Genome-Wide Association Analysis and Candidate Gene Prediction of Wheat Grain Copper Concentration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Materials and Field Planting
2.2. Determination of Copper Content in Mature Grains of Wheat
2.3. Statistical Analysis
2.4. Multi-Loci Association Analysis
2.5. Candidate Gene Prediction
3. Results
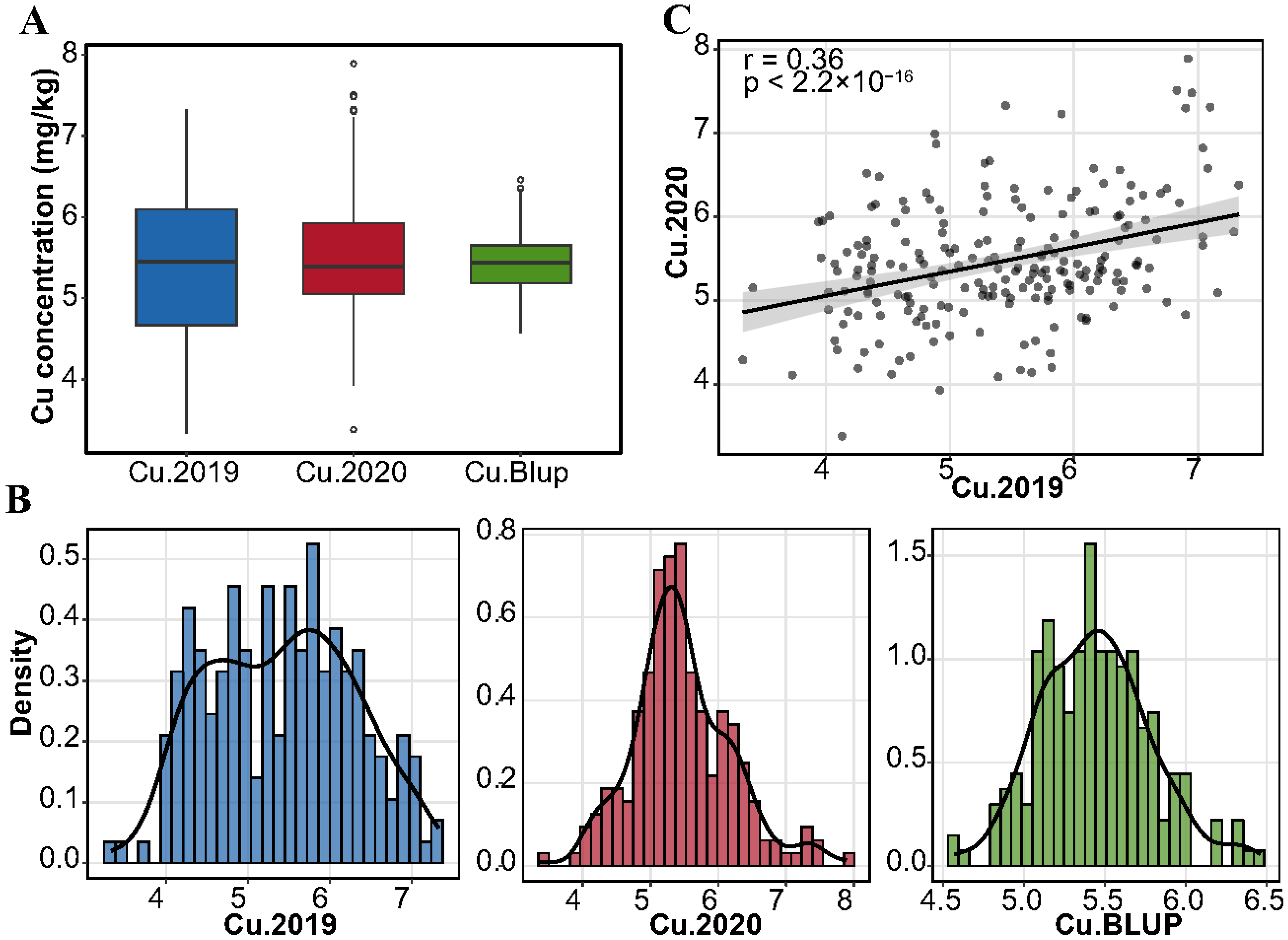
3.1. Variation Analysis of Copper Content in Mature Grains of Wheat
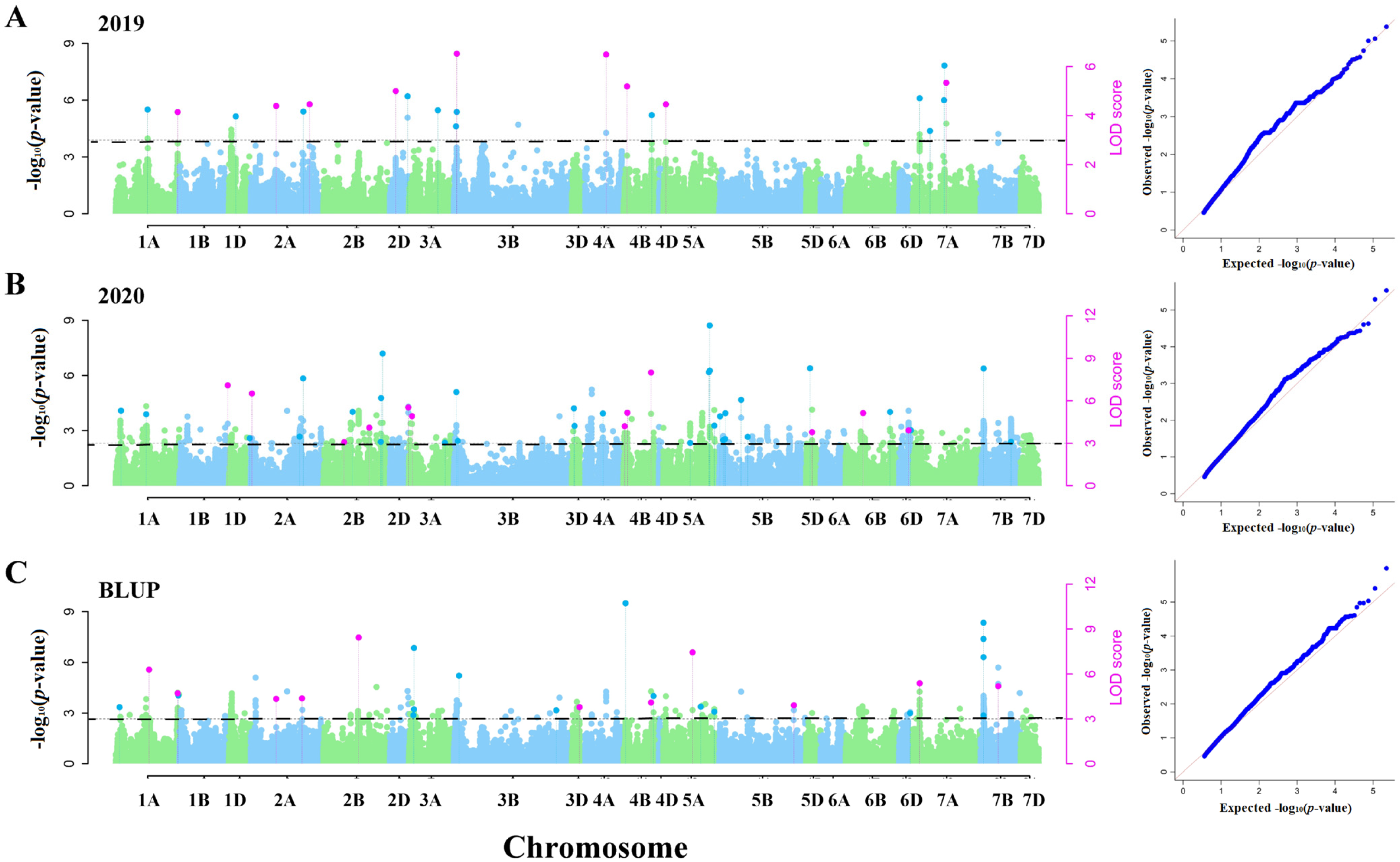
3.2. Genome-Wide Association Analysis Based on Multiple Models
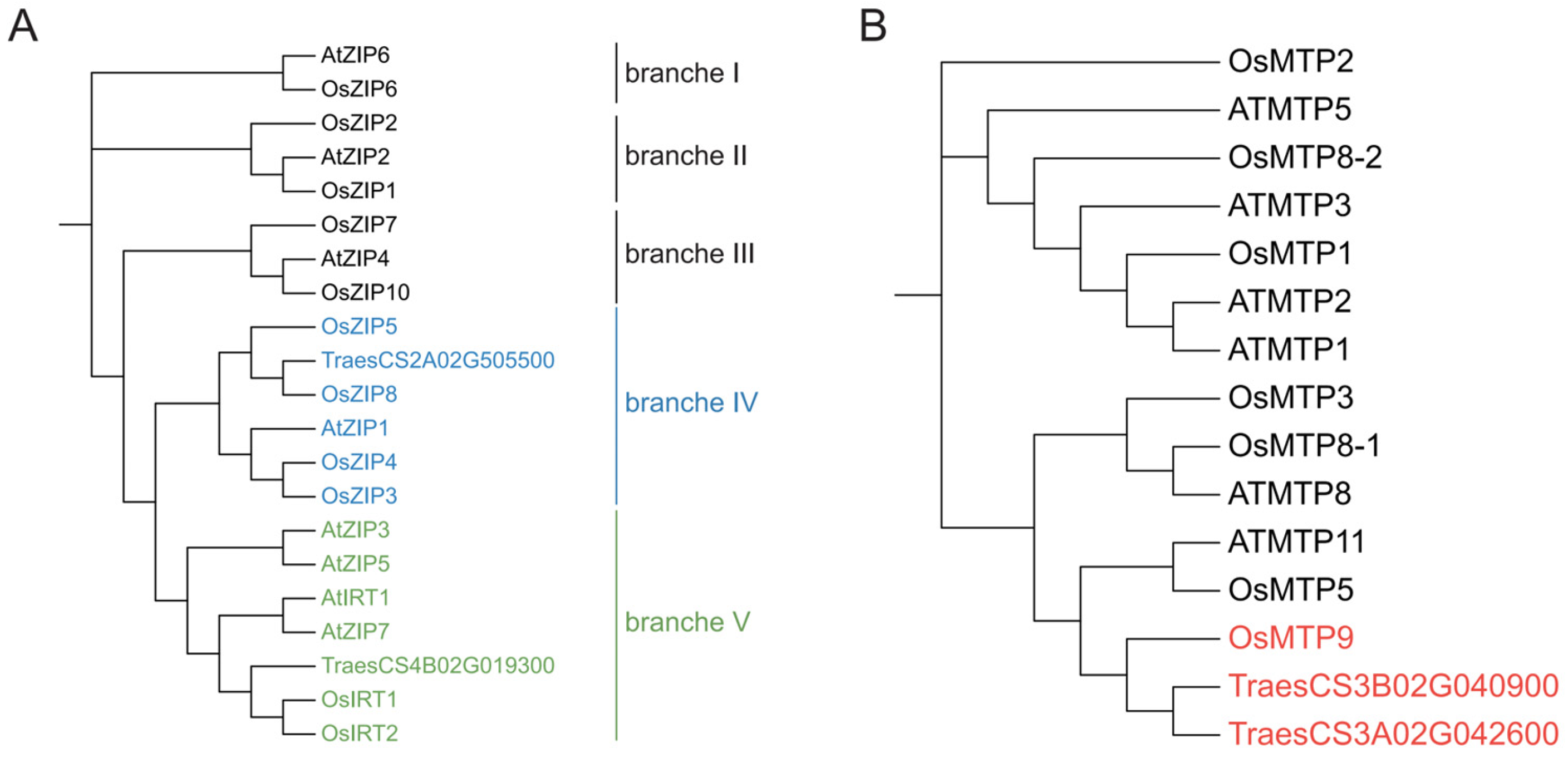
3.3. Candidate Gene Analysis
4. Discussion
4.1. Natural Variation of Copper Content in Wheat Grain
4.2. Advantages of Multi-Loci Model
4.3. Candidate Gene Prediction of Copper Content in Wheat Grain
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Poudel, R.; Bhatta, M. Review of Nutraceuticals and Functional Properties of Whole Wheat. J. Nutr. Food Sci. 2017, 7, 571. [Google Scholar] [CrossRef]
- Burkhead, J.L.; Gogolin Reynolds, K.A.; Abdel-Ghany, S.E.; Cohu, C.M.; Pilon, M. Copper Homeostasis. New Phytol. 2009, 182, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Ravet, K.; Pilon, M. Copper and Iron Homeostasis in Plants: The Challenges of Oxidative Stress. Antioxid. Redox Signal. 2013, 19, 919–932. [Google Scholar] [CrossRef]
- Binesh, A.; Venkatachalam, K. Copper in Human Health and Disease: A Comprehensive Review. J. Biochem. Mol. Tox 2024, 38, e70052. [Google Scholar] [CrossRef]
- Chhetri, S.K.; Mills, R.J.; Shaunak, S.; Emsley, H.C.A. Copper Deficiency. BMJ 2014, 348, g3691. [Google Scholar] [CrossRef]
- Cobbett, C.; Goldsbrough, P. Phytochelatins and metallothioneins: Roles in Heavy Metal Detoxification and Homeostasis. Annu. Rev. Plant Biol. 2002, 53, 159–182. [Google Scholar] [CrossRef]
- Andrés-Colás, N.; Sancenón, V.; Rodríguez-Navarro, S.; Mayo, S.; Thiele, D.J.; Ecker, J.R.; Puig, S.; Peñarrubia, L. The Arabidopsis Heavy Metal P-type ATPase HMA5 Interacts with Metallochaperones and Functions in Copper Detoxification of Roots. Plant J. 2006, 45, 225–236. [Google Scholar]
