Minichromosome Maintenance Proteins: From DNA Replication to the DNA Damage Response
Abstract
1. Introduction
2. MCMs and the Pre-Replication Complex (Pre-RC) in DNA Replication
2.1. The MCM-Associated Proteins in DNA Replication
2.2. MCM2–7, MCM8/9 and MCM10 Function in DNA Replication
3. MCMs and DDR
3.1. MCM2–7 and DDR
3.1.1. MCM2 and MCM3 in the ATM/ATR Pathway
3.1.2. MCM4 and MCM6 in the ATM/ATR Pathway
3.1.3. MCM7 in the ATM/ATR Pathway
3.1.4. MCMs and the Fanconi Anemia Pathway and Homologous Recombination Repair
3.2. MCM8/9 and DDR
Auxiliary MCM8/MCM9 Helicases in DDR
3.3. MCM10 and DDR
4. Intersection of MCMs, R-Loop Resolution and DDR
5. Acetylation/Deacetylation of MCMs
6. Targeting MCMs in Cancer Therapeutics
7. Open-Ended Questions for the MCMs and DDR Relationship and How to Address These Knowledge Gaps
8. Conclusions and Perspective
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maine, G.T.; Sinha, P.; Tye, B.K. Mutants of S. Cerevisiae defective in the maintenance of minichromosomes. Genetics 1984, 106, 365–385. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Schwacha, A. The Mcm Complex: Unwinding the Mechanism of a Replicative Helicase. Microbiol. Mol. Biol. Rev. 2009, 73, 652–683. [Google Scholar] [CrossRef] [PubMed]
- Forsburg, S.L. Eukaryotic MCM Proteins: Beyond Replication Initiation. Microbiol. Mol. Biol. Rev. 2004, 68, 109–131. [Google Scholar] [CrossRef]
- Mei, L.; Cook, J.G. Efficiency and equity in origin licensing to ensure complete DNA replication. Biochem. Soc. Trans. 2021, 49, 2133–2141. [Google Scholar] [CrossRef] [PubMed]
- Coster, G.; Frigola, J.; Beuron, F.; Morris, E.P.; Diffley, J.F. Origin Licensing Requires ATP Binding and Hydrolysis by the MCM Replicative Helicase. Mol. Cell 2014, 55, 666–677. [Google Scholar] [CrossRef]
- Iyer, L.M.; Leipe, D.D.; Koonin, E.V.; Aravind, L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2003, 146, 11–31. [Google Scholar] [CrossRef]
- Zhang, S.; Mao, Y. AAA+ ATPases in Protein Degradation: Structures, Functions and Mechanisms. Biomolecules 2020, 10, 629. [Google Scholar] [CrossRef] [PubMed]
- Tye, B.K. MCM Proteins in DNA Replication. Annu. Rev. Biochem. 1999, 68, 649–686. [Google Scholar] [CrossRef] [PubMed]
- Forsburg, S.L. The MCM helicase: Linking checkpoints to the replication fork. Biochem. Soc. Trans. 2008, 36 Pt 1, 114–119. [Google Scholar] [CrossRef]
- Passmore, S.; Elble, R.; Tye, B.K. A protein involved in minichromosome maintenance in yeast binds a transcriptional enhancer conserved in eukaryotes. Genes Dev. 1989, 3, 921–935. [Google Scholar] [CrossRef]
- McInerny, C.J.; Patridge, J.F.; Mikesell, G.E.; Creemer, D.P.; Breeden, L.L. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6 and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 1997, 11, 1277–1288. [Google Scholar] [CrossRef] [PubMed]
- Griffin, W.C.; Trakselis, M.A. The MCM8/9 complex: A recent recruit to the roster of helicases involved in genome maintenance. DNA Repair 2019, 76, 1–10. [Google Scholar] [CrossRef]
- Nishitani, H.; Lygerou, Z. Control of DNA replication licensing in a cell cycle. Genes Cells 2002, 7, 523–534. [Google Scholar] [CrossRef]
- Ticau, S.; Friedman, L.J.; Ivica, N.A.; Gelles, J.; Bell, S.P. Single-Molecule Studies of Origin Licensing Reveal Mechanisms Ensuring Bidirectional Helicase Loading. Cell 2015, 161, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Bailis, J.M.; Luche, D.D.; Hunter, T.; Forsburg, S.L. Minichromosome Maintenance Proteins Interact with Checkpoint and Recombination Proteins To Promote S-Phase Genome Stability. Mol. Cell. Biol. 2008, 28, 1724–1738. [Google Scholar] [CrossRef] [PubMed]
