Abstract
Large and distinct families of receptor-like kinases (RLKs) play elemental roles in many fundamental processes of plants. The proline-rich extensin-like receptor kinase (PERK) family is one of the most pivotal classes of RLKs. To date, there have been no comprehensive or published studies conducted on the PERK gene family in Glycine max. This research aimed to characterize the role of the PERK gene family in cultivated soybean using a systematic array of bioinformatic and experimental approaches. We identified 16 PERK members in G. max through local BLASTp, using PERK members from Arabidopsis thaliana as a query. Tissue expression of genes, predicted via tissue specific expression analysis from the soybean database “SoyBase”, revealed that these PERK genes exhibit differentiated expression patterns in various plant organs. The gene structure was predicted via Gene Structure Display Server (GSDS). Phylogeny was demonstrated through an evolutionary tree employing the neighbor-joining method. Subcellular localization of proteins was identified via “Softberry” and cis-acting elements were identified through PlantCARE. The KASP (Kompetitive Allele Specific PCR (KASP)) marker was developed for the GmPERK-1-C and GmPERK-1-T allele, targeting position 167 nt in the CDS region. Genotyping results indicated that GmPERK-1 exhibits promising potential for utilization in molecular breeding programs for soybean to increase crop yield. Collectively, our findings indicate that G. max accessions harboring the GmPERK-1-C allele exhibit significantly higher thousand grain weight compared to accessions carrying the GmPERK-1-T allele. This research enhances the understanding of the molecular roles of PERK genes in G. max, providing valuable insights for the utilization of favorable genetic variations in soybean molecular breeding programs.
1. Introduction
Soybean (Glycine max (L.) Merr.) is an annual oilseed crop belonging to the Leguminosae family. Soybean plays a crucial role in food systems as it is a significant source of both calorie and protein intake [1]. Soybean protein, in particular, is an essential component for living organisms, prompting the sequencing of the soybean genome [2]. Soybean processed products, such as tofu, soy sauce, soy milk, and other soybean-processed products are gaining popularity as a dairy substitute. Additionally, soybean is a common ingredient in chicken feed due to its high protein content, making it an important component in poultry nutrition [3]. The first Receptor-like Kinase (RLK) was isolated in maize more than thirty years ago, by John C. Walker and Ren Zhang in 1990. A typical RLK is composed of an extracellular domain, a single-pass transmembrane domain, and a cytoplasmic kinase domain [4]. These RLKs play pivotal roles in diverse cellular processes, such as signal transduction [5,6], organismal development [7,8,9], and immune responses [10,11,12], thereby serving as crucial elements of cellular communication networks.
Recent research has elucidated the presence of several distinguished members of the RLK family in Arabidopsis thaliana (L.) Heynh, Brassica napus (L. supsb. napus), and Oryza sativa (L.) [8,13,14] and its typical subfamilies include proline-rich extensin-like receptor kinases (PERKs) and leucine-rich repeat receptor-like kinases (LRR-RLKs). These families play elemental roles in many underlying and inherent cellular processes, including signal transduction, external stimulus response, and homeostasis [6,15].
PERKs are a prominent subclass of RLKs and feature an N-terminal extracellular proline-rich domain and a C-terminal serine/threonine kinase domain [16,17,18,19,20]. The first PERK gene initially identified in Brassica napus, which was swiftly expanded into a PERK family as further research, including 15 PERK genes in Arabidopsis thaliana, 33 in Gossypium hirsutum (L.), and 25 in Brassica rapa (L. var. rapa) [14,19,21]. Nevertheless, their functions are currently not thoroughly understood. The proline-rich domain is involved in protein–protein interactions and is believed to be implicated in cell wall integrity, environmental stress sensors, as well as physiological processes including growth and development in plants. PERKs are rapidly activated upon injury and ubiquitously expressed in various parts of the plant. Additionally, they play a crucial role in regulating root and stem branching [22].
Numerous compelling examples demonstrate the functional significance of RLK family genes, like HAR1, CLAVATA1, Xa21, GmSARK in the LRR-RLK subfamily, and AtPERK4 in the PERK subfamily. The HAR1 gene has been reported to be involved in legume root growth, nodulation, and nitrate sensitivity during symbiotic development [23]. CLAVATA1 (CLV1) is responsible for meristem differentiation in Arabidopsis [24] and Xa21 for disease resistance in rice [25]. GmSARK (Glycine max senescence-associated receptor-like kinase) plays a part in regulating leaf senescence [26]. Furthermore, AtPERK4, a Ca2+ signaling regulator, is implicated in root growth and seed germination and is highly expressed in mature pollen and pollen tubes [27]. However, to date, the precise role of the PERK gene family in plant growth and development in Glycine max remains largely unexplored.
In this study, we have initiated a systematic investigation of the GmPERK members by conducting a comprehensive analysis on this family using both bioinformatics and experimental approaches. The aim of this study was to identify the individual genes in the PERK family implicated in soybean drought stress resistance. Ultimately, 16 members of the PERK family in G. max. were identified. Promoter and gene sequences were used to predict sequence logos, subcellular localization, gene duplication, chromosomal distribution, Ka/Ks ratio, promoter cis-element, exon/intron gene structure, and protein domain structure. The findings of this study will serve as the foundation for future investigations into the regulatory roles of PERK genes in the soybean.
2. Results
2.1. Sequence Identification Reveals PERKs to Be Widely Present in Plants
To examine the function of PERK genes in Glycine max, we first carried out BLAST search using the known Arabidopsis PERK gene and protein sequences as queries. We found 110 PERK genes with high sequence similarity in 8 distinct plant species, including 15 members from Arabidopsis thaliana, 16 from Gossypium raimondii (Ulbr.), 16 from Glycine max; 14 from Solanum tuberosum (L.), 8 from Oryza sativa, from Vitis vinifera (L.), 23 from Zea mays (L.), and 5 from Physcomitrella patens (Hedw) (Table S1). Using the bioinformatics tools “InterProscan 63.0”, “SMART”, and “PROSITE”, we further validated that these members belong to the PERK gene family.
We observed that all studied plants have at least 4 PERK genes. G. max, G. soja, and G. raimondii have 16 genes each, Zea mays has the most, 23 genes, and V. vinifera has the least, only 4 genes, indicating that PERK genes have undergone a large-scale expansion.
2.2. PERK mRNA Expression Profiling in Different Tissues Reveals Tissue Specific mRNA Expression
We next examined the expression pattern of GmPERK genes in different soybean tissues (Figure 1A). The GmPERK mRNA expression levels in young leaf, flower, pod (1 cm), pod shell (10 DAF, days after flowering), pod shell (14 DAF), nodule, root, seed (10 DAF), and seed (25 DAF) were retrieved from the ePlant/Soybase database. These values were then used to generate a heat map and the genes with identical expression patterns were clustered together, depicting the mRNA expression profile of 16 GmPERK genes in various tissues (Figure 1A and Table S2). GmPERK-8, 10, 16 exhibited expression in multiple plant organs, while GmPERK-6, 12, 15 showed nearly no expression in these tissues. In particular, GmPERK-16 has a greater mRNA expression level than the other GmPERK genes.
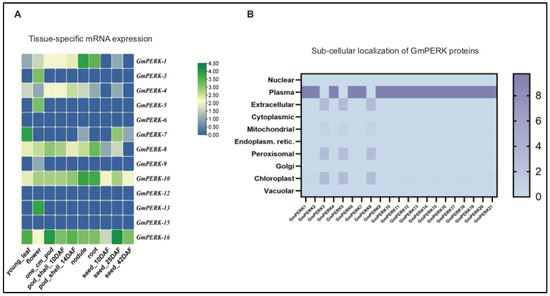
Figure 1.
GmPERK tissue-specific mRNA expression and cellular localization of proteins. (A) Heatmap, generated via the data from Soybase, visualizes the expression of 13 GmPERK mRNAs in different plant tissues. (B) The subcellular localization of GmPERK proteins was predicted via “Softberry”. The PERK proteins are mostly localized in plasma membrane. DAF: days after flowering.
2.3. GmPERK Proteins Are Predicted to Be Localized in the Plasma
GmPERK protein sequences were retrieved from the Plant Comparative Genomics portal “phytozome” (Table S3). The protein sequences were next subjected to analysis through “Softberry” to examine the localization of these proteins. The data was then further processed via TBtool to generate a protein localization map (Figure 1B). “Softberry” predicted that GmPERK-3, 5, 8 proteins are localized in the extracellular region, mitochondria, chloroplast, peroxisome, and other GmPERK proteins are possibly in the plasma membrane, consistent with the known roles of RLK family genes working as receptor kinases (Figure 1B). Another software, “CELLO V2”, predicted that GmPERK proteins can be localized in plasma membrane and the nucleus (Table S4). Although it is unclear whether these proteins have potential for functioning in nucleus as nuclear receptors, AtPERK1 has been reported to bind to nuclear shuttle proteins (NSPs) to facilitate viral DNA export from the nucleus to the cytoplasm [28,29]. Thus, we could not rule out the possibility that GmPERKs are present in the nucleus.