- Lu, K.; Li, L.; Zheng, X.; Zhang, Z.; Mou, T.; Hu, Z. Quantitative Trait Loci Controlling Cu, Ca, Zn, Mn and Fe Content in Rice Grains. J. Genet. 2008, 87, 305–310. [Google Scholar] [CrossRef]
- Hu, B.-L.; Huang, D.-R.; Xiao, Y.-Q.; Fan, Y.-Y.; Chen, D.-Z.; Zhuang, J.-Y. Mapping QTLs for Mineral Element Contents in Brown and Milled Rice Using an Oryza Sativa × O. Rufipogon Backcross Inbred Line Population. Cereal Res. Commun. 2016, 44, 57–68. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, J.; Jin, T.; Huang, Y.; Chen, J.; Zhu, L.; Zhao, Y.; Guo, J. Correction to: Identification of Quantitative Trait Locus and Prediction of Candidate Genes for Grain Mineral Concentration in Maize across Multiple Environments. Euphytica 2018, 214, 71. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Y.; Yang, Y.; Zhang, Q.; Fu, B.; Cai, J.; Guo, W.; Shi, L.; Wu, J.; Chen, Y. A Thorough Screening Based on QTLs Controlling Zinc and Copper Accumulation in the Grain of Different Wheat Genotypes. Environ. Sci. Pollut. Res. 2021, 28, 15043–15054. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Qi, S.; Yuan, M.; Zhao, D.; Zhang, D.; Feng, J.; Wang, J.; Li, W.; Song, C.; Wang, T.; et al. A Genome-Wide Association Study Revealed the Genetic Variation and Candidate Genes for Grain Copper Content in Bread Wheat (Triticum aestivum L.). Food Funct. 2022, 13, 5177–5188. [Google Scholar] [CrossRef] [PubMed]
- Bálint, A.F.; Röder, M.S.; Hell, R.; Galiba, G.; Börner, A. Mapping of QTLs Affecting Copper Tolerance and the Cu, Fe, Mn and Zn Contents in the Shoots of Wheat Seedlings. Biol. Plant 2007, 51, 129–134. [Google Scholar] [CrossRef]
- Zhao, L.; Pan, Y.; Dong, Z.; Zheng, Y.; Liu, J.; Geng, J.; Ren, Y.; Zhang, N.; Chen, F. Investigation and Genome-Wide Association Study of Grain Copper Content in Chinese Common Wheat. J. Cereal Sci. 2020, 95, 102991. [Google Scholar] [CrossRef]
