Identification of PIF Gene Family and Functional Study of PbPIF3a/PbPIF4 in Anthocyanin Biosynthesis of Pear
Abstract
1. Introduction
2. Materials and Methods
2.1. The Plant Materials
2.2. Identification of PbPIFs, Conservation Domain Check, Physicochemical Property Prediction
2.3. Chromosome Location, Phylogenetic, Motif, Gene Structure, Promoter Cis-Element Analysis
2.4. RNA Extraction and Quantitative Real-Time Fluorescent PCR (qRT-PCR)
2.5. Subcellular Localization
2.6. Transient Injection of Pear Fruits
2.7. Yeast One-Hybrid Assays
2.8. Dual-Luciferase Reporter Assays and Luminescence Detection
2.9. Determination of Anthocyanin Content
2.10. Statistical Analyses
2.11. Accession Numbers
3. Results
3.1. Identification of PbPIFs
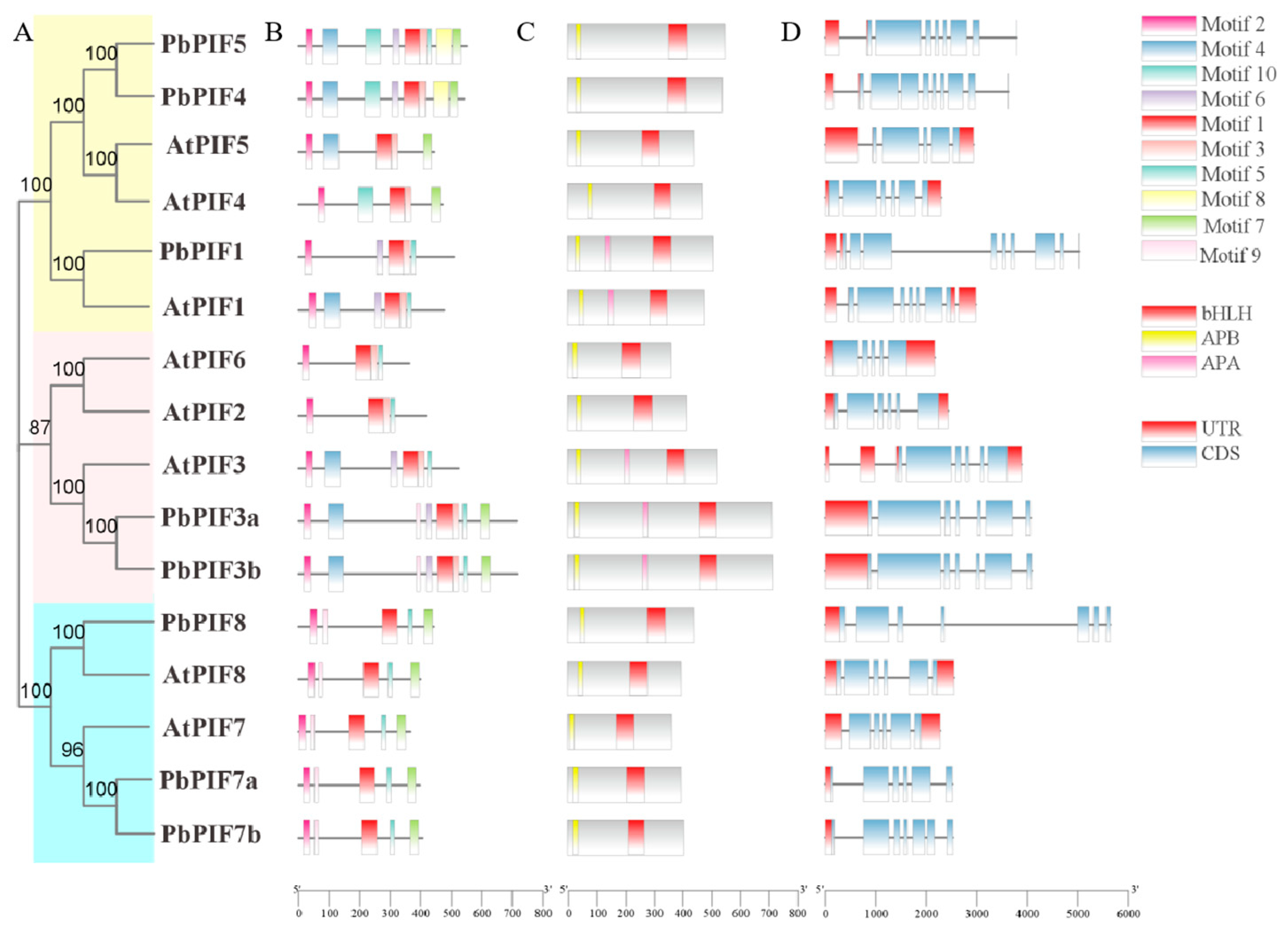
3.2. Chromosomal Location and Phylogenetic Analysis of PbPIFs
3.3. Motifs, Conserved Domains, and Gene Structures Analysis of PIFs
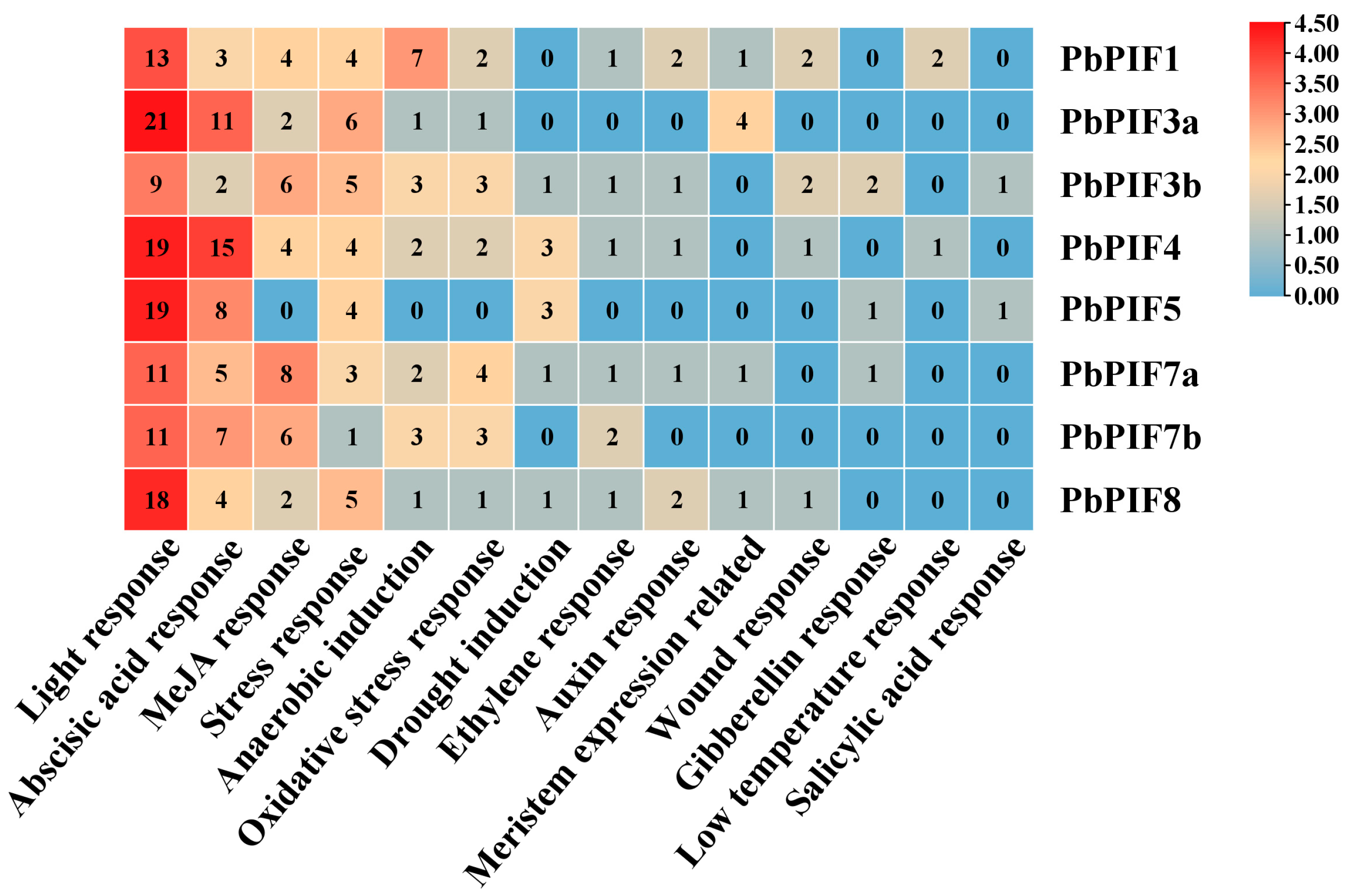
3.4. Analysis of Cis-Elements in the Promoter of PbPIFs
3.5. Subcellular Localization of PbPIFs
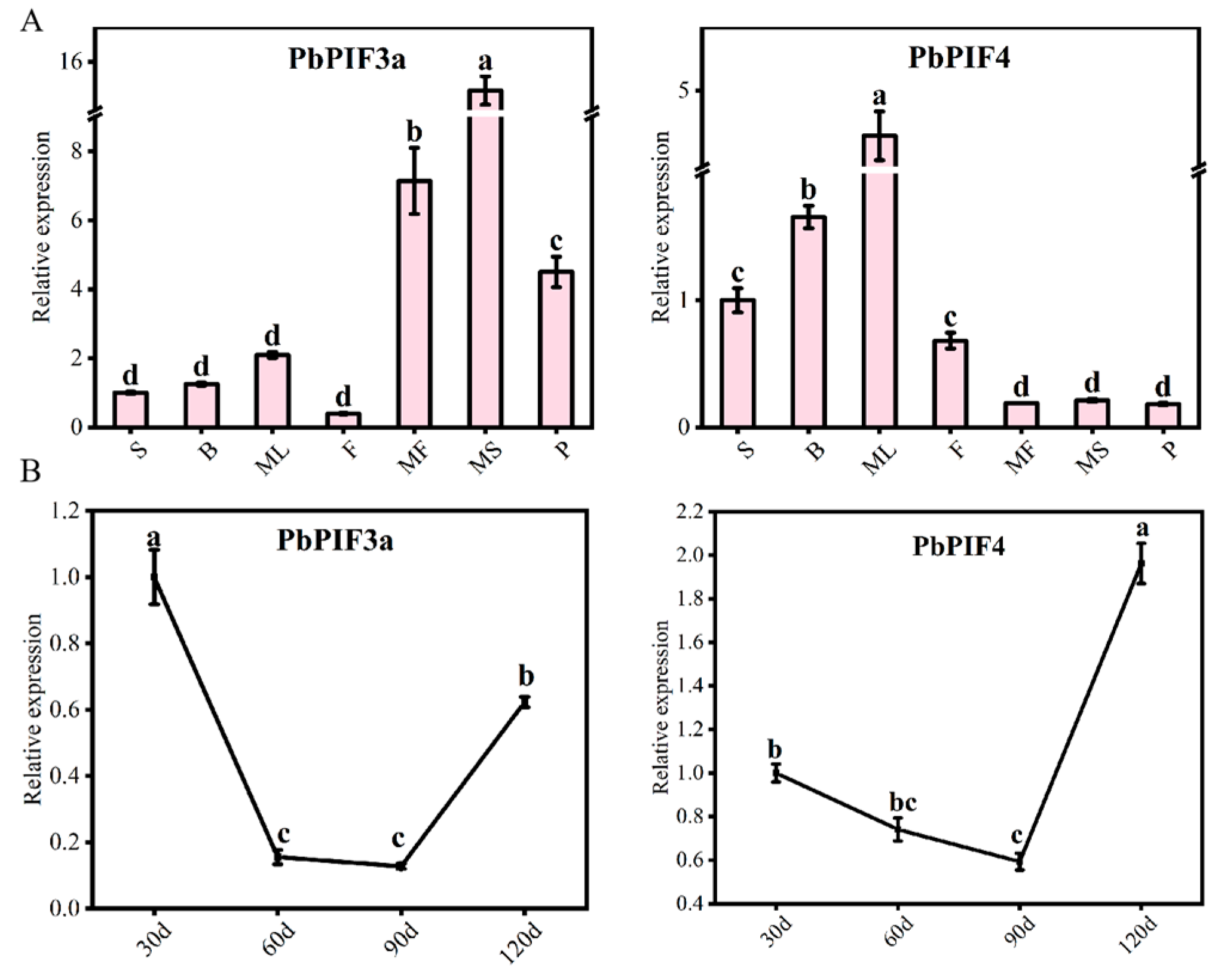
3.6. Expression Pattern Analysis of PbPIFs
3.7. Overexpression of PbPIFs Inhibits Anthocyanin Accumulation
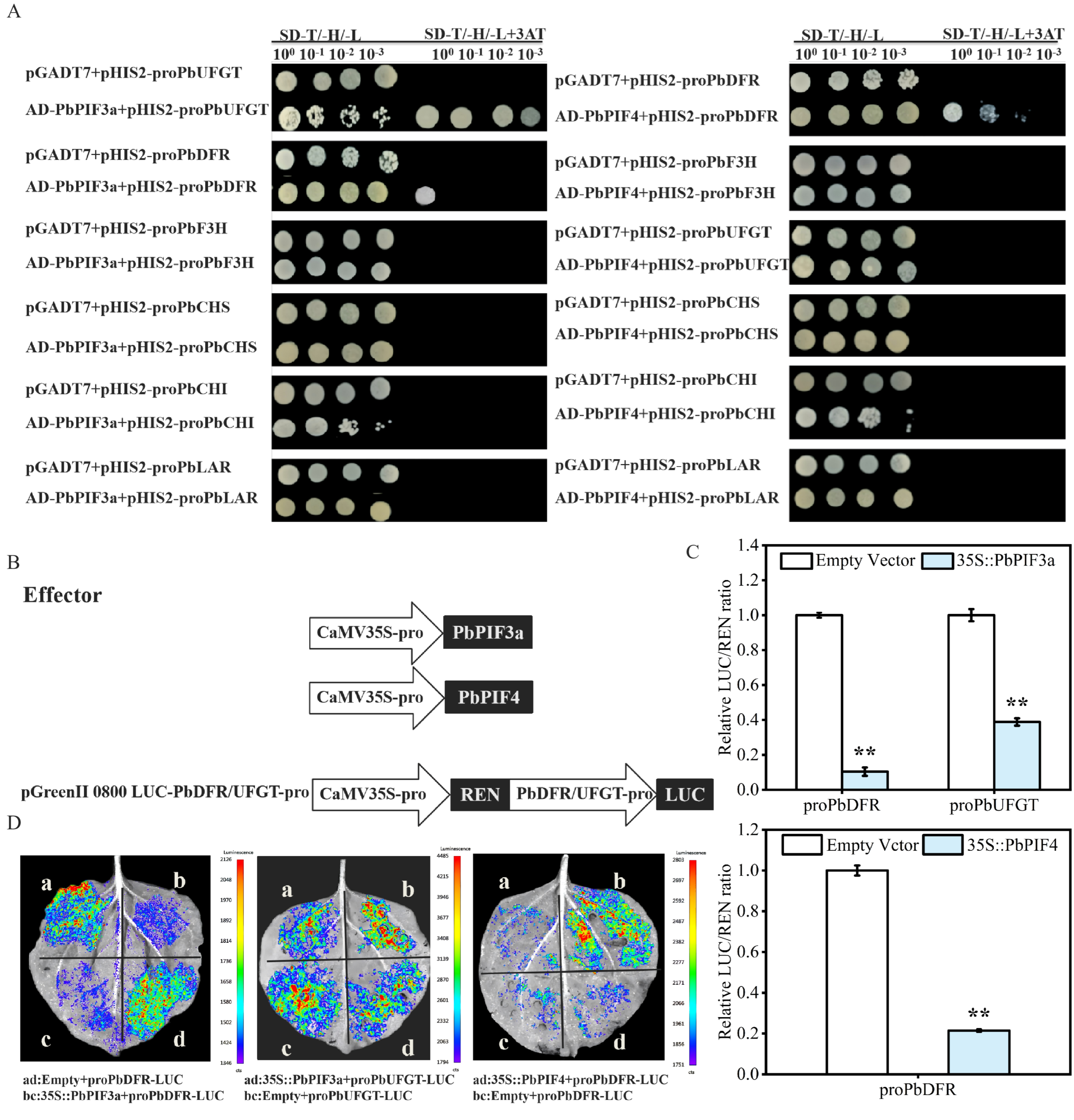
3.8. PbPIFs Inhibits the Activity of Anthocyanin Biosynthetic Genes
4. Discussion
4.1. Characteristics and Evolutionary Conservation of the PbPIF Family
4.2. A Multi-Level Regulatory Network Revealed by Promoter Cis-Elements
4.3. Functional Correlation of Subcellular Localization and Tissue-Specific Expression Patterns
4.4. Molecular Mechanism of PbPIFs Negatively Regulating Anthocyanin Biosynthesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, M.; Liu, J.; Song, L.; Li, X.; Cong, L.; Yue, R.; Yang, C.; Liu, Z.; Xu, L.; Wang, Z. Differences among the anthocyanin accumulation patterns and related gene expression levels in red pears. Plants 2019, 8, 100. [Google Scholar] [CrossRef] [PubMed]