2.4. Visualized GmPERKs Gene Structure Reveals Evolution Similarities
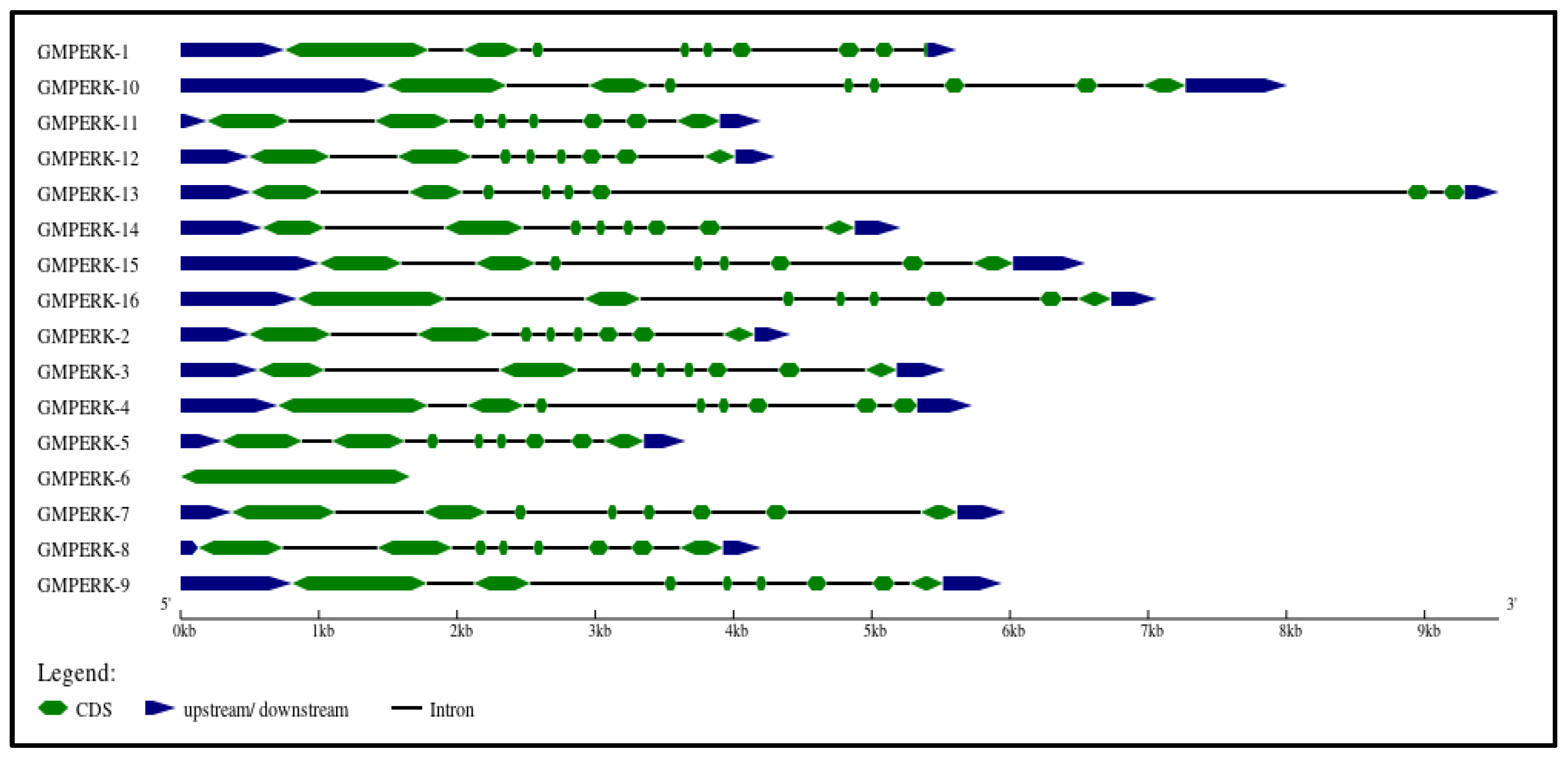
Genomic and complementary DNA sequences was retrieved from NCBI, and subsequently Phytozome (https://phytozome.jgi.doe.gov/) was employed to cross-check sequences. “Gene Structure Display Server 2.0” was used to visualize the gene structure of GmPERKs (Figure 2). Gene structure analysis and phylogenetic linkages were used to determine the phylogenetic relationship between GmPERK genes. This approach is based on the understanding that gene structure is associated with the evolution of different plant species [30]. All members of the GmPERK gene family exhibit a similar number of exons and introns, with the main difference being length. GmPERK-10 and GmPERK-13 are the longest in length, while GmPERK-5, 6, 2, 11, and 12 are among the shortest. All genes, with the exception of GmPERK-1 and 6, contain seven introns and eight exons.
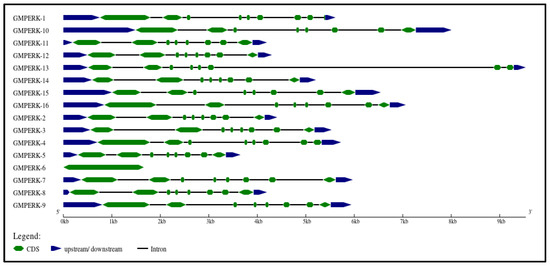
Figure 2.
The gene structure of all GmPERK genes. The coding sequence (CDS) is shown in green; the 5′ untranslated region (5′ UTR) and 3′ untranslated region (3′ UTR) is in blue; the introns are in black.
2.5. Sequence Logos Reveal Consistency in 59 Amino Acids
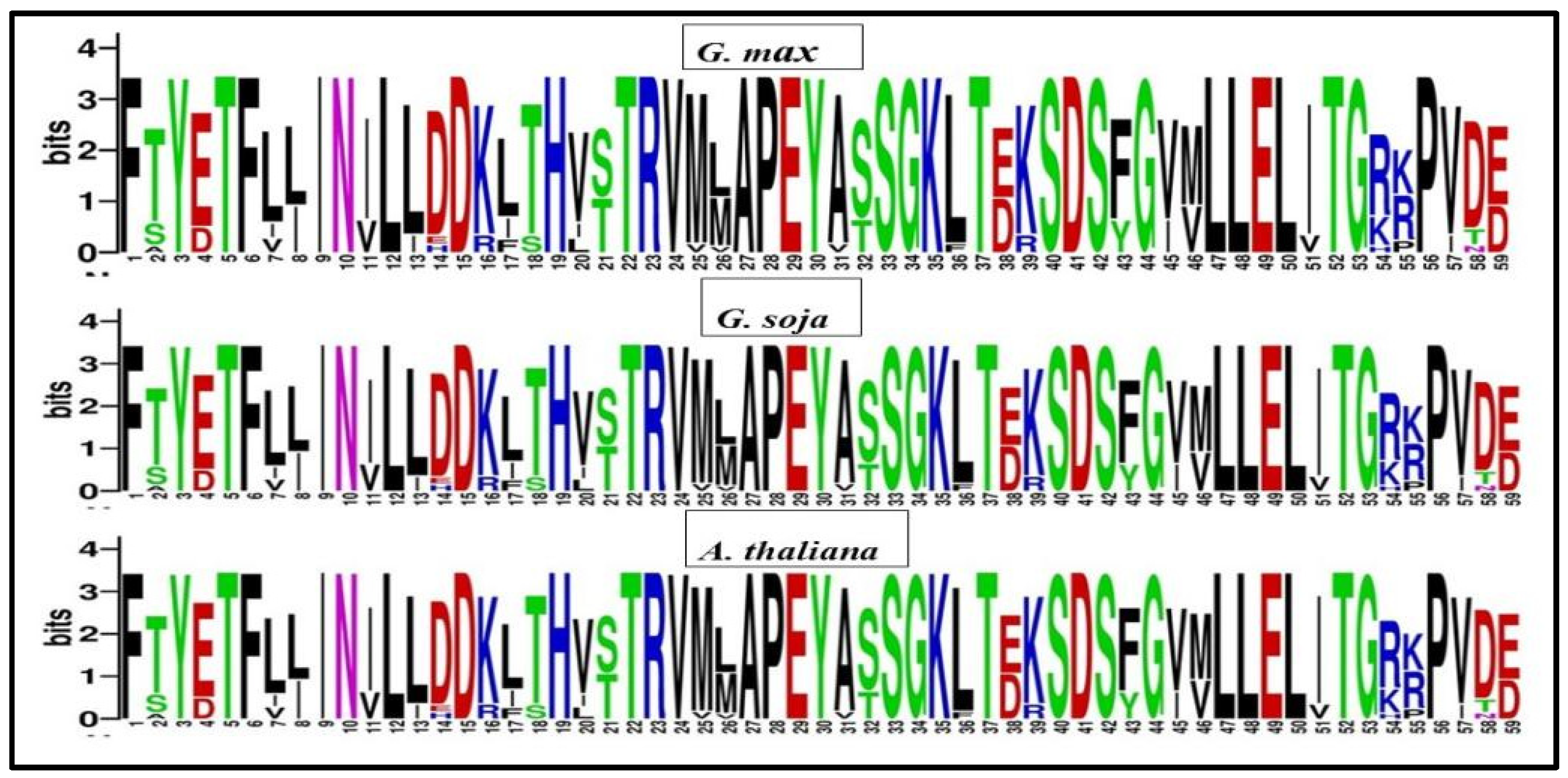
Sequence logos were generated using multiple sequence alignments from Arabidopsis thaliana, Glycine soja, and Glycine max to identify comparable domain sequences and examine the conservation of each amino acid residue within the domains of GmPERK. The results revealed no significant similarity in the amino acid residue patterns across most loci among three plant species studies. Nonetheless, we detected a very consistent region including 59 amino acids, among which F [1], T [5], F [6], N [10], L [12], D [15], D [19], T [22], R [23], A [27], P [28], E [29], L [47], L [48], E [49] were highly conserved, implicating functional significance of these residues (Figure 3). On the other hand, certain amino acids, namely TS [2], ED [4], LI [8], ED [38], MV [46], RK [54], and VI [57] were observed to have variation in their composition across different homolog genes and species (Figure 3). The identified sequence logo will help in defining new homolog genes in other species.
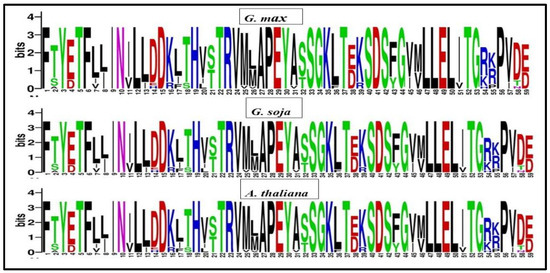
Figure 3.
Sequence logos of conserved amino acids in PERK proteins were created in Glycine max (top), Glycine soja (middle), and Arabidopsis thaliana (bottom).
2.6. Motif Analysis
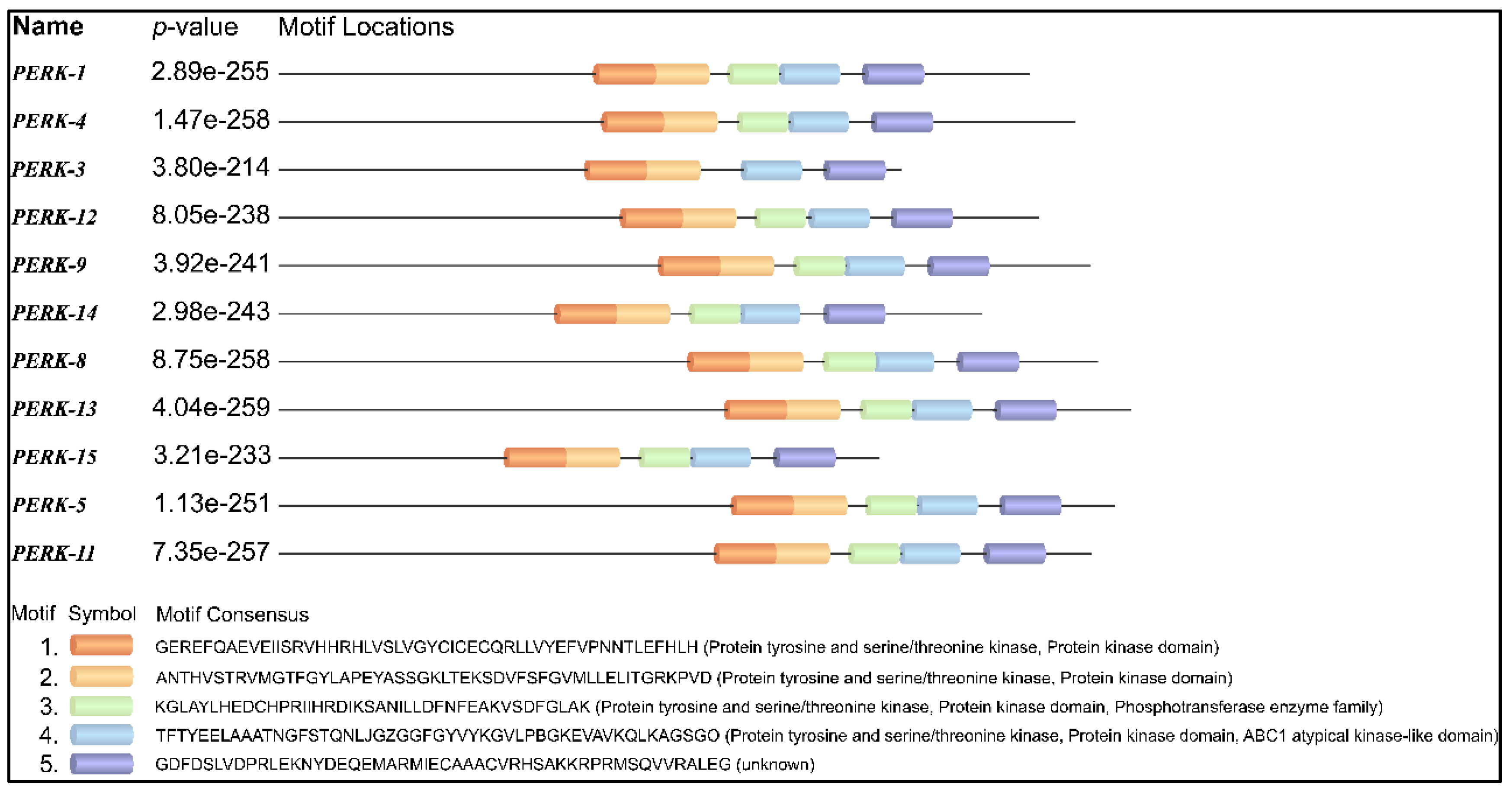
A protein motif refers to a short consensus amino acid sequence that holds a specific biological function. Motif analysis revealed that motifs often occurring in the protein tyrosine and serine/threonine kinase domain were observed in the PERK family (Figure 4). It is shown that certain motifs are not only conserved in all sequences, but are also arranged in a similar way, while the distance varies between motifs. Most members of GmPERK except GmPERK-3 carry similar motif types and arrangement. Conserved motifs are important for the proper functioning of the genes within the family. Interestingly, GmPERK-3 possesses one less motif compared to the other members of the gene family, implying it may vary in protein kinase activity.