- Zhou, G.; Zhu, Q.; Mao, Y.; Chen, G.; Xue, L.; Lu, H.; Shi, M.; Zhang, Z.; Song, X.; Zhang, H.; et al. Multi-Locus Genome-Wide Association Study and Genomic Selection of Kernel Moisture Content at the Harvest Stage in Maize. Front. Plant Sci. 2021, 12, 697688. [Google Scholar] [CrossRef]
- Zhong, H.; Liu, S.; Sun, T.; Kong, W.; Deng, X.; Peng, Z.; Li, Y. Multi-Locus Genome-Wide Association Studies for Five Yield-Related Traits in Rice. BMC Plant Biol. 2021, 21, 364. [Google Scholar] [CrossRef]
- Vikas, V.K.; Pradhan, A.K.; Budhlakoti, N.; Mishra, D.C.; Chandra, T.; Bhardwaj, S.C.; Kumar, S.; Sivasamy, M.; Jayaprakash, P.; Nisha, R.; et al. Multi-Locus Genome-Wide Association Studies (ML-GWAS) Reveal Novel Genomic Regions Associated with Seedling and Adult Plant Stage Leaf Rust Resistance in Bread Wheat (Triticum aestivum L.). Heredity 2022, 128, 434–449. [Google Scholar] [CrossRef]
- Peng, Y.; Liu, H.; Chen, J.; Shi, T.; Zhang, C.; Sun, D.; He, Z.; Hao, Y.; Chen, W. Genome-Wide Association Studies of Free Amino Acid Levels by Six Multi-Locus Models in Bread Wheat. Front. Plant Sci. 2018, 9, 1196. [Google Scholar] [CrossRef]
- Zhou, Z.; Guan, H.; Liu, C.; Zhang, Z.; Geng, S.; Qin, M.; Li, W.; Shi, X.; Dai, Z.; Lei, Z.; et al. Identification of Genomic Regions Affecting Grain Peroxidase Activity in Bread Wheat Using Genome-Wide Association Study. BMC Plant Biol. 2021, 21, 52. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using Lme4. J. Stat. Soft. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Tamba, C.; Zhang, Y.M. A fast mrMLM algorithm for multi-locus genome-wide association studies. bioRxiv 2018, 7, 341784. [Google Scholar]
- Tamba, C.L.; Ni, Y.L.; Zhang, Y.M. Iterative sure independence screening EM-Bayesian LASSO algorithm for multi-locus genome-wide association studies. PLoS Comput. Biol. 2017, 13, e1005357. [Google Scholar]