- Havsteen, B.H. The biochemistry and medical significance of the flavonoids. Pharmacol. Ther. 2002, 96, 67–202. [Google Scholar]
- Pervaiz, T.; Songtao, J.; Faghihi, F.; Haider, M.S.; Fang, J. Naturally occurring anthocyanin, structure, functions and biosynthetic pathway in fruit plants. J. Plant Biochem. Physiol. 2017, 5, 1–9. [Google Scholar] [CrossRef]
- Willige, B.C.; Zander, M.; Yoo, C.Y.; Phan, A.; Garza, R.M.; Wanamaker, S.A.; He, Y.; Nery, J.R.; Chen, H.; Chen, M.; et al. Phytochrome-Interacting factors trigger environmentally responsive chromatin dynamics in plants. Nat. Genet. 2021, 53, 955–961. [Google Scholar] [CrossRef]
- Nakamura, Y.; Kato, T.; Yamashino, T.; Murakami, M.; Mizuno, T. Characterization of a set of phytochrome-interacting factor-like bHLH proteins in Oryza sativa. Biosci. Biotechnol. Biochem. 2007, 71, 1183–1191. [Google Scholar] [CrossRef]
- Wu, G.; Zhao, Y.; Shen, R.; Wang, B.; Xie, Y.; Ma, X.; Zheng, Z.; Wang, H. Characterization of maize phytochrome-interacting factors in light signaling and photomorphogenesis. Plant Physiol. 2019, 181, 789–803. [Google Scholar] [CrossRef]
- Han, Y.; Yang, J.; Zhang, N.; Gong, Y.; Liu, M.; Qiao, R.; Jiao, X.; Zhu, F.; Li, X.; Si, H. Genome-wide identification of phytochrome-interacting factor (PIF) gene family in potatoes and functional characterization of StPIF3 in regulating shade-avoidance syndrome. Agronomy 2024, 14, 873. [Google Scholar] [CrossRef]
- Rosado, D.; Gramegna, G.; Cruz, A.; Lira, B.S.; Freschi, L.; de Setta, N.; Rossi, M. Phytochrome interacting factors (PIFs) in Solanum lycopersicum: Diversity, evolutionary history and expression profiling during different developmental processes. PLoS ONE 2016, 11, e0165929. [Google Scholar] [CrossRef]