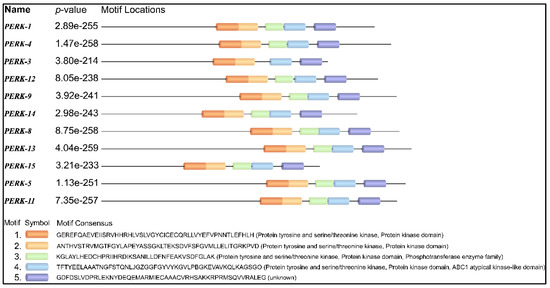
Figure 4.
Conserved motifs in GmPERK proteins.
2.7. Sequence Alignment and Phylogenetic Analysis
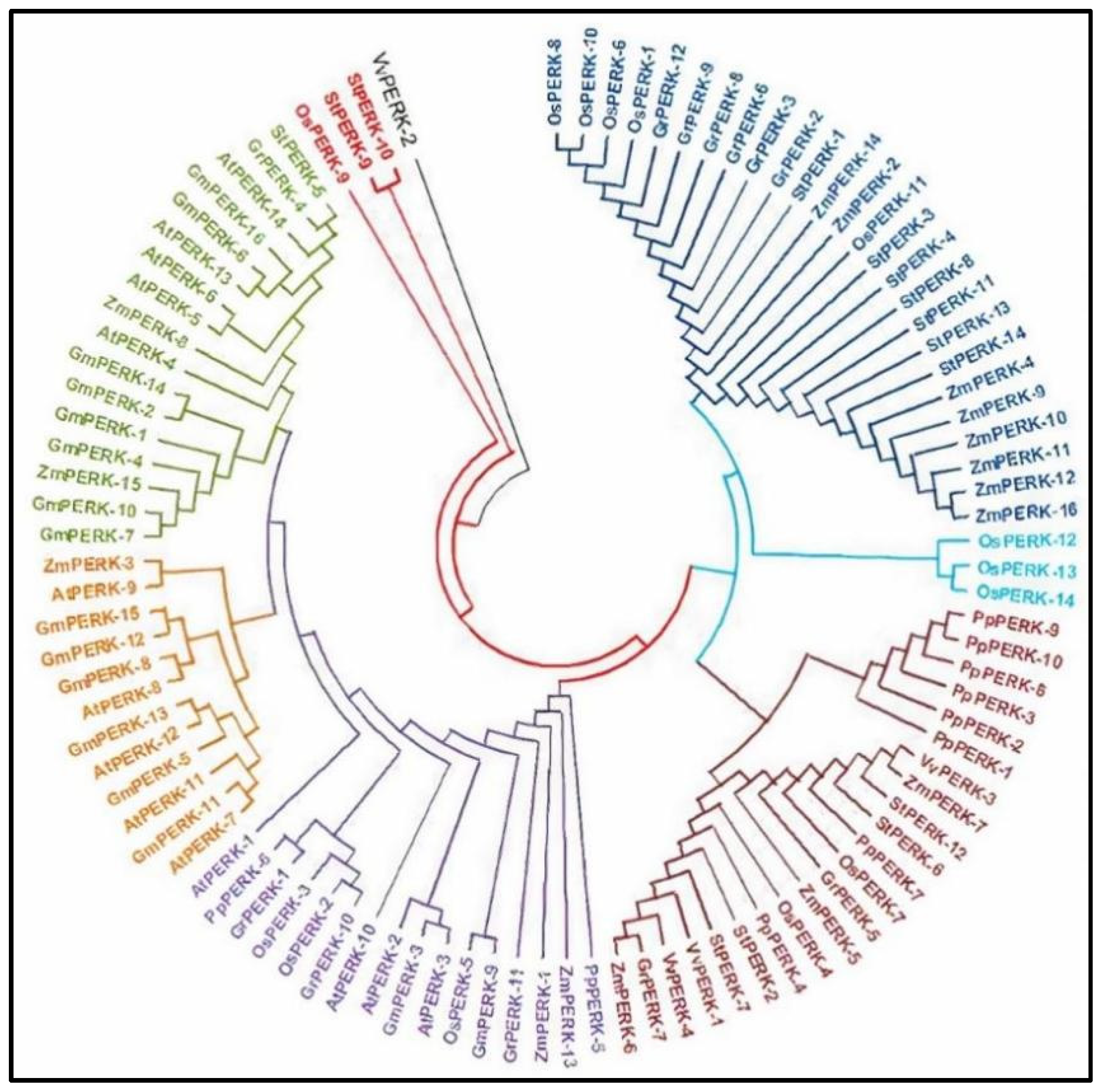
PERK genes from different plant species, including Arabidopsis thaliana, Gossypium raimondii, Solanum tuberosum (Solanaceae) and Glycine max (dicotyledon), Oryza sativa, Zea mays (monocotyledon), Physcomitrella patens (streptophyta), and Vitis vinifera (angiosperm), were investigated to examine whether a closer relationship existed between species. Hence, the phylogenetic tree was generated with mega software and protein sequences of different crops were then aligned using the mega alignment tool to create a neighbor-joining tree (Figure 5).
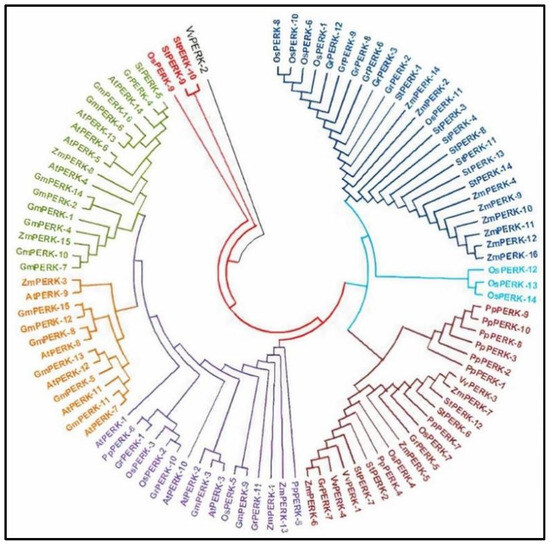
Figure 5.
Phylogenetic tree visualizes the evolutionary link among PERK genes from 8 different plant species. At: A. thaliana; Gm: G. max; Gr: G. raimondii; St: S. tuberosum; Os: Oryza sativa; Zm: Zea mays; Pp: Physcomitrella patens; Vv: Vitis vinifera.
The phylogenetic tree shows that all PERK genes from 8 plant species were naturally divided into seven lineages. Clade-1 in this tree contains most of the PERK genes (26 genes), followed by clade-2 (3 genes), clade-3 (22 genes), clade-4 (16 genes), clade-5 (12 genes), clade-6 (17 genes), and the last 4 genes are separate. Clade-1 contains PERK genes from S. tuberosum, Z. mays, O. sativa, and G. raimondii and lacks ones from G. max, P. patens, V. vinifera, and A. thaliana. Clade-2 contains 3 PERK members all from O. sativa. Clade-3 contains maximum genes from P. patens, V. vinifera, S. tuberosum, Z. mays, O. sativa, and G. raimondii and lacks genes from A. thaliana and G. max. Clade-4 contains 16 members from O. sativa, A. thaliana, G. max, P. patens, and G. raimondii and lacks members from S. tuberosum, Z. mays, and V. vinifera. Clade-5 contains 12 members from G. max and A. thaliana, and 1 gene from Z. mays. Clade-6 contains 17 members from G. max, A. thaliana, Z. mays, and 1 gene from both S. tuberosum and G. raimondii. Members of the PERK gene family in Glycine max were found to be distributed across all clades in our phylogenetic analysis.