- Wang, S.B.; Feng, J.Y.; Ren, W.L.; Huang, B.; Zhou, L.; Wen, Y.J.; Jin, Z.; Dunwell, J.M.; Xu, S.; Zhang, Y.M. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci. Rep. 2016, 6, 19444. [Google Scholar]
- Ren, W.L.; Wen, Y.J.; Dunwell, J.M.; Zhang, Y.M. pKWmEB: Integration of kruskal-wallis test with empirical bayes under polygenic background control for multi-locus genome-wide association study. Heredity 2018, 120, 208–218. [Google Scholar]
- Zhang, J.; Feng, J.Y.; Ni, Y.L.; Wen, Y.J.; Niu, Y.; Tamba, C.L.; Yue, C.; Song, Q.; Zhang, Y.M. pLARmEB: Integration of least angle regression with empirical Bayes for multilocus genome-wide association studies. Heredity 2017, 118, 517–524. [Google Scholar]
- Alaux, M.; Rogers, J.; Letellier, T.; Flores, R.; Alfama, F.; Pommier, C.; Mohellibi, N.; Durand, S.; Kimmel, E.; Michotey, C.; et al. Linking the International Wheat Genome Sequencing Consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol. 2018, 19, 111. [Google Scholar] [CrossRef]
- Alaux, M.; Dyer, S.; Sen, T.Z. Wheat Data Integration and FAIRification: IWGSC, GrainGenes, Ensembl and Other Data Repositories. In The Wheat Genome. Compendium of Plant Genomes; Appels, R., Eversole, K., Feuillet, C., Gallagher, D., Eds.; Springer: Cham, Switzerland, 2024. [Google Scholar]
- Yuan, M.; Li, X.; Xiao, J.; Wang, S. Molecular and Functional Analyses of COPT/Ctr-Type Copper Transporter-like Gene Family in Rice. BMC Plant Biol. 2011, 11, 69. [Google Scholar] [CrossRef]
- Lee, S.; Kim, S.A.; Lee, J.; Guerinot, M.L.; An, G. Zinc Deficiency-Inducible OsZIP8 Encodes a Plasma Membrane-Localized Zinc Transporter in Rice. Mol. Cells 2010, 29, 551–558. [Google Scholar]
- Tan, L.; Qu, M.; Zhu, Y.; Peng, C.; Wang, J.; Gao, D.; Chen, C. ZINC TRANSPORTER5 and ZINC TRANSPORTER9 Function Synergistically in Zinc/Cadmium Uptake. Plant Physiol. 2020, 183, 1235–1249. [Google Scholar]
