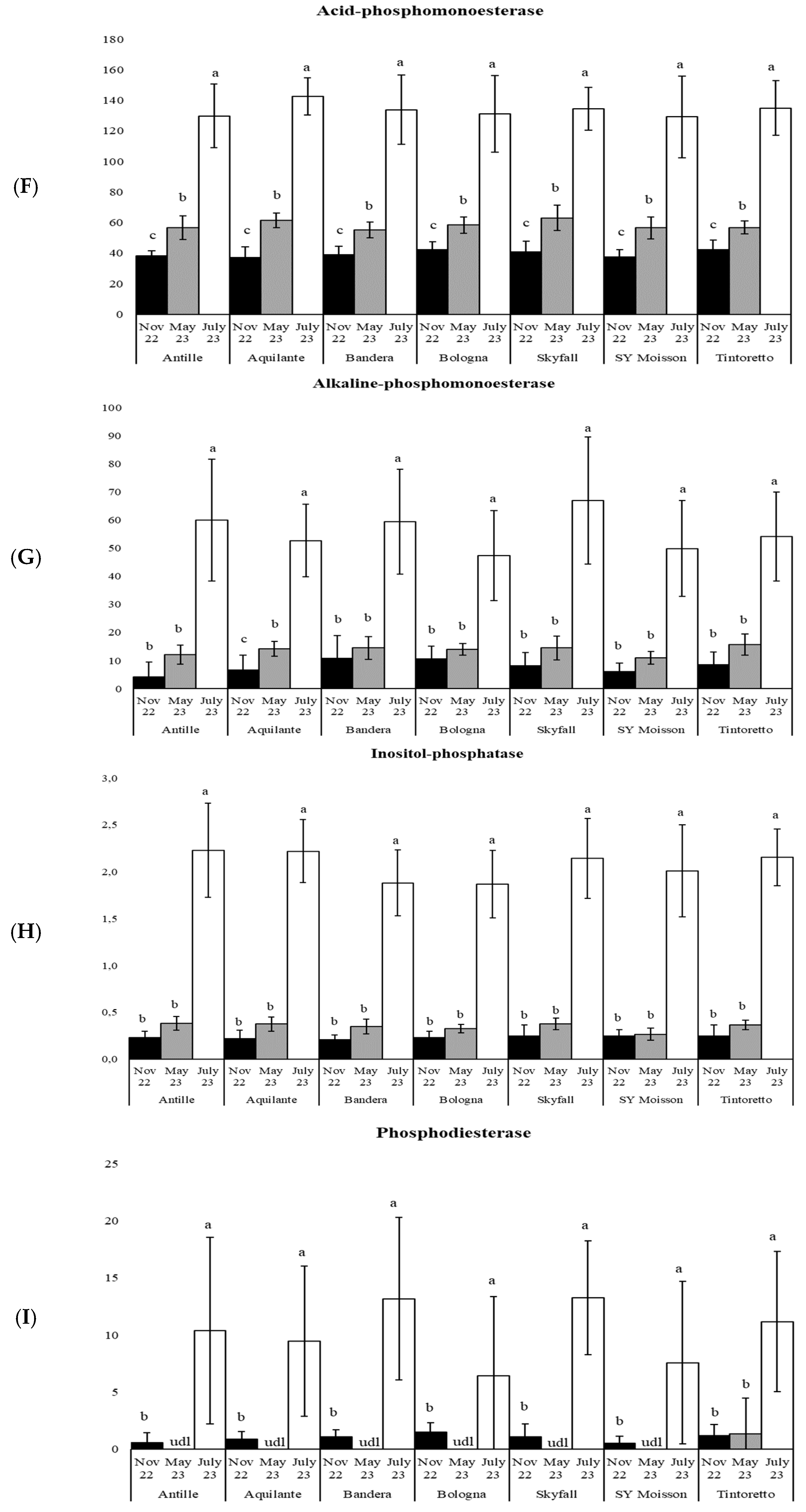
| Parameters | November 2022 | May 2023 | July 2023 |
|---|---|---|---|
| Air temperature (°C) | 8.8 ± 3.97 | 17.7 ± 4.47 | 24.7 ± 5.15 |
| Air humidity (%) | 87.0 ± 0.09 | 73,2 ± 0.13 | 75.7 ± 0.15 |
| Precipitation (mm) | 21.5 ± 0.15 | 58.7 ± 0.23 | 145.8 ± 6.40 |
| Soil T at 5 cm depth (°C) | 11.0 ± 3.01 | 20.2 ± 2.38 | 25.8 ± 1.42 |
| Soil moisture content (%) | 25.2 ± 0.05 | 24.1 ± 0.04 | 23.8 ± 0.06 |
| Soil oxygen content (%) | 19.8 ± 0.35 | 18.2 ± 0.49 | 14.3 ± 1.39 |
| Year | Date | Type | Commercial Name | Manufacturer | Active Substances | Application Rate (L/ha) | Plot Treatment |
|---|---|---|---|---|---|---|---|
| 2022–2023 | 9 November 2022 | Herbicide | Zodiac DFF® | Bayer CropScience S.r.l. Milan, Italy | Diflufenican | 2 | Treated and untreated |
| Chlortoluron | |||||||
| 16 March 2023 | Herbicide | Traxos® Pronto 60 | Pinoxaden | 1 | Treated and untreated | ||
| SYNGENTA ITLIA S.p.A. Milan, Italy | Clodinafop-propargyl | ||||||
| Cloquintocet-mexyl | |||||||
| Herbicide | Zypar™ | Dow AgroSciences Italy s.r.l. Milan, Italy | Florasulam | 1 | |||
| Cloquintocet-mexyl | |||||||
| Adjuvant | Codacide | DU PONT DE NEMOURS ITALIANA Srl Milan, Italy | Rapeseed oil | 1 | |||
| 29 March 2023 | PGR | Trimaxx® | ADAMA Italia S.r.l. Bergamo, Italy | Trinexapac-ethyl | 0.5 | Treated | |
| Fungicide | Priaxor® | Fluxapyroxad | 1.5 | ||||
| BASF Italia S.p.A. Cesano Maderno, Italy | Pyraclostrobin | ||||||
| 5 May 2023 | Fungicide | Prosaro® | Prothioconazole | 1 | Treated | ||
| Bayer CropScience S.r.l. Milan, Italy | Tebuconazole |
| Parameter | Period | Treatment | |
|---|---|---|---|
| Untreated | Treated | ||
| SOM | November 2022 | 2.61 (0.15) a | 2.60 (0.12) a |
| May 2023 | 2.45 (0.14) a | 2.52 (0.37) a | |
| July 2023 | 2.59 (0.16) a | 2.59 (0.14) a | |
| Microbial Biomass | November 2022 | 32.10 (9.08) a | 33.34 (10.67) a |
| May 2023 | 26.52 (3.29) a | 25.20 (4.32) b | |
| July 2023 | 28.30 (3.72) a | 27.85 (4.20) a | |
| Antille | Aquilante | Bandera | Bologna | Skyfall | Sy Moisson | Tintoretto | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameters | F | p | F | p | F | p | F | p | F | p | F | p | F | p |
| SOM | 8 | ** | 5 | * | 4 | * | 2 | ns | 2 | ns | 9 | *** | 3 | * |
| dsDNA | 1 | ns | 3 | ns | 9 | *** | 13 | *** | 1 | ns | 2 | ns | 8 | ** |