2.8. Domain Structure Analysis
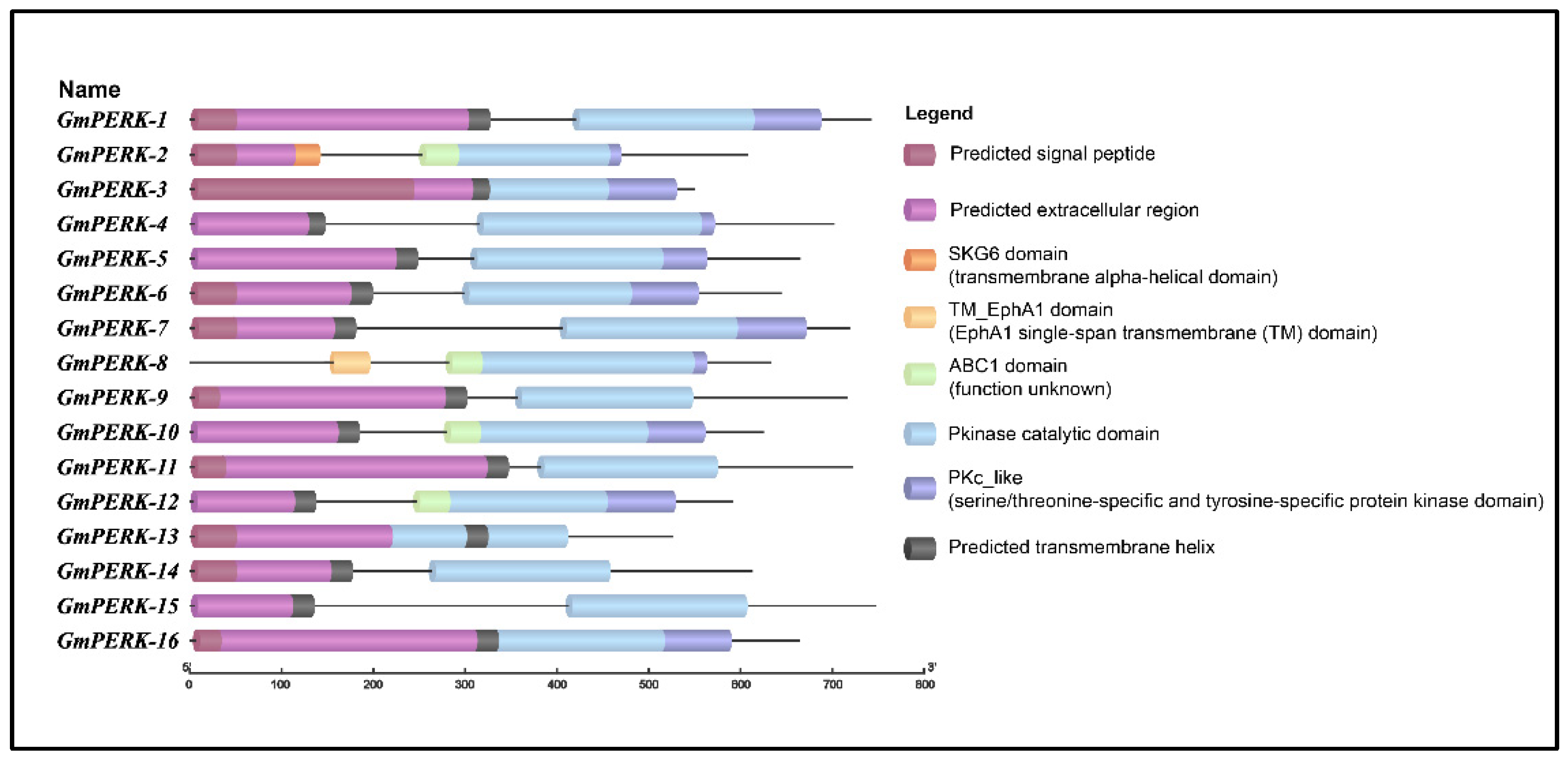
The conserved domains in GmPERK proteins were determined using the NCBI Conserved Domain Database (CDD) search. The full-length protein sequences were submitted to TB tools to investigate the presence of specific domains in all members of the gene family. The results indicated that all members of the PERK gene family possess a single Pkinase domain (Figure 6). Furthermore, the Protein Kinase C (PKc-like) superfamily domain is also common with the exception of GmPERK-9, 11, 13, 14, 15. In addition, conserved transmembrane domains were also detected in GmPERK-2 and GmPERK-8. We further predicted signal peptides, extracellular regions and transmembrane regions using MEMPACK in the PSIPRED workbench. Ten GmPERK proteins were predicted to possess all three regions, indicating that they are highly likely to function as receptor kinases (Figure 6 and Figure S1).
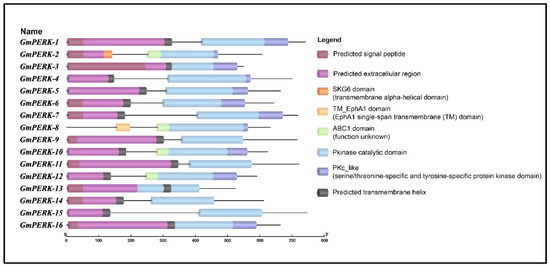
Figure 6.
Domain structure analysis of PERK proteins in G. max shows that these proteins have the common Pkinase domain and most of them include signal peptides, extracellular regions, and single-transmembrane domains.
2.9. DNA Cis-Element Analysis
Cis-acting DNA elements regulate gene expression, which allows us to gain insights into the transcriptional regulatory mechanism. The cis-elements are specific DNA sequences that are recognized via transcription factors and other regulatory proteins, and the specific DNA–protein interactions play crucial roles in determining when and where genes are expressed. By analyzing cis-elements, the transcriptional regulatory network can be identified [31].
Cis-elements were observed in promoter regions of GmPERK genes and categorized on the basis of their functional relevance. GmPERK genes all have different cis-elements for diverse responses in growth and development. CAAT-Box, CCAAT-Box, and TATA-Box were present; light-response element AE-box, AT1-motif, Box-4, BOX-II, G-Box, GA-motif, GT1-motif, LAMP-element, and TCT-motif were observed; and stress-response element, ABRE, ARE, GARE-motif, P-box, TC-rich repeats, TGACG-motif, and CGTCA-motif were also identified in the soybean PERK gene family (Table S5).
2.10. Chromosomal Localization, Ka/Ks Ratio, and Collinearity Analysis
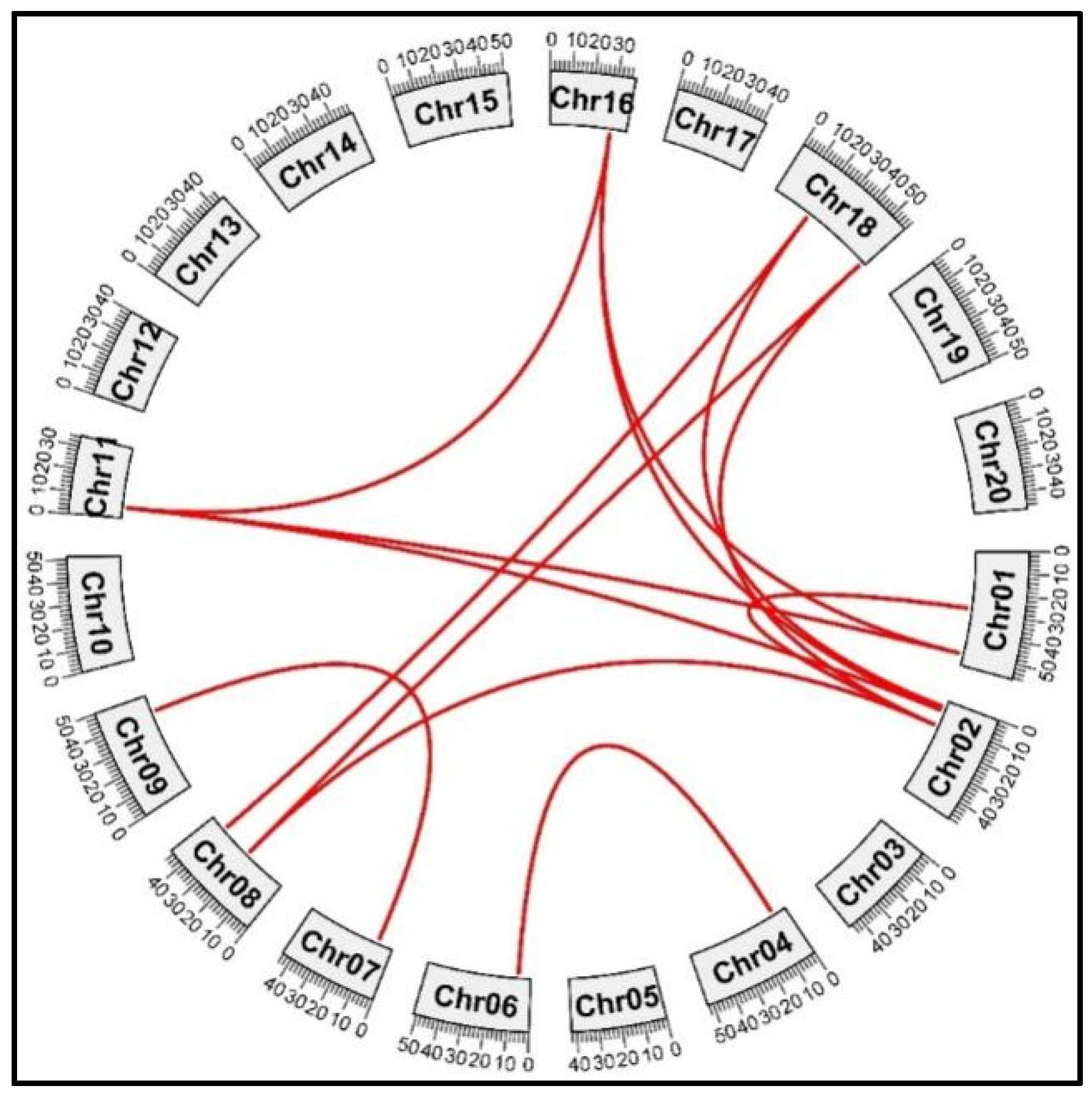
The GmPERK genes were mapped to respective chromosomal locations. We identified soybean paralogous gene pairs to investigate the locus connections among GmPERK genes. Synteny analysis revealed that several gene loci are extremely conserved (Figure 7 and Table S6). This suggests that during evolution, these genes hold important biological roles, or are under strong selection pressure, resulting in their high conservation across different species.
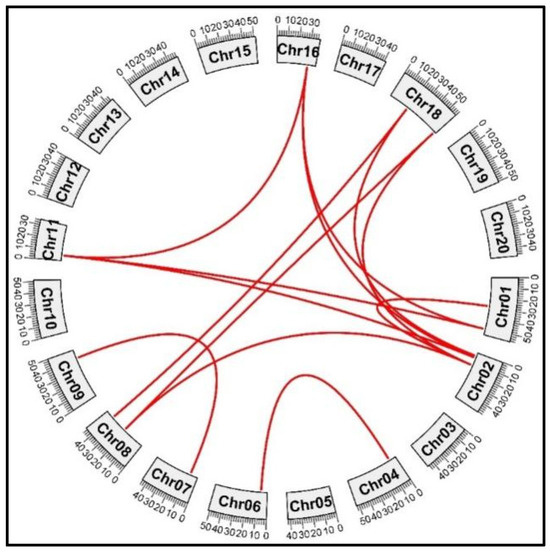
Figure 7.
The evolutionary duplication events of GmPERK genes on different chromosomes in G. max. GmPERK-1 and GmPERK-2 are located on chromosome 1; GmPERK-3 and GmPERK-4 on chromosome 2; GmPERK-6 on chromosome 4; GmPERK-7 on chromosome 6; GmPERK-8 on chromosome 7, GmPERK-9 and GmPERK-10 on chromosome 8; GmPERK-11 and GmPERK-12 on chromosomes 9 and 11 respectively; GmPERK-14 on chromosome 16; GmPERK-15 and GmPERK-16 on chromosome 18. Red lines connect the paralogous gene pairs.
Paralogous gene pairs usually come from gene duplication events, which include whole genome duplication (WGD), segmental duplication, and dispersed duplication. WGD and segmental duplication involve whole/a large part of chromosome duplication, while dispersed duplication includes a restricted region duplication. During evolution, these duplicated genes can become functionally divergent, leading to nonfunctionalization (loss of gene function), subfunctionalization (leaving part of the original function), or neofunctionalization (gaining new/beneficial function). Our analysis indicated that GmPERK-5 and GmPERK-13 formed through dispersed duplication while the remaining were attributed to segmental or whole genome duplication (WGD) (Table S7). WGD gives plants great advantages to adapt to severe environmental changes, possibly through the neofunctionalization of genes. Dispersed duplication events accumulating over time can contribute to the nonfunctionalization or loss of gene function.