- Ishimaru, Y.; Suzuki, M.; Tsukamoto, T.; Suzuki, K.; Nakazono, M.; Kobayashi, T.; Wada, Y.; Watanabe, S.; Matsuhashi, S.; Takahashi, M.; et al. Rice Plants Take up Iron as an Fe3+ -phytosiderophore and as Fe2+. Plant J. 2006, 45, 335–346. [Google Scholar]
- Yu, E.; Yamaji, N.; Mao, C.; Wang, H.; Ma, J.F. Lateral Roots but Not Root Hairs Contribute to High Uptake of Manganese and Cadmium in Rice. J. Exp. Bot. 2021, 72, 7219–7228. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W. Systemic Impact of Trace Elements on Human Health and Diseases: Nutrition, Toxicity, and Beyond. J. Trace Elem. Med. Biol. 2020, 62, 126634. [Google Scholar]
- Li, J.; Martin, C.; Fernie, A. Biofortification’s Contribution to Mitigating Micronutrient Deficiencies. Nat. Food 2024, 5, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Hao, B.; Ma, J.; Chen, P.; Jiang, L.; Wang, X.; Li, C.; Wang, Z. Wheat Breeding in China over the Past 80 Years Has Increased Grain Zinc but Decreased Grain Iron Concentration. Field Crops Res. 2021, 271, 108253. [Google Scholar]
- Murphy, K.M.; Reeves, P.G.; Jones, S.S. Relationship between Yield and Mineral Nutrient Concentrations in Historical and Modern Spring Wheat Cultivars. Euphytica 2008, 163, 381–390. [Google Scholar] [CrossRef]
- Bhatta, M.; Baenziger, P.S.; Waters, B.M.; Poudel, R.; Belamkar, V.; Poland, J.; Morgounov, A. Genome-Wide Association Study Reveals Novel Genomic Regions Associated with 10 Grain Minerals in Synthetic Hexaploid Wheat. Int. J. Mol. Sci. 2018, 19, 3237. [Google Scholar] [CrossRef]
- Cu, S.T.; Guild, G.; Nicolson, A.; Velu, G.; Singh, R.; Stangoulis, J. Genetic Dissection of Zinc, Iron, Copper, Manganese and Phosphorus in Wheat (Triticum aestivum L.) Grain and Rachis at Two Developmental Stages. Plant Sci. 2020, 291, 110338. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, F.; Zhou, Y. The Application of Multi-Locus GWAS for the Detection of Salt-Tolerance Loci in Rice. Front. Plant Sci. 2018, 9, 1464. [Google Scholar] [CrossRef]