- Zheng, P.F.; Wang, X.; Yang, Y.Y.; You, C.X.; Zhang, Z.L.; Hao, Y.J. Identification of phytochrome-interacting factor family members and functional analysis of MdPIF4 in Malus domestica. Int. J. Mol. Sci. 2020, 21, 7350. [Google Scholar] [CrossRef]
- Khanna, R.; Huq, E.; Kikis, E.A.; Al-Sady, B.; Lanzatella, C.; Quail, P.H. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 2004, 16, 3033–3044. [Google Scholar] [CrossRef]
- Shen, H.; Zhu, L.; Castillon, A.; Majee, M.; Downie, B.; Huq, E. Light-induced phosphorylation and degradation of the negative regulator phytochrome-interacting factor1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 2008, 20, 1586–1602. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Park, E.; Choi, G. Pif3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 2007, 49, 981–994. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Y.; Wang, J.; Li, P.; Zhao, C.; Chen, Y.; Bi, Y. Phytochrome-interacting factors PIF4 and PIF5 negatively regulate anthocyanin biosynthesis under red light in Arabidopsis Seedlings. Plant Sci. 2015, 238, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, Y.; Fan, K.; Li, Z.; Jia, Q.; Lin, W.; Zhang, Y. Phytochrome-interacting factor 4 (PIF4) negatively regulates anthocyanin accumulation by inhibiting PAP1 transcription in Arabidopsis seedlings. Plant Sci. 2021, 303, 110788. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Yang, F.; Zhou, X.; Jia, W.; Zhu, X.; Mu, J.; Wang, Y.; Zhang, Y.; Mi, Z.; Zhang, S.; et al. Genome-wide identification of the bHLH gene family and the mechanism regulation of anthocyanin biosynthesis by ChEGL1 in Cerasus humilis. Int. J. Biol. Macromol. 2025, 288, 138783. [Google Scholar] [CrossRef]
- Li, P.; Chen, B.; Zhang, G.; Chen, L.; Dong, Q.; Wen, J.; Mysore, K.S.; Zhao, J. Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 2016, 210, 905–921. [Google Scholar] [CrossRef]