To assess the magnitude and nature of selection pressure on these duplicated gene pairs, Ka/Ks values were calculated. Ka/Ks = 1 indicates neutral selection, Ka/Ks > 1 indicates positive selection or Darwinian selection (driving change) as a result of rapid evolution, and Ka/Ks < 1 indicates purifying or stabilizing selection (acting against change). We discovered that all GmPERK genes had a Ka/Ks value of less than one in this analysis. Because of segmental and WGD duplication, we believe soybean PERK genes were subjected to strong selection pressure with a moderate operative variation and stably maintained their function.
2.11. Sequence Polymorphism Assay and the Development of Kompetitive Allele Specific PCR (KASP) Markers
Soybean PAN-genome was used to explore the polymorphic sites for all GmPERK genes. Except for GmPERK-1, the other PERK genes did not show sequence polymorphism, and hence were excluded from further marker trait association analyses. In the exons of GmPERK-1, an SNP was discovered at 167 nt (T/C), which resulted in an amino acid alteration. The clustering of soybean accessions is shown in the scatter plot for the developed KASP assay on X-HEX and Y-FAM axes. Accessions colored in blue have the GmPERK-T allele, whereas accessions shown in orange have the GmPERK-C allele.
2.12. Association Analysis of GmPERK-1 Allelic Variations and Morphological Traits
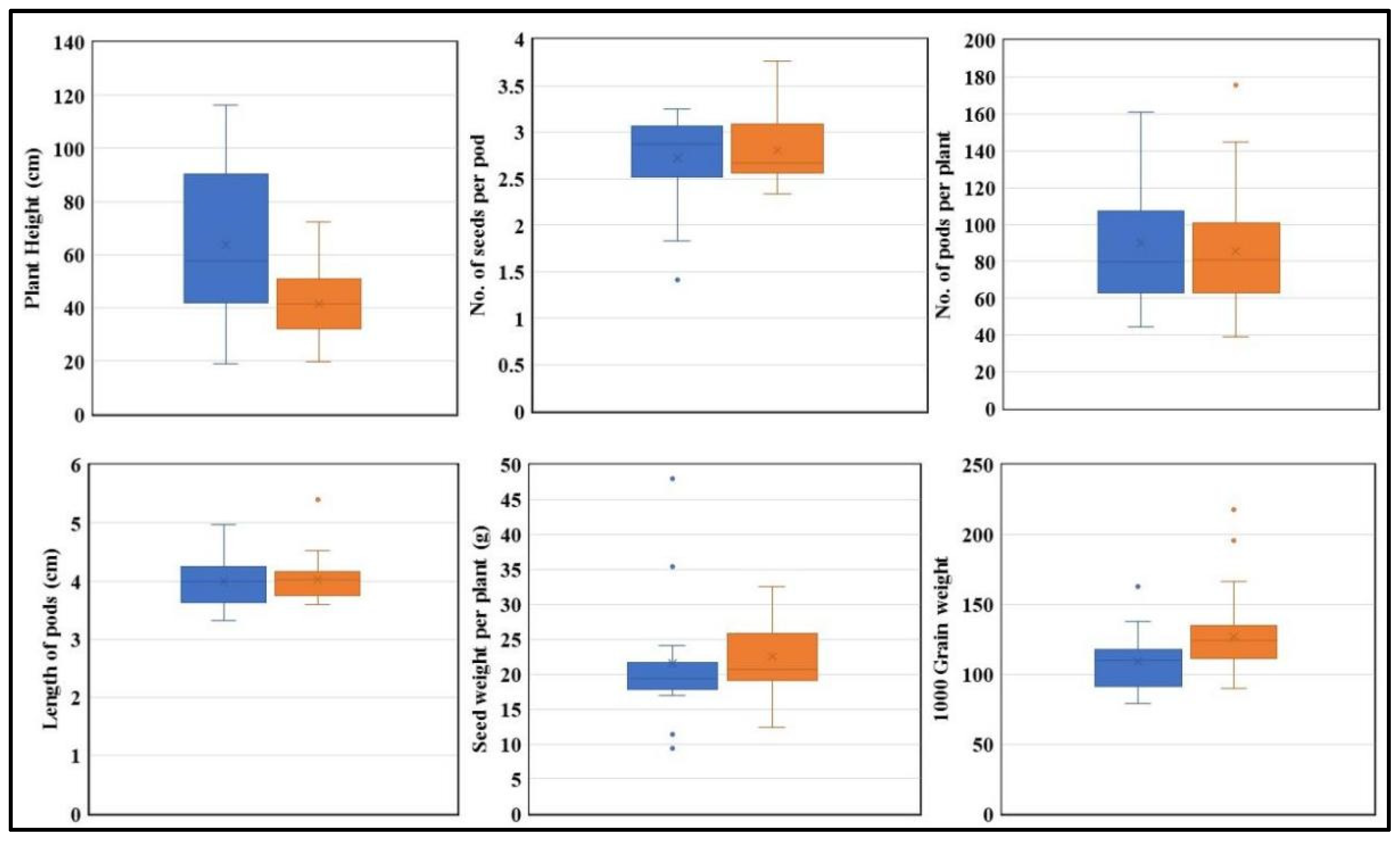
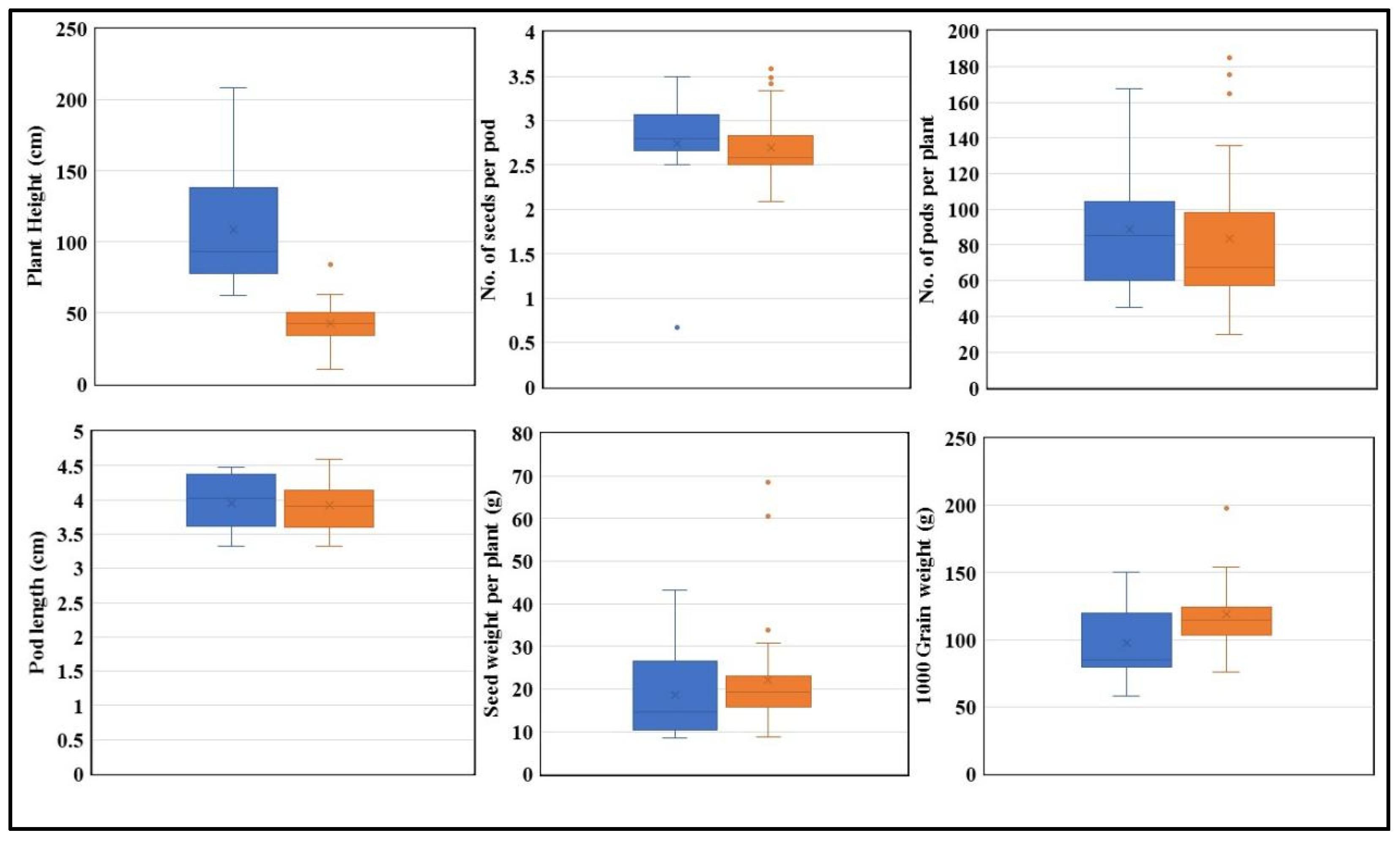
For GmPERK-1, 30% of the studied germplasm possessed “T” while 70% possessed GmPERK-1-C. Association analysis between allele types and traits was performed for plant height, no. of seeds per pod, no. of pods per plant, pod length, seed weight per plant, and 1000 grain weight in both optimal (Figure 8) and drought conditions (Figure 9).
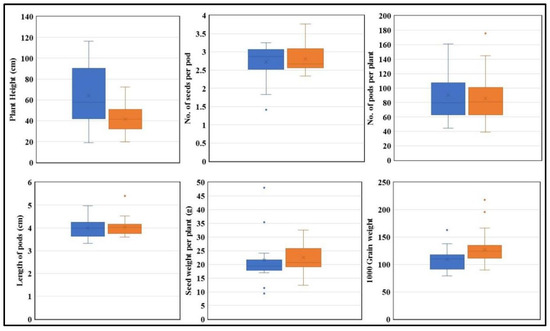
Figure 8.
The performance of the “T” and “C” alleles under optimal condition. “T” is represented in blue, and “C” is represented in orange. Plant height, no. of seeds per pod, no. of pods per plant, pod length, seed weight per plant, and 1000 grain weight are plotted (n = 3). (p < 0.05).
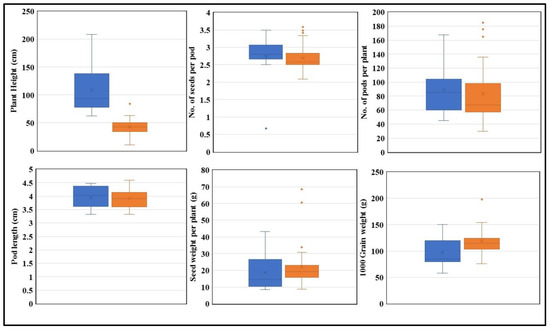
Figure 9.
The performance of “T” and “C” in drought. The “T” allele is shown in blue and the “C” allele is shown in orange. Plant height, no. of seeds per pod, no. of pods per plant, pod length, seed weight per plant, and 1000 grain weight are plotted (n = 3). (p < 0.05).