- Segura, V.; Vilhjálmsson, B.J.; Platt, A.; Korte, A.; Seren, Ü.; Long, Q.; Nordborg, M. An Efficient Multi-Locus Mixed-Model Approach for Genome-Wide Association Studies in Structured Populations. Nat. Genet. 2012, 44, 825–830. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Shakoor, N.; Ziegler, G.; Dilkes, B.P.; Brenton, Z.; Boyles, R.; Connolly, E.L.; Kresovich, S.; Baxter, I. Integration of Experiments across Diverse Environments Identifies the Genetic Determinants of Variation in Sorghum Bicolor Seed Element Composition. Plant Physiol. 2016, 170, 1989–1998. [Google Scholar] [PubMed]
- Pandey, A.; Khan, M.K.; Hakki, E.E.; Thomas, G.; Hamurcu, M.; Gezgin, S.; Gizlenci, O.; Akkaya, M.S. Assessment of Genetic Variability for Grain Nutrients from Diverse Regions: Potential for Wheat Improvement. SpringerPlus 2016, 5, 1912. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Yang, S.; Liu, B.; Zhang, M.; Wu, K. Molecular Characterization of a Rice Metal Tolerance Protein, OsMTP1. Plant Cell Rep. 2012, 31, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fujii, Y.; Yamaji, N.; Masuda, S.; Takemoto, Y.; Kamiya, T.; Yusuyin, Y.; Iwasaki, K.; Kato, S.; Maeshima, M.; et al. Mn Tolerance in Rice Is Mediated by MTP8.1, a Member of the Cation Diffusion Facilitator Family. J. Exp. Bot. 2013, 64, 4375–4387. [Google Scholar] [CrossRef]
| Program | Step 1 | Step 2 | Step 3 |
|---|---|---|---|
| Temperature/°C | RT *–120 | 120–180 | 180–RT * |
| climbing time/min | 12 | 10 | 15 |
| settling time/min | 5 | 35 | 10 |
| Traits | Environment * | Min | Max | Mean | Kurtosis | Skewness | CV (%) |
|---|---|---|---|---|---|---|---|
| Copper | E1 | 3.33 | 7.33 | 5.41 | −0.86 | 0.04 | 16.39 |
| E2 | 3.38 | 7.89 | 5.47 | 0.69 | 0.46 | 13.29 | |
| BLUP | 4.57 | 6.46 | 5.44 | 0.16 | 0.31 | 6.34 |
| QTN | Chromosome | Position (Mb) | −log10(p) | r2 (%) | Model | Environment * |
|---|---|---|---|---|---|---|
| AX-110905625 | 1A | 473.8 | 6.6–7.7 | 2.1–3.9 | FASTmrMLM, pLARmEB | BLUP |
| AX-111542470 | 1A | 580.1 | 4.4–6.8 | 2.3–4.7 | FASTmrMLM, pLARmEB, pKWmEB | E1, BLUP |