- Giglia-Mari, G.; Zotter, A.; Vermeulen, W. DNA damage response. Cold Spring Harb. Perspect. Biol. 2011, 3, a000745. [Google Scholar] [CrossRef]
- Das, M.; Singh, S.; Pradhan, S.; Narayan, G. MCM Paradox: Abundance of Eukaryotic Replicative Helicases and Genomic Integrity. Mol. Biol. Int. 2014, 2014, 1–11. [Google Scholar] [CrossRef]
- Hyrien, O.; Marheineke, K.; Goldar, A. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. BioEssays 2003, 25, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, A.; Schwob, E.; Méndez, J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl. Acad. Sci. USA 2008, 105, 8956–8961. [Google Scholar] [CrossRef] [PubMed]
- Polasek-Sedlackova, H.; Miller, T.C.R.; Krejci, J.; Rask, M.-B.; Lukas, J. Solving the MCM paradox by visualizing the scaffold of CMG helicase at active replisomes. Nat. Commun. 2022, 13, 6090. [Google Scholar] [CrossRef] [PubMed]
- Laskey, R.A.; Madine, M.A. A rotary pumping model for helicase function of MCM proteins at a distance from replication forks. Embo Rep. 2003, 4, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Yankulov, K.; Todorov, I.; Romanowski, P.; Licatalosi, D.; Cilli, K.; McCracken, S.; Laskey, R.; Bentley, D.L. MCM Proteins Are Associated with RNA Polymerase II Holoenzyme. Mol. Cell. Biol. 1999, 19, 6154–6163. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bodmer-Glavas, M.; Edler, K.; Barberis, A. RNA polymerase II and III transcription factors can stimulate DNA replication by modifying origin chromatin structures. Nucleic Acids Res. 2001, 29, 4570–4580. [Google Scholar] [CrossRef]
- DaFonseca, C.J.; Shu, F.; Zhang, J.J. Identification of two residue in MCM5 critical for the assembly of MCM complexes and Stat1 mediated transcription activation in response to IFN gamma. Proc. Natl. Acad. Sci. USA 2001, 98, 3034–3039. [Google Scholar] [CrossRef]
- Fitch, M.J.; Donato, J.J.; Tye, B.K. Mcm7, a Subunit of the Presumptive MCM Helicase, Modulates Its Own Expression in Conjunction with Mcm1. J. Biol. Chem. 2003, 278, 25408–25416. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, L.; Dziak, R.; Kramer, D.J.H.; Leishman, D.; Song, X.; Ho, J.; Radovic, M.; Bentley, D.; Yankulov, K. The Role of the Carboxyterminal Domain of RNA Polymerase II in Regulating Origins of DNA Replication in Saccharomyces cerevisiae. Genetics 2002, 162, 1117–1129. [Google Scholar] [CrossRef]
- Iizuka, M.; Stillman, B. Histone Acetyltransferase HBO1 Interacts with the ORC1 Subunit of the Human Initiator Protein. J. Biol. Chem. 1999, 274, 23027–23034. [Google Scholar] [CrossRef] [PubMed]
- Burke, T.W.; Cook, J.G.; Asano, M.; Nevins, J.R. Replication Factors MCM2 and ORC1 Interact with the Histone Acetyltransferase HBO1. J. Biol. Chem. 2001, 276, 15397–15408. [Google Scholar] [CrossRef] [PubMed]
- Sterner, J.M.; Dew-Knight, S.; Musahl, C.; Kornbluth, S.; Horowitz, J.M. Negative Regulation of DNA Replication by the Retinoblastoma Protein Is Mediated by Its Association with MCM7. Mol. Cell. Biol. 1998, 18, 2748–2757. [Google Scholar] [CrossRef] [PubMed]
- Cortez, D.; Glick, G.; Elledge, S.J. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. USA 2004, 101, 10078–10083. [Google Scholar] [CrossRef]