Under optimal growth conditions, the presence of the “T” allele is associated with increased height, as there is a major difference in height between genotypes carrying “C” and “T” alleles (Figure 8). Genotypes carrying the “T” allele have an average height of 63.7 cm while genotypes with “C” allele have an average height of 41.4 cm. Furthermore, the average number of pods is 90.1 for the “T” allele genotype whereas it is 86.4 for the “C” allele genotype. However, seed weight per plant is higher in genotypes with the “C” allele, with 22.7 g and only 21.5 g in genotypes with “T” allele. The 1000 grain weight is also higher in genotypes with the “C” allele (127.2 g) than that with the ‘T” allele (109.28 g) (Figure 8).
Further morphological data analysis demonstrated a significant difference in plant height and thousand grain weight between genotypes grown under both optimal and drought conditions (Figure 8 and Figure 9). The KASP marker results revealed that G. max accessions carrying the “T” allele were associated with increased plant height compared to accessions carrying the “C” allele. Additionally, G. max accessions carrying the “C” allele exhibited an association with higher thousand grain weight compared to accessions carrying the “T” allele according to association analysis, indicating that the “C” allele has a superiority over “T” and hence can be used for future soybean breeding programs (Figure 8 and Figure 9).
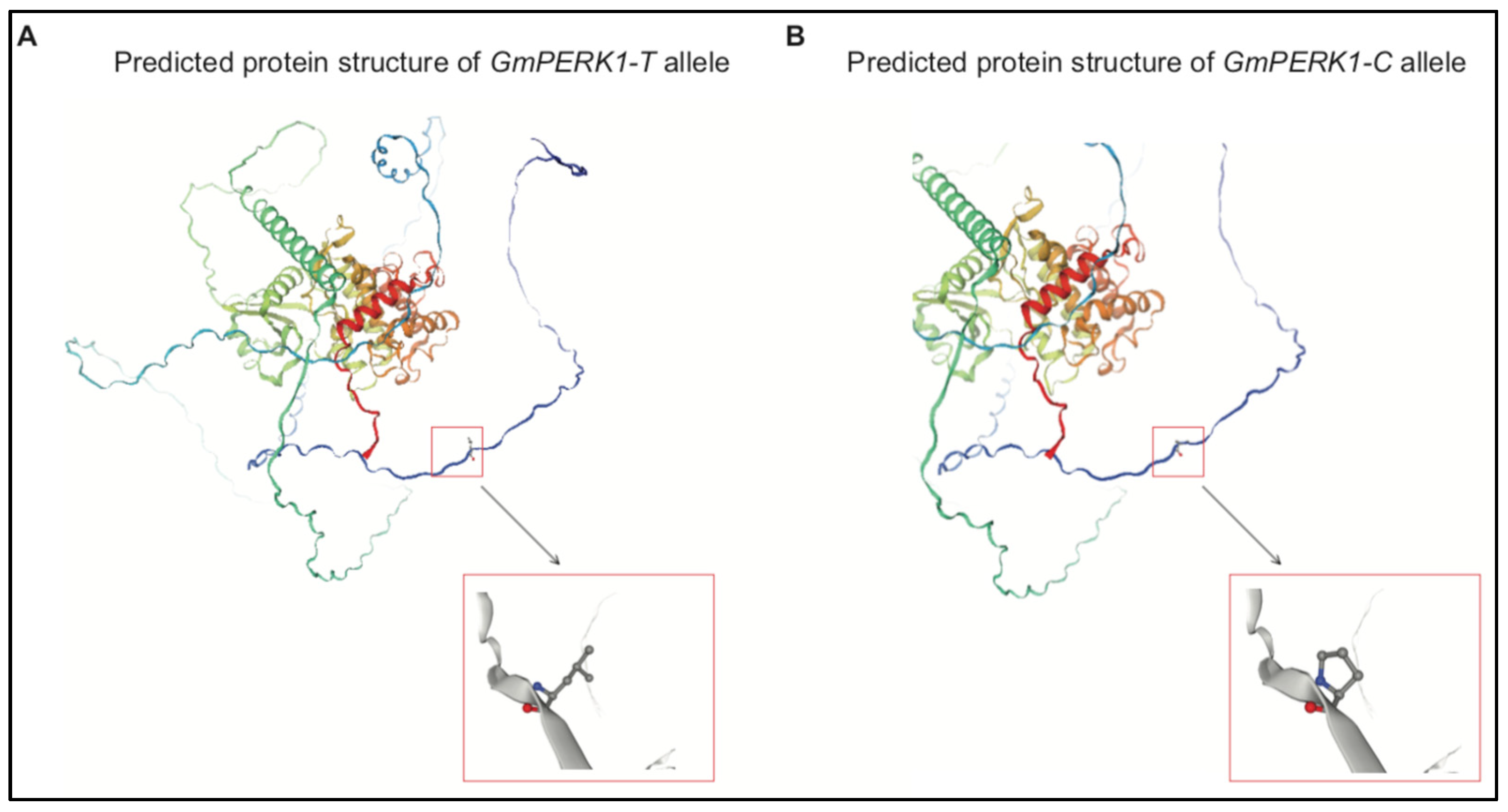
2.13. The Protein Structure Prediction of Two GmPERK-1 Alleles
In GmPERK1, the T to C substitution at 167 nt results in leucine to proline substitution at 56 aa. To find the molecular mechanism for how this substitution is significant, we predicted protein secondary structure with “PSIPRED” (Figure 6 and Figure S1), and tertiary structure with “SWISS-MODEL”. Predicted secondary structure revealed that the 56 aa position in the GmPERK1 protein lies outside of the cell membrane, in the extracellular domain (Figure 6 and Figure S1). Tertiary structure prediction shows multiple α helices (Figure 10A,B) presumed to be part of the transmembrane region and protein kinase region; the substitution is located outside of these helices. The amino acid sequences of GmPERK1-T and GmPERK1-C revealed that the region between 41 aa and 294 aa, located within the extracellular domain, is rich in Ser/Pro (EXT-) motifs. These motifs are proposed to be implicated in EXT-network sensing [16]. More recent research has shown that this extracellular domain is a binding site for signal peptides [32]. Our data support the importance of maintaining proline residue at the 56 aa position which resides in GmPERK1 extracellular Ser/Pro-rich motifs.
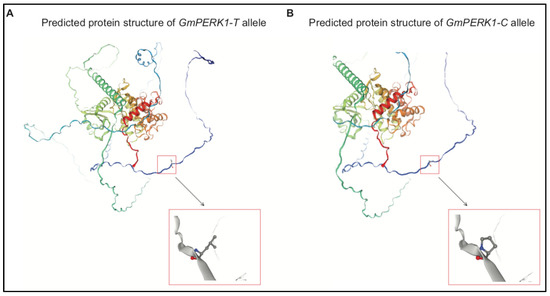
Figure 10.
“SWISS-MODEL” predicted protein tertiary structure of GmPERK1-T allele (A) and GmPERK1-C allele (B).
3. Materials and Methods
3.1. PERK Gene Sequence Identification
The PERK genes were discovered via BLAST search in diverse plant species, such as Glycine max, Glycine soja, Gossyium hirsutum, Zea mays, and Brassica rapa utilizing the Arabidopsis PERK gene and protein sequences as queries provided by NCBI (http://planttfdb.gao-lab.org/blast.php, accessed on 2 May 2023). Arabidopsis data was downloaded from the Arabidopsis database TAIR (http://www.arabidopsis.org/, accessed on 2 May 2023). The bioinformatics tools, including “InterProscan 63.0” (http://www.ebi.ac.uk/interpro/, accessed on 5 May 2023), SMART (http://smart.embl-heidelberg.de/, accessed on 8 May 2023), and PROSITE (http://prosite.expasy.org, accessed on 8 May 2023) were used to confirm the members of GmPERK gene family.
3.2. Tissue Specific Expression of GmPERK mRNAs
To examine the mRNA expression of GmPERK in different tissues at different development stages, the expression patterns were obtained from the ePlant/SoyBase database (https://bar.utoronto.ca/eplant_soybean/, accessed on 8 May 2023). The heatmap was generated via “TBtools” to express the transcript levels of GmPERK in different tissues and stages.
3.3. Subcellular Localization of GmPERK Proteins
GmPERK protein sequences were retrieved from phytozome (https://phytozome-next.jgi.doe.gov/pz/portal.html, accessed on 8 May 2023). Subcellular localization of sixteen GmPERK proteins was identified in “Softberry” (http://www.softberry.com/, accessed on 8 May 2023) and the “Protein structure and functions” module from the “ProtComp version 9.0” software was used to predict the subcellular localization for plant proteins. Another online tool “CELLOv.2.5” (http://cello.life.nctu.edu.tw/, accessed on 17 December 2023) was also used to predict eukaryote protein localization.
3.4. Identification of Gene Structure
Genomic and conserved DNA sequences of the soybean PERK genes were retrieved from NCBI. Gene structure display server 2.0 (GSDS 2.0) (http://gsds.gao-lab.org/) was used to identify the exon and intron regions. Potential cis-acting DNA elements of PERK genes were identified from promoter regions (2 kb upstream of transcription start sites) via “PlantCARE” (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 27 August 2023).
3.5. Evolutionary Analysis to Generate Phylogeny Tree and Sequence Logos of PERK Proteins
To implement evolutionary analysis and determine sequence logos, PERK protein sequences from different species were first downloaded from NCBI databases (https://www.ncbi.nlm.nih.gov/). Next, ClustalW was used to perform multiple sequence alignment of PERK family protein sequences.
The phylogeny tree for evolutionary analysis was established through the neighbor joining method (NJ) using “MEGA6.0” following parameters as previously reported [33]. The sequence logos were constructed using an online tool “WebLogo”. Lastly, protein sequences of PERK for Glycine max, Glycine soja, and Arabidopsis thaliana were used to create sequence logos (https://weblogo.berkeley.edu/logo.cgi, accessed on 28 August 2023).
3.6. Conserved Protein Motif, Domain, and Synteny Analysis
For conserved protein motif analysis, “MEME” suite (https://meme-suite.org/meme/, accessed on 30 August 2023) was employed to analyze the motif architecture in full-length proteins. The NCBI Conserved Domain Database “CDD” (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 30 August 2023) was used to identify conserved domains in PERK proteins. MEMPACK (TM Topology and Helix Packing) in PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/, accessed on 31 August 2023) was used for prediction of signal peptides, extracellular regions, and transmembrane regions. Synteny analysis was performed using Fa and GFF3 files from the soybean genome. Gene duplication analysis was performed following Yang et al., 2017 [34]. CIRCOS was used to generate the figure and Ka/Ks values were calculated using PAL2NAL.
3.7. Isolation of Single Nucleotide Polymorphism (SNP) Sites in GmPERK Genes from Soybean PAN-Genome
PAN-genome was used to identify SNP sites in GmPERKs. For this, whole genome sequences of three cultivars of soybean (Willliam-82, Lee, and Zhonghuang-13) were downloaded from the soybean database “SoyBase” (https://soybase.org/). Local BLAST was used to identify GmPERK sequences in the three cultivars mentioned above. The specific sequence of each gene was generated using the DNAstar Lasergene’s software (https://www.dnastar.com/software/lasergene/, accessed on 8 May 2023) package “SeqMan” function [35,36] and SNP was identified (Table S8).