| AX-94651674 | 1B | 685.1 | 4.2–8.3 | 1.4–3.6 | FASTmrMLM, ISIS EM-BLASSO, pLARmEB, pKWmEB | E2 |
| AX-110129246 | 2A | 19.8 | 6.0–8.7 | 8.0–11.5 | FASTmrMLM, mrMLM | E2 |
| AX-110386266 | 2A | 226.5 | 4.9–6.2 | 6.2–14.8 | FASTmrMLM, mrMLM, pLARmEB, pKWmEB | E1, BLUP |
| AX-108874892 | 2A | 691.1 | 4.8–6.3 | 1.9–5.8 | FASTmrMLM, mrMLM, pLARmEB | BLUP |
| AX-109319457 | 2A | 729.3 | 5.2–5.3 | 3.9–4.7 | FASTmrMLM, pKWmEB | E1 |
| AX-110676161 | 2B | 172.8 | 3.8–3.8 | 0.5–1.6 | FASTmrMLM, pLARmEB | E2 |
| AX-110456252 | 2B | 449.1 | 3.8–10.1 | 3.0–6.2 | FASTmrMLM, pLARmEB, pKWmEB | BLUP |
| AX-110707064 | 2B | 603.8 | 3.8–5.7 | 0.3–2.2 | FASTmrMLM, ISIS EM-BLASSO, pLARmEB | E2 |
| AX-110192731 | 2D | 75.6 | 5.6–5.9 | 3.3–4.2 | FASTmrMLM, pKWmEB | E1 |
| AX-111211824 | 2D | 647.5 | 4.1–8.6 | 5.5–17.0 | FASTmrMLM, pKWmEB | E2 |
| AX-111231642 | 3A | 20.3 | 5.2–6.2 | 1.4–10.9 | mrMLM, pLARmEB | E2 |
| AX-110092804 | 3B | 18.8 | 4.9–7.4 | 2.2–4.9 | FASTmrMLM, pLARmEB, pKWmEB | E1 |
| AX-110816744 | 3D | 594.5 | 4.2–4.9 | 2.5–4.9 | FASTmrMLM, pLARmEB | BLUP |
| AX-110710058 | 4A | 606.6 | 3.8–7.6 | 1.2–7.3 | ISIS EM-BLASSO, pLARmEB, pKWmEB | E1 |
| AX-110630308 | 4B | 10.6 | 3.8–6.1 | 1.4–8.1 | FASTmrMLM, mrMLM | E2 |
| AX-110984751 | 4B | 32.3 | 5.2–6.8 | 5.9–6.1 | ISIS EM-BLASSO, pKWmEB | E1 |
| AX-109407721 | 4B | 38.8 | 5.7–6.3 | 1.1–3.3 | ISIS EM-BLASSO, pLARmEB | E2 |
| AX-108752003 | 4B | 610.2 | 4.0–9.5 | 2.9–7.8 | mrMLM, ISIS EM-BLASSO, pLARmEB | E2, BLUP |
| AX-108801851 | 5A | 17.4 | 3.9–6.5 | 3.4–5.2 | ISIS EM-BLASSO, pKWmEB | E1 |
| AX-110975044 | 5A | 512.6 | 6.2–11.1 | 4.0–11.3 | FASTmrMLM, pLARmEB, pKWmEB | BLUP |
| AX-111073739 | 5A | 681.5 | 4.2–5.0 | 1.2–3.7 | pLARmEB, pKWmEB | E2, BLUP |
| AX-111822227 | 5B | 634.1 | 4.5–4.8 | 1.2–1.8 | FASTmrMLM, pLARmEB | BLUP |
| AX-109737823 | 5D | 443.9 | 4.3–4.8 | 2.4–16.2 | pLARmEB, pKWmEB | E2 |
| AX-109330452 | 6B | 202.9 | 5.4–6.4 | 3.0–6.0 | FASTmrMLM, mrMLM | E2 |