- Jiang, L.; Yue, M.; Liu, Y.; Zhang, N.; Lin, Y.; Zhang, Y.; Wang, Y.; Li, M.; Luo, Y.; Zhang, Y.; et al. A novel R2R3-MYB transcription factor FaMYB5 positively regulates anthocyanin and proanthocyanidin biosynthesis in cultivated strawberries (Fragaria × ananassa). Plant Biotechnol. J. 2023, 21, 1140–1158. [Google Scholar] [CrossRef]
- Martínez-Rivas, F.J.; Blanco-Portales, R.; Serratosa, M.P.; Ric-Varas, P.; Guerrero-Sánchez, V.; Medina-Puche, L.; Moyano, L.; Mercado, J.A.; Alseekh, S.; Caballero, J.L.; et al. FaMYB123 Interacts with FabHLH3 to regulate the late steps of anthocyanin and flavonol biosynthesis during ripening. Plant J. 2023, 114, 683–698. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the myb transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef]
- Li, C.; Wu, J.; Hu, K.D.; Wei, S.W.; Sun, H.Y.; Hu, L.Y.; Han, Z.; Yao, G.F.; Zhang, H. PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Hortic. Res. 2020, 7, 37. [Google Scholar] [CrossRef]
- Tao, R.; Yu, W.; Gao, Y.; Ni, J.; Yin, L.; Zhang, X.; Li, H.; Wang, D.; Bai, S.; Teng, Y. Light-induced basic/helix-loop-helix64 enhances anthocyanin biosynthesis and undergoes constitutively photomorphogenic1-mediated degradation in pear. Plant Physiol. 2020, 184, 1684–1701. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.J.; Wang, Y.; Wang, Y.; Song, A.; Jiang, J.; Chen, S.; Ding, B.; Guan, Z.; Chen, F. Transcription factor CmbHLH16 regulates petal anthocyanin homeostasis under different lights in Chrysanthemum. Plant Physiol. 2022, 190, 1134–1152. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.W.; Liu, X.; Zheng, P.F.; Su, L.; Wang, G.L.; Wang, X.F.; Li, Y.Y.; You, C.X.; An, J.P. Phytochrome interacting factor MdPIF7 modulates anthocyanin biosynthesis and hypocotyl growth in apple. Plant Physiol. 2022, 188, 2342–2363. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, Z.Y.; Zhang, F.J.; Zheng, P.F.; Ma, N.; Li, L.; Li, H.; Sun, P.; Zhang, S.; Wang, X.F.; et al. A viroid-derived small interfering rna targets bHLH transcription factor MdPIF1 to regulate anthocyanin biosynthesis in Malus domestica. Plant Cell Environ. 2024, 47, 4664–4682. [Google Scholar] [CrossRef]
- Liu, H.; Shu, Q.; Lin-Wang, K.; Allan, A.C.; Espley, R.V.; Su, J.; Pei, M.; Wu, J. The PyPIF5-PymiR156a-PySPL9-PyMYB114/MYB10 module regulates light-induced anthocyanin biosynthesis in red pear. Mol. Hortic. 2021, 1, 14. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Wei, C.; Cheng, Y.; Shang, Z.; Guo, X.; Guan, J. RNA-seq analysis identifies transcription factors involved in anthocyanin biosynthesis of ‘red zaosu’ pear peel and functional study of PpPIF8. Int. J. Mol. Sci. 2022, 23, 4798. [Google Scholar] [CrossRef]
- Sun, S.; Wu, P.; Gao, F.; Yu, X.; Liu, Y.; Zheng, C. Genome-wide identification and expression analysis of phytochrome-interacting factor genes during abiotic stress responses and secondary metabolism in the tea plant. Plant Physiol. Biochem. 2024, 215, 108988. [Google Scholar] [CrossRef]
- Zhang, X.; Xiong, L.; Luo, Y.; Wen, B.; Wang, K.; Liu, Z.; Huang, J.A.; Li, J. Identification, molecular characteristic, and expression analysis of PIFs related to chlorophyll metabolism in tea plant (Camellia sinensis). Int. J. Mol. Sci. 2021, 22, 10949. [Google Scholar] [CrossRef]
- Zhang, K.; Zheng, T.; Zhu, X.; Jiu, S.; Liu, Z.; Guan, L.; Jia, H.; Fang, J. Genome-wide identification of PIFs in grapes (Vitis vinifera L.) and their transcriptional analysis under lighting/shading conditions. Genes 2018, 9, 451. [Google Scholar] [CrossRef]