- Drissi, R.; Chauvin, A.; McKenna, A.; Lévesque, D.; Blais-Brochu, S.; Jean, D.; Boisvert, F.-M. Destabilization of the MiniChromosome Maintenance (MCM) complex modulates the cellular response to DNA double strand breaks. Cell Cycle 2018, 17, 2593–2609. [Google Scholar] [CrossRef] [PubMed]
- Drissi, R.; Dubois, M.-L.; Douziech, M.; Boisvert, F.-M. Quantitative Proteomics Reveals Dynamic Interactions of the Minichromosome Maintenance Complex (MCM) in the Cellular Response to Etoposide Induced DNA Damage. Mol. Cell. Proteom. 2015, 14, 2002–2013. [Google Scholar] [CrossRef] [PubMed]
- Borlado, L.R.; Méndez, J. CDC6: From DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis 2008, 29, 237–243. [Google Scholar] [CrossRef]
- Lee, D.G.; Makhov, A.M.; Klemm, R.D.; Griffith, J.D.; Bell, S.P. Regulation of origin recognition complex conformation and ATPase activity: Differential effects of single-stranded and double-stranded DNA binding. EMBO J. 2000, 19, 4774–4782. [Google Scholar] [CrossRef]
- Duncker, B.P.; Chesnokov, I.N.; McConkey, B.J. The origin recognition complex protein family. Genome Biol. 2009, 10, 1–8. [Google Scholar] [CrossRef]
- Pozo, P.N.; Cook, J.G. Regulation and Function of Cdt1; A Key Factor in Cell Proliferation and Genome Stability. Genes 2016, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Ishimi, Y.; Komamura-Kohno, Y.; Karasawa-Shimizu, K.; Yamada, K. Levels of MCM4 phosphorylation and DNA synthesis in DNA replication block checkpoint control. J. Struct. Biol. 2004, 146, 234–241. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Ishimi, Y.; Masai, H.; Hanoka, F. Roles of MCM7 and MCM4 subunits in the DNA helicase activity of the mouse MCM4/6/7 complex. J. Biol. Chem. 2002, 277, 42471–42479. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Bielinsky, A.-K. Human Mcm10 Regulates the Catalytic Subunit of DNA Polymerase-α and Prevents DNA Damage during Replication. Mol. Biol. Cell 2007, 18, 4085–4095. [Google Scholar] [CrossRef]
- Ishimi, Y.; Komamura-Kohno, Y.; Arai, K.-I.; Masai, H. Biochemical Activities Associated with Mouse Mcm2 Protein. J. Biol. Chem. 2001, 276, 42744–42752. [Google Scholar] [CrossRef] [PubMed]
- Maiorano, D.; Cuvier, O.; Danis, E.; Méchali, M. MCM8 Is an MCM2-7-Related Protein that Functions as a DNA Helicase during Replication Elongation and Not Initiation. Cell 2005, 120, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Gambus, A.; Blow, J.J. Mcm8 and Mcm9 form a dimeric complex in Xenopus laevis egg extract that is not essential for DNA replication initiation. Cell Cycle 2013, 12, 1225–1232. [Google Scholar] [CrossRef]
- Wohlschlegel, J.A.; Dhar, S.K.; Prokhorova, T.A.; Dutta, A.; Walter, J.C. Xenopus Mcm10 Binds to Origins of DNA Replication after Mcm2-7 and Stimulates Origin Binding of Cdc45. Mol. Cell 2002, 9, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Homesley, L.; Lei, M.; Kawasaki, Y.; Sawyer, S.; Christensen, T.; Tye, B.K. Mcm10 and the MCM2–7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev. 2000, 14, 913–926. [Google Scholar] [CrossRef]
- Lõoke, M.; Maloney, M.F.; Bell, S.P. Mcm10 regulates DNA replication elongation by stimulating the CMG replicative helicase. Genes Dev. 2017, 31, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Langston, L.D.; Mayle, R.; Schauer, G.D.; Yurieva, O.; Zhang, D.; Yao, N.Y.; Georgescu, R.E.; O’Donnell, M.E.; States, U. Mcm10 promotes rapid isomerization of CMG-DNA for replisome bypass of lagging strand DNA blocks. eLife 2017, 6, e29118. [Google Scholar] [CrossRef]