| β-gluc | 22 | *** | 23 | *** | 19 | *** | 15 | *** | 27 | *** | 7 | ** | 6 | ** |
| xylo | 54 | *** | 38 | *** | 28 | *** | 19 | *** | 51 | *** | 17 | *** | 29 | *** |
| uroni | 122 | *** | 238 | *** | 240 | *** | 168 | *** | 119 | *** | 207 | *** | 270 | *** |
| chit | 2 | ns | 3 | ns | 0.1 | ns | 1 | ns | 3 | * | 1 | ns | 0.2 | ns |
| leu | 2 | ns | 1 | ns | 3 | ns | 2 | ns | 1 | ns | 2 | ns | 0.3 | ns |
| acP | 166 | *** | 502 | *** | 162 | *** | 118 | *** | 276 | *** | 107 | *** | 237 | *** |
| alkP | 64 | *** | 109 | *** | 61 | *** | 53 | *** | 68 | *** | 68 | *** | 76 | *** |
| inositP | 172 | *** | 350 | *** | 236 | *** | 220 | *** | 205 | *** | 148 | *** | 389 | *** |
| bisP | 18 | *** | 23 | *** | 38 | *** | 8 | ** | 74 | *** | 13 | *** | 24 | *** |
| piroP | 8 | ** | 10 | *** | 14 | *** | 5 | * | 10 | *** | 6 | ** | 7 | ** |
| aryS | 19 | *** | 22 | *** | 26 | *** | 20 | *** | 15 | *** | 12 | *** | 24 | *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bardelli, T.; Fornasier, F.; Novarina, E.; Donniacuo, A.; Romano, E.; Bianchi, P.G.; Giulini, A.P.M. Changes in the Rhizosphere Biome Depending on the Variety of Wheat, Timing of Its Growing Season, and Agrochemical Components in the Soils of Italy. Agronomy 2024, 14, 832. https://doi.org/10.3390/agronomy14040832
Bardelli T, Fornasier F, Novarina E, Donniacuo A, Romano E, Bianchi PG, Giulini APM. Changes in the Rhizosphere Biome Depending on the Variety of Wheat, Timing of Its Growing Season, and Agrochemical Components in the Soils of Italy. Agronomy. 2024; 14(4):832. https://doi.org/10.3390/agronomy14040832
Chicago/Turabian StyleBardelli, Tommaso, Flavio Fornasier, Elena Novarina, Antonella Donniacuo, Elio Romano, Pier Giacomo Bianchi, and Anna Pia Maria Giulini. 2024. "Changes in the Rhizosphere Biome Depending on the Variety of Wheat, Timing of Its Growing Season, and Agrochemical Components in the Soils of Italy" Agronomy 14, no. 4: 832. https://doi.org/10.3390/agronomy14040832
APA StyleBardelli, T., Fornasier, F., Novarina, E., Donniacuo, A., Romano, E., Bianchi, P. G., & Giulini, A. P. M. (2024). Changes in the Rhizosphere Biome Depending on the Variety of Wheat, Timing of Its Growing Season, and Agrochemical Components in the Soils of Italy. Agronomy, 14(4), 832. https://doi.org/10.3390/agronomy14040832