- Nie, N.; Huo, J.; Sun, S.; Zuo, Z.; Chen, Y.; Liu, Q.; He, S.; Gao, S.; Zhang, H.; Zhao, N.; et al. Genome-wide characterization of the PIFs family in sweet potato and functional identification of IbPIF3.1 under drought and Fusarium wilt stresses. Int. J. Mol. Sci. 2023, 24, 4092. [Google Scholar] [CrossRef]
- Liu, H.; Su, J.; Zhu, Y.; Yao, G.; Allan, A.C.; Ampomah-Dwamena, C.; Shu, Q.; Lin-Wang, K.; Zhang, S.; Wu, J. The involvement of PybZIPa in light-induced anthocyanin accumulation via the activation of PyUFGT through binding to tandem G-boxes in its promoter. Hortic. Res. 2019, 6, 134. [Google Scholar] [CrossRef]
- Li, Z.; Liu, W.; Chen, Q.; Zhang, S.; Mei, Z.; Yu, L.; Wang, C.; Mao, Z.; Chen, Z.; Chen, X.; et al. Mdm-miR858 targets MdMYB9 and MdMYBPA1 to participate anthocyanin biosynthesis in red-fleshed apple. Plant J. 2023, 113, 1295–1309. [Google Scholar] [CrossRef]
- Alabd, A.; Ahmad, M.; Zhang, X.; Gao, Y.; Peng, L.; Zhang, L.; Ni, J.; Bai, S.; Teng, Y. Light-responsive transcription factor PpWRKY44 induces anthocyanin accumulation by regulating PpMYB10 expression in pear. Hortic. Res. 2022, 9, uhac199. [Google Scholar] [CrossRef]
- Wang, P.; Abid, M.A.; Qanmber, G.; Askari, M.; Zhou, L.; Song, Y.; Liang, C.; Meng, Z.; Malik, W.; Wei, Y.; et al. Photomorphogenesis in plants: The central role of phytochrome interacting factors (PIFs). Environ. Exp. Bot. 2022, 194, 104704. [Google Scholar] [CrossRef]
- Ni, W.; Xu, S.L.; González-Grandío, E.; Chalkley, R.J.; Huhmer, A.F.R.; Burlingame, A.L.; Wang, Z.Y.; Quail, P.H. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 2017, 8, 15236. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Jia, M.; Wang, S.; Han, S.; Jiang, J. Identification and characterization of cotton phytochrome-interacting factors in temperature-dependent flowering. J. Exp. Bot. 2023, 74, 3765–3780. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Bo, K.; Pan, Y.; Li, Y.; Yu, D.; Li, C.; Chang, J.; Wu, S.; Wang, Z.; Zhang, X.; et al. Phytochrome-interacting factor PIF3 integrates phytochrome B and UV-B signaling pathways to regulate gibberellin- and auxin-dependent growth in cucumber hypocotyls. J. Exp. Bot. 2023, 74, 4520–4539. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, L.; Chen, Y.; Xiao, N.; Zhang, D.; Zhang, M.; Wang, W.; Zhang, C.; Zhang, A.; Li, H.; et al. Phytochrome interacting factor regulates stomatal aperture by coordinating red light and abscisic acid. Plant Cell 2022, 34, 4293–4312. [Google Scholar] [CrossRef]
- Wang, L.; Wu, X.; Xing, Q.; Zhao, Y.; Yu, B.; Ma, Y.; Wang, F.; Qi, H. PIF8-WRKY42-mediated salicylic acid synthesis modulates red light induced powdery mildew resistance in oriental melon. Plant Cell Environ. 2023, 46, 1726–1742. [Google Scholar] [CrossRef]
- Lin, L.; Liu, X.; Yin, R. PIF3 integrates light and low temperature signaling. Trends Plant Sci. 2018, 23, 93–95. [Google Scholar] [CrossRef]
- Ni, M.; Tepperman, J.M.; Quail, P.H. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 1998, 95, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Park, E.; Song, K.; Bae, G.; Choi, G. Phytochrome interacting factor 8 inhibits phytochrome A-mediated far-red light responses in Arabidopsis. Plant Cell 2020, 32, 186–205. [Google Scholar] [CrossRef] [PubMed]
- Bogs, J.; Jaffé, F.W.; Takos, A.M.; Walker, A.R.; Robinson, S.P. The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 2007, 143, 1347–1361. [Google Scholar] [CrossRef]
- Feng, S.; Sun, S.; Chen, X.; Wu, S.; Wang, D.; Chen, X. PyMYB10 and PyMYB10.1 interact with bHLH to enhance anthocyanin accumulation in pears. PLoS ONE 2015, 10, e0142112. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Gene ID | CDS Length (bp) | Amino Acids Length (aa) | Molecular Weight (Da) | Theoretical pI | Instability Index | Grand Average of Hydropathicity | Best Hits |