3.8. Phenotyping and Genotyping
A set of 46 G. max accessions was collected from the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences. These accessions were planted in two different environments i.e., under “well water” and “water limited” conditions following augmented design (check = UAM-SB-200) at the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences in 2021. The “well water” samples were irrigated after an interval of ~15 days, whereas “water limited” samples were subjected to water stress, especially at the flowering stage. Each accession was planted on two beds on either side. The dimensions of each bed were 15 ft (length) × 2.5 ft (width). The seeds were planted with a plant-to-plant distance of 1 ft. Two seeds were placed in each spot, and during thinning, the unhealthy seedling was removed, and only the healthy one was retained. Morphological data were collected for plant height (cm), number of pods plant−1, pod length (cm), number of seeds pod−1, seed weight plant−1 (g), thousand grain weight (g), seed length (mm), seed width (mm), and seed thickness (mm) from both water regime conditions. The genomic DNA of the studied soybean germplasm was extracted from leaves using the CTAB method in the School of Life Sciences, Beijing Institute of Technology [37]. The quality of the extracted DNA was initially assessed using a NANO-Drop (K5800C Micro-Spectrophotometer). Subsequently, the extracted DNA was run on a 1% agarose gel for further analysis. Among 16 GmPERKs, only GmPERK-1 possessed a polymorphic site. Table S8 includes the nucleotide sequences of GmPERK-1 near the SNP sites from three cultivars. A KASP assay [38] was specifically designed to determine the SNPs.
Analysis of the PAN-genome revealed that Williams-82 harbored the GmPERK-1-C, “C” allele at position 167 nt. Furthermore, LEE and Zhonghuang-13 also carried the GmPERK-1-T, “T” allele at the same position. Therefore, a KASP assay on the SNP was applied at 167 nt C/T. Two allele specific reverse primers and one common forward primer were designed for allele calling (Table S9).
The “T” allele-specific reverse primer was labeled via FAM fluorophore while the “C” allele-specific reverse primer was labeled via HEX fluorophore (Table S9). Initially, to verify the reliability of molecular markers, target genes that had been previously sequenced were used. The primer mixture consisted of 12 μL of each tailed primer (100 μM), 30 μL common primer (100 μM), and 46 μL double distilled water (DDW). KASP assay was conducted in 96-well PCR plates with approximately 5 μL reaction mixture in each well. The recipe for 1× mixture is provided (Table S10).
Genotyping PCR was performed in the CFX Connect Real Time PCR detection system (Bio-Rad® laboratories Inc., Hercules, CA, USA).
3.9. Association Analyses
Microsoft Excel 2019 was used to perform descriptive statistics and variance estimations. The measure of probability p < 0.05 was used to determine statistically significant correlation. Student’s t-test at p < 0.05 was used to examine the impact of each allele of GmPERK-1 at 167 nt position in its coding region.
4. Discussion
Soybean is a significant leguminous oilseed crop that holds great value for both human and livestock consumption. Previous studies have extensively investigated the morphological and cellular characteristics of PERK genes in diverse plant species [8,19,22,39], however, there is currently a lack of reported systematic research on the involvement of PERK genes in soybeans. Therefore, the purpose of this study was to characterize the Proline-rich Extensin-like Receptor Kinase (PERK) gene family in soybean and examine their vital role in soybean growth and development. The findings of this study offer insights and serve as a valuable reference for further research on the functional roles of PERK gene in soybeans.
In this study, GmPERK-1-C showed significant association with thousand grain weight in both environments, indicating that the use of this allele could be beneficial for higher thousand grain weight selection (Figure 9 and Figure 10). In soybean breeding programs, gel-free KASP tests (high throughput) can considerably enhance selection speed and efficiency. Several studies have also proposed that the use of SNP-based functional markers for yield improvement is more successful [35,40,41]. The PERK genes described in this paper, as well as the KASP/SNP marker used to identify allelic variation, could be valuable in marker-assisted breeding for the increase in thousand grain weight. This functional marker can be used independently or in combination with other functional markers.
Sequences of 16 PERK genes were retrieved and subjected to various bioinformatic analyses. Gene structure display revealed that all except one gene are consistent in their intron and exon numbers (Figure 2). This means that the gene family has experienced relatively low selection pressure. Introns have been previously reported to be important in the evolution of many plant genes [42,43]. The structural discrepancies between exons and introns could be related to insertion and deletion (indel) occurrences and thus be used to identify the course of evolution [44]. The number of introns present earlier in the evolution of a gene family has gradually reduced through time, resulting in gene families with fewer or no introns [45]. Gene families with more or longer introns are thought to have acquired extra roles during evolution. Previous reports show that tandem duplication can increase the number of introns and result in the production of new genes [46].
Tissue expression analysis via “Soybase” suggests various PERK genes have distinguished expression patterns in different plant organs, demonstrating the versatile roles of PERK genes (Figure 1A). Consistently, GmPERK genes have different cis-acting DNA elements such as CAAT-Box, TATA-Box, ABRE etc., for patterning their differentiated expression and triggering varying responses to light exposure and drought stress. The importance of light in plant growth and development has been demonstrated in previous research [47]. It has also been shown that gene families in cotton and other plant species with the same cis-elements display functional relevance [48,49]. The characterized cis-elements, specific for plant growth, reproduction, light sensitivity, and adversity can be found in the promoter regions of the GmPERK genes. Thus, it can be expected that GmPERK transcription is regulated by diverse transcription factors binding to these cis-acting regulatory sites. Cellular localization and prediction of signal peptides and transmembrane regions indicate that most GmPERK proteins are present in the cellular membrane and likely function as receptor kinases (Figure 1B and Figure 6). Sequence logos have determined the conserved regions of this family proteins among Glycine max, Glycine soja, and Arabidopsis thaliana (Figure 3).
This study demonstrated that amino acid residues were highly conserved in both the N and C termini. This finding provides valuable insights into the conservation patterns of PERK protein sequences across different plant species. Synteny analyses revealed that GmPERK genes have undergone both segmental and whole genome duplication events (Figure 7). In the Brassicaceae lineage, Arabidopsis has had two complete genome duplications [50]. Domain structure analysis showed that GmPERK proteins have N-terminal extracellular proline-rich domain, transmembrane domain, and C-terminal Protein kinase domain (Figure 6). The phylogenetic tree demonstrated that PERK genes originated in phylostratum, a type of previous land plant, and that PERK orthologous genes are present throughout the plant kingdom (Figure 5). Because the PERK gene family is substantially preserved in cotton, this phylogenetic tree suggests that it is conserved and present in all crops [21,51].
Currently, conventional breeding methods remain insufficient and unsatisfactory [52], therefore, the utilization of high-throughput SNP-based markers has emerged as a promising approach to accelerate and enhance breeding programs [40,53]. The PAN-genome of soybean will revolutionize the approach for molecular and genomic studies in soybean. Since 1923, soybean genetic gain is estimated to be ~0.34 bu/ac [54]. Grain yield improvements have accounted for the majority of genetic gain in the past. The application of advanced molecular breeding tools is expected to accelerate the genetic gain.
Future research could possibly focus on a more thorough comparison and analysis of the tertiary and quaternary structure of GmPERK-1-C and GmPERK-1-T, and investigate the impacts of the SNP in GmPERK-1 more in depth.
Collectively, these results revealed that PERK genes are globally expressed throughout plant tissues, highly conserved through evolution and across different species, with alleles that could potentially be used to optimize crop yield.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy14081679/s1, Table S1: PERK genes identified from different plant species; Table S2: Tissue-specific expression of PERK transcripts in soybean; Table S3: GmPERK protein sequence; Table S4: PERK protein localization predicted by “CELLO V2” in soybean; Table S5: Predicted cis-acting DNA elements; Table S6: Collinearity/synteny and Ka/Ks analysis of GmPERK gene family; Table S7: Gene Duplication Analysis in GmPERK gene family; Table S8: The identified SNP after alignment of PERK-1; Table S9: KASP marker sequence developed for screening of GmPERK-1 in soybean; Table S10: Recipe of GmPERK-3 KASP assay for 1×; Figure S1: GmPERK protein regions/domains including signal peptide, extracellular region and transmembrane helix. Predicted by http://bioinf.cs.ucl.ac.uk/ using PSIPRED MEMPACK (accessed on 8 May 2023). Below are gene names and their respective predictions shown in amino acid sequences.
Author Contributions
Conceptualization, H.J., Y.L. and A.I.; methodology, S.U.R.; software, A.I. and S.U.R.; validation, A.I. and S.U.R.; formal analysis, Y.W.; investigation, Y.W. and S.U.R.; data curation, S.U.R.; writing—original draft preparation, Y.L. and Y.W.; writing—review and editing, H.J., B.Z. and A.I.; visualization, Y.L. and S.U.R.; supervision, H.J.; project administration, H.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The original data presented in the study are openly available on the databases mentioned in Section 3.