| AX-110276099 | 6D | 463.9 | 4.6–4.9 | 0.9–3.1 | FASTmrMLM, pLARmEB, pKWmEB | E2 |
| AX-109925765 | 7A | 37.3 | 5.1–7.2 | 2.1–4.6 | FASTmrMLM, mrMLM, pLARmEB, pKWmEB | E1, BLUP |
| AX-111012263 | 7A | 261.7 | 4.9–6.4 | 5.2–9.7 | FASTmrMLM, pLARmEB, pKWmEB | E1 |
| AX-110506329 | 7B | 336.4 | 4.2–7.1 | 6.7–29.3 | mrMLM, ISIS EM-BLASSO, pLARmEB, pKWmEB | BLUP |
| QTN | Chromosome | Candidate Gene | Annotation | Distance to QTN (Mb) |
|---|---|---|---|---|
| AX-109319457 | 2A | TraesCS2A02G505500 | Zinc transporter | 4.50 |
| AX-110456252 | 2B | TraesCS2B02G313200 | Copper transporter family protein | −0.40 |
| AX-111231642 | 3A | TraesCS3A02G042600 | Metal tolerance protein | 2.49 |
| AX-110092804 | 3B | TraesCS3B02G040900 | Metal tolerance protein | 1.52 |
| AX-110630308 | 4B | TraesCS4B02G019300 | Zinc transporter | 3.38 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, Z.; Liu, X.; Li, F.; Hou, J.; Zhou, Z.; Jing, X.; Peng, Y.; Man, J.; Lei, Z. Genome-Wide Association Analysis and Candidate Gene Prediction of Wheat Grain Copper Concentration. Agronomy 2025, 15, 792. https://doi.org/10.3390/agronomy15040792
Zou Z, Liu X, Li F, Hou J, Zhou Z, Jing X, Peng Y, Man J, Lei Z. Genome-Wide Association Analysis and Candidate Gene Prediction of Wheat Grain Copper Concentration. Agronomy. 2025; 15(4):792. https://doi.org/10.3390/agronomy15040792
Chicago/Turabian StyleZou, Zhaojun, Xiaofei Liu, Fengfeng Li, Jinna Hou, Zhengfu Zhou, Xiaojing Jing, Yanchun Peng, Jianguo Man, and Zhensheng Lei. 2025. "Genome-Wide Association Analysis and Candidate Gene Prediction of Wheat Grain Copper Concentration" Agronomy 15, no. 4: 792. https://doi.org/10.3390/agronomy15040792
APA StyleZou, Z., Liu, X., Li, F., Hou, J., Zhou, Z., Jing, X., Peng, Y., Man, J., & Lei, Z. (2025). Genome-Wide Association Analysis and Candidate Gene Prediction of Wheat Grain Copper Concentration. Agronomy, 15(4), 792. https://doi.org/10.3390/agronomy15040792