|---|---|---|---|---|---|---|---|---|
| PbPIF1 | gene2064 | 1552 | 508 | 55,214.18 | 9.39 | 68.07 | −0.784 | AtPIF1 |
| PbPIF3a | gene16013 | 2177 | 713 | 76,404.91 | 6.09 | 53.64 | −0.679 | AtPIF3 |
| PbPIF3b | gene15524 | 2186 | 716 | 76,555.15 | 6.12 | 55.57 | −0.614 | AtPIF3 |
| PbPIF4 | gene2521 | 1653 | 541 | 59,342.29 | 6.32 | 57.52 | −0.733 | AtPIF4 |
| PbPIF5 | gene8334 | 1680 | 550 | 60,367.33 | 6.47 | 54.5 | −0.722 | AtPIF5 |
| PbPIF7a | gene31629 | 1213 | 397 | 43,872.68 | 9.2 | 70.85 | −0.709 | AtPIF7 |
| PbPIF7b | gene3216 | 1238 | 406 | 44,850.85 | 9.02 | 64.49 | −0.671 | AtPIF7 |
| PbPIF8 | gene35851 | 1351 | 442 | 47,545.76 | 7.14 | 54.29 | −0.499 | AtPIF8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Lei, D.; Zhou, X.; Li, S.; Zhang, Y.; Lin, Y.; Chen, Q.; Luo, Y.; Tang, H.; Zhang, Y. Identification of PIF Gene Family and Functional Study of PbPIF3a/PbPIF4 in Anthocyanin Biosynthesis of Pear. Agronomy 2025, 15, 959. https://doi.org/10.3390/agronomy15040959
Wang H, Lei D, Zhou X, Li S, Zhang Y, Lin Y, Chen Q, Luo Y, Tang H, Zhang Y. Identification of PIF Gene Family and Functional Study of PbPIF3a/PbPIF4 in Anthocyanin Biosynthesis of Pear. Agronomy. 2025; 15(4):959. https://doi.org/10.3390/agronomy15040959
Chicago/Turabian StyleWang, Haiyan, Diya Lei, Xuan Zhou, Shangyun Li, Yunting Zhang, Yuanxiu Lin, Qing Chen, Ya Luo, Haoru Tang, and Yong Zhang. 2025. "Identification of PIF Gene Family and Functional Study of PbPIF3a/PbPIF4 in Anthocyanin Biosynthesis of Pear" Agronomy 15, no. 4: 959. https://doi.org/10.3390/agronomy15040959
APA StyleWang, H., Lei, D., Zhou, X., Li, S., Zhang, Y., Lin, Y., Chen, Q., Luo, Y., Tang, H., & Zhang, Y. (2025). Identification of PIF Gene Family and Functional Study of PbPIF3a/PbPIF4 in Anthocyanin Biosynthesis of Pear. Agronomy, 15(4), 959. https://doi.org/10.3390/agronomy15040959