Acknowledgments
We thank the Biological and Medical Engineering Core Facilities of Beijing Institute of Technology for supporting experimental equipment.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Okumu, O.O.; Otieno, H.M.; Okeyo, G.O. Production systems and contributions of grain legumes to soil health and sustainable agriculture: A review. Arch. Agric. Environ. Sci. 2023, 8, 259–267. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Bressani, R. The role of soybeans in food systems. J. Am. Oil Chem. Soc. 1981, 58, 392–400. [Google Scholar] [CrossRef]
- Li, J.; Tax, F.E. Receptor-Like Kinases: Key Regulators of Plant Development and Defense. J. Integr. Plant Biol. 2013, 55, 1184–1187. [Google Scholar] [CrossRef] [PubMed]
- Gish, L.; Clark, S. The RLK/Pelle family of kinases. Plant J. 2011, 176, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Dievart, A.; Clark, S.E. LRR-containing receptors regulating plant development and defense. Development 2004, 131, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wang, K.; Li, X.; Sun, C.; Yin, R.; Wang, Y.; Wang, Y.; Zhang, M. Evolution, functional differentiation, and co-expression of the RLK gene family revealed in Jilin ginseng, Panax ginseng C.A. Meyer. Mol. Genet. Genom. 2018, 293, 845–859. [Google Scholar] [CrossRef]
- Morris, E.R.; Walker, J.C. Receptor-like protein kinases: The keys to response. Curr. Opin. Plant Biol. 2003, 6, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.; Kui, H.; Li, J. Receptor-like Kinases in Root Development: Current Progress and Future Directions. Mol. Plant 2021, 14, 166–185. [Google Scholar] [CrossRef]
- Soltabayeva, A.; Dauletova, N.; Serik, S.; Sandybek, M.; Omondi, J.O.; Kurmanbayeva, A.; Srivastava, S. Receptor-like Kinases (LRR-RLKs) in Response of Plants to Biotic and Abiotic Stresses. Plants 2022, 11, 2660. [Google Scholar] [CrossRef]
- Greeff, C.; Roux, M.; Mundy, J.; Petersen, M. Receptor-like kinase complexes in plant innate immunity. Front. Plant Sci. 2012, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhou, J.M. Receptor-like kinases in plant innate immunity. J. Integr. Plant Biol. 2013, 55, 1271–1286. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.-H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.-H.; Mayer, K.F.X.; Li, W.-H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef]
- Chen, G.; Wang, J.; Wang, H.; Wang, C.; Tang, X.; Li, J.; Zhang, L.; Song, J.; Hou, J.; Yuan, L. Genome-wide analysis of proline-rich extension-like receptor protein kinase (PERK) in Brassica rapa and its association with the pollen development. BMC Genom. 2020, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.-H.; Bleecker, A.B. Plant Receptor-Like Kinase Gene Family: Diversity, Function, and Signaling. Sci. STKE 2001, 2001, re22. [Google Scholar] [CrossRef] [PubMed]
- Borassi, C.; Sede, A.R.; Mecchia, M.A.; Salgado Salter, J.D.; Marzol, E.; Muschietti, J.P.; Estevez, J.M. An update on cell surface proteins containing extensin-motifs. J. Exp. Bot. 2016, 67, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, T.V.; Bonetta, D.T.; Goring, D.R. Sentinels at the wall: Cell wall receptors and sensors. New Phytol. 2007, 176, 7–21. [Google Scholar] [CrossRef]
- Humphrey, T.V.; Haasen, K.E.; Aldea-Brydges, M.G.; Sun, H.; Zayed, Y.; Indriolo, E.; Goring, D.R. PERK-KIPK-KCBP signalling negatively regulates root growth in Arabidopsis thaliana. J. Exp. Bot. 2015, 66, 71–83. [Google Scholar] [CrossRef]
- Nakhamchik, A.; Zhao, Z.; Provart, N.J.; Shiu, S.-H.; Keatley, S.K.; Cameron, R.K.; Goring, D.R. A comprehensive expression analysis of the Arabidopsis proline-rich extensin-like receptor kinase gene family using bioinformatic and experimental approaches. Plant Cell Physiol. 2004, 45, 1875–1881. [Google Scholar] [CrossRef]
- Lee, H.K.; Santiago, J. ScienceDirect Plant Biology Structural insights of cell wall integrity signaling during development and immunity. Curr. Opin. Plant Biol. 2023, 76, 102455. [Google Scholar] [CrossRef]
- Qanmber, G.; Liu, J.; Yu, D.; Liu, Z.; Lu, L.; Mo, H.; Ma, S.; Wang, Z.; Yang, Z. Genome-wide identification and characterization of the PERK gene family in Gossypium hirsutum reveals gene duplication and functional divergence. Int. J. Mol. Sci. 2019, 20, 1750. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.F.; Goring, D.R. The proline-rich, extensin-like receptor kinase-1 (PERK1) gene is rapidly induced by wounding. Plant Mol. Biol. 2002, 50, 667–685. [Google Scholar] [CrossRef] [PubMed]
- Krusell, L.; Madsen, L.H.; Sato, S.; Aubert, G.; Genua, A.; Szczyglowski, K.; Duc, G.; Kaneko, T.; Tabata, S.; de Bruijn, F.; et al. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 2002, 420, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.E.; Williams, R.W.; Meyerowitz, E.M. The CLAVATA1 Gene Encodes a Putative Receptor Kinase That Controls Shoot and Floral Meristem Size in Arabidopsis. Otolaryngol. Pol. 2008, 62, 117. [Google Scholar]
- Song, W.-Y.; Wang, G.-L.; Chen, L.-L.; Kim, H.-S.; Pi, L.-Y.; Holsten, T.; Gardner, J.; Wang, B.; Zhai, W.-X.; Zhu, L.-H.; et al. A Receptor Kinase-Like Protein Encoded by the Rice Disease Resistance Gene, Xa21. Science 1995, 270, 1804–1806. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-P.; Gan, R.; Li, P.-L.; Ma, Y.-Y.; Zhang, L.-W.; Zhang, R.; Wang, Y.; Wang, N.N. Identification and functional characterization of a leucine-rich repeat receptor-like kinase gene that is involved in regulation of soybean leaf senescence. Plant Mol. Biol. 2006, 61, 829–844. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Zhou, Y.; Song, C. Arabidopsis proline-rich extensin-like receptor kinase 4 modulates the early event toward abscisic acid response in root tip growth. Plant Signal. Behav. 2009, 4, 1075–1077. [Google Scholar] [CrossRef] [PubMed]
- Roby, D.; Invernizzi, M.; Hanemian, M.; Libourel, C.; Roby, D. PERKing up our understanding of the proline-rich extensin-like receptor kinases, a forgotten plant receptor kinase family. New Phytol. 2022, 235, 875–884. [Google Scholar]
- Florentino, L.H.; Santos, A.A.; Fontenelle, M.R.; Pinheiro, G.L.; Zerbini, F.M.; Baracat-Pereira, M.C.; Fontes, E.P.B. A PERK-Like Receptor Kinase Interacts with the Geminivirus Nuclear Shuttle Protein and Potentiates Viral Infection. J. Virol. 2006, 80, 6648–6656. [Google Scholar] [CrossRef]
- Roy, S.W.; Gilbert, W. Complex early genes. Proc. Natl. Acad. Sci. USA 2005, 102, 1986–1991. [Google Scholar] [CrossRef]
- Weirauch, M.T.; Yang, A.; Albu, M.; Cote, A.G.; Montenegro-Montero, A.; Drewe, P.; Najafabadi, H.S.; Lambert, S.A.; Mann, I.; Cook, K.; et al. Determination and Inference of Eukaryotic Transcription Factor Sequence Specificity. Cell 2015, 158, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Feng, Y.; Xue, J.; Chen, P.; Zhang, A.; Yu, Y. Advances in Receptor-like Protein Kinases in Balancing Plant Growth and Stress Responses. Plants 2023, 12, 427. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Gong, Q.; Qin, W.; Yang, Z.; Cheng, Y.; Lu, L.; Ge, X.; Zhang, C.; Wu, Z.; Li, F. Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol. 2017, 17, 113. [Google Scholar] [CrossRef] [PubMed]
- Zafer, M.Z.; Tahir, M.H.N.; Khan, Z.; Sajjad, M.; Gao, X.; Bakhtavar, M.A.; Waheed, U.; Siddique, M.; Geng, Z.; Rehman, S.U. Genome-Wide Characterization and Sequence Polymorphism Analyses of Glycine max Fibrillin (FBN) Revealed Its Role in Response to Drought Condition. Genes 2023, 14, 1188. [Google Scholar] [CrossRef] [PubMed]
- Swindell, S.R. Sequence Data Analysis Guidebook; Springer: Berlin/Heidelberg, Germany, 1997. [Google Scholar]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol. 2000, 132, 71–91. [Google Scholar]
- Alvarez-Fernandez, A.; Bernal, M.J.; Fradejas, I.; Ramírez, A.M.; Yusuf, N.A.M.; Lanza, M.; Hisam, S.; de Ayala, A.P.; Rubio, J.M. KASP: A genotyping method to rapid identification of resistance in Plasmodium falciparum. Malar. J. 2021, 20, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Zhang, G.; Zhou, Y.; Zhang, Z.; Wang, W.; Du, Y.; Wu, Z.; Song, C.P. Plasma membrane-associated proline-rich extensin-like receptor kinase 4, a novel regulator of Ca2+ signalling, is required for abscisic acid responses in Arabidopsis thaliana. Plant J. 2009, 60, 314–327. [Google Scholar] [CrossRef]
- Rasheed, A.; Hao, Y.; Xia, X.; Khan, A.; Xu, Y.; Varshney, R.K.; He, Z. Crop Breeding Chips and Genotyping Platforms: Progress, Challenges, and Perspectives. Mol. Plant 2017, 10, 1047–1064. [Google Scholar] [CrossRef]
- Semagn, K.; Babu, R.; Hearne, S.; Olsen, M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol. Breed. 2014, 33, 1–14. [Google Scholar] [CrossRef]
- Mehmood Rana, R.; Saeed, S.; Masoud Wattoo, F.; Waqas Amjid, M.; Azam Khan, M. Protein modeling and evolutionary analysis of Calmodulin Binding Transcription Activator (CAMTA) gene family in Sorghum bicolor. Asian J. Agric. Biol. 2019, 7, 27–38. [Google Scholar]
- Roy, S.W.; Gilbert, W. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat. Rev. Genet. 2006, 7, 211–221. [Google Scholar]
- Lecharny, A.; Boudet, N.; Gy, I.; Aubourg, S.; Kreis, M. Introns in, introns out in plant gene families: A genomic approach of the dynamics of gene structure. J. Struct. Funct. Genom. 2003, 3, 111–116. [Google Scholar] [CrossRef]
- Roy, S.W.; Penny, D. A very high fraction of unique intron positions in the intron-rich diatom Thalassiosira pseudonana indicates widespread intron gain. Mol. Biol. Evol. 2007, 24, 1447–1457. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Maekawa, M.; Saito, A.; Higo, H.; Higo, K. Evolutionary relationship of plant catalase genes inferred from exon-intron structures: Isozyme divergence after the separation of monocots and dicots. Theor. Appl. Genet. 1998, 97, 9–19. [Google Scholar] [CrossRef]
- Fankhauser, C.; Chory, J. Light Control of Plant. Annu. Rev. Cell Dev. Biol. 1997, 13, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Qanmber, G.; Yu, D.; Li, J.; Wang, L.; Ma, S.; Lu, L.; Yang, Z.; Li, F. Genome-wide identification and expression analysis of Gossypium RING-H2 finger E3 ligase genes revealed their roles in fiber development, and phytohormone and abiotic stress responses. J. Cott. Res. 2018, 1, 1–17. [Google Scholar] [CrossRef]
- Pandey, A.; Misra, P.; Alok, A.; Kaur, N.; Sharma, S.; Lakhwani, D.; Asif, M.H.; Tiwari, S.; Trivedi, P.K. Genome-wide identification and expression analysis of homeodomain leucine zipper subfamily IV (HDZ IV) gene family from Musa accuminata. Front. Plant Sci. 2016, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1040. [Google Scholar] [CrossRef]
- Li, F.; Fan, G.; Wang, K.; Sun, F.; Yuan, Y.; Song, G.; Li, Q.; Ma, Z.; Lu, C.; Zou, C.; et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 2014, 46, 567–572. [Google Scholar] [CrossRef]
- Rehman, S.U.; Wang, J.; Chang, X.; Zhang, X.; Mao, X.; Jing, R. A wheat protein kinase gene TaSnRK2.9-5A associated with yield contributing traits. Theor. Appl. Genet. 2019, 132, 907–919. [Google Scholar] [CrossRef] [PubMed]
- Rauf, A.; Sher, M.A.; Farooq, U.; Rasheed, A.; Sajjad, M.; Jing, R.; Khan, Z.; Attia, K.A.; Mohammed, A.A.; Fiaz, S.; et al. An SNP based genotyping assay for genes associated with drought tolerance in bread wheat. Mol. Biol. Rep. 2024, 51, 527. [Google Scholar] [CrossRef] [PubMed]
- Rincker, K.; Nelson, R.L.; Specht, J.; Sleper, D.; Cary, T.; Cianzio, S.; Casteel, S.; Conley, S.; Chen, P.; Davis, V.; et al. Genetic Improvement of, U.S. Soybean in Maturity Groups II, III, and IV. Crop Sci. 2014, 54, 1419–1432. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).