- Cabello-Lobato, M.J.; González-Garrido, C.; Cano-Linares, M.I.; Wong, R.P.; Yáñez-Vílchez, A.; Morillo-Huesca, M.; Roldán-Romero, J.M.; Vicioso, M.; González-Prieto, R.; Ulrich, H.D.; et al. Physical interactions between MCM and Rad51 facilitate replication fork lesion bypass and ssDNA gap filling by non-recombinogenic functions. Cell Rep. 2021, 36, 109440. [Google Scholar] [CrossRef]
- Martínez, T.F.; Phillips, J.W.; Karanja, K.K.; Polaczek, P.; Wang, C.-M.; Li, B.C.; Campbell, J.L.; Dervan, P.B. Replication stress by Py–Im polyamides induces a non-canonical ATR-dependent checkpoint response. Nucleic Acids Res. 2014, 42, 11546–11559. [Google Scholar] [CrossRef] [PubMed]
- Charych, D.H.; Coyne, M.; Yabannavar, A.; Narberes, J.; Chow, S.; Wallroth, M.; Shafer, C.; Walter, A.O. Inhibition of Cdc7/Dbf4 kinase activity affects specific phosphorylation sites on MCM2 in cancer cells. J. Cell. Biochem. 2008, 104, 1075–1086. [Google Scholar] [CrossRef]
- Tanaka, T.; Nasmyth, K. Association of RPA with chromosomal replication origins requires MCM protein and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 1998, 17, 5182–5191. [Google Scholar] [CrossRef] [PubMed]
- Weinreich, M.; Stillman, B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 1999, 18, 5334–5346. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, K.; Morisaki, H.; Kaneko, Y.; Fujimoto, A.; Tanaka, T.; Ohtsubo, M.; Hirai, M.; Okayama, H.; Ikeda, K.; Nakanishi, M. Role of Human Cds1 (Chk2) Kinase in DNA Damage Checkpoint and Its Regulation by p53. J. Biol. Chem. 1999, 274, 31463–31467. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Pozo, F.M.; Wisotsky, J.N.; Wang, B.; Jacobberger, J.W.; Zhang, Y. Phosphorylation of Minichromosome Maintenance 3 (MCM3) by Checkpoint Kinase 1 (Chk1) Negatively Regulates DNA Replication and Checkpoint Activation. J. Biol. Chem. 2015, 290, 12370–12378. [Google Scholar] [CrossRef]
- Li, J.; Deng, M.; Wei, Q.; Liu, T.; Tong, X.; Ye, X. Phosphorylation of MCM3 Protein by Cyclin E/Cyclin-dependent Kinase 2 (Cdk2) Regulates Its Function in Cell Cycle. J. Biol. Chem. 2011, 286, 39776–39785. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.Y.; Shevchenko, A.; Shevchenko, A.; Dunphy, W.G. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. J Biol Chem. 2004, 279, 53353–53364. [Google Scholar] [CrossRef]
- Montagnoli, A.; Valsasina, B.; Brotherton, D.; Troiani, S.; Rainoldi, S.; Tenca, P.; Molinari, A.; Santocanale, C. Identification of Mcm2 Phosphorylation Sites by S-phase-regulating Kinases. J. Biol. Chem. 2006, 281, 10281–10290. [Google Scholar] [CrossRef] [PubMed]
- Stead, B.E.; Brandl, C.J.; Sandre, M.K.; Davey, M.J. Mcm2 phosphorylation and the response to replicative stress. BMC Genet. 2012, 13, 36. [Google Scholar] [CrossRef] [PubMed]
- Tsai, F.-L.; Vijayraghavan, S.; Prinz, J.; MacAlpine, H.K.; MacAlpine, D.M.; Schwacha, A. Mcm2-7 Is an Active Player in the DNA Replication Checkpoint Signaling Cascade via Proposed Modulation of Its DNA Gate. Mol. Cell. Biol. 2015, 35, 2131–2143. [Google Scholar] [CrossRef]
- Vijayraghavan, S.; Tsai, F.L.; Schwacha, A. A Checkpoint-Related Function of the MCM Replicative Helicase Is Required to Avert Accumulation of RNA:DNA Hybrids during S-phase and Ensuing DSBs during G2/M. PLoS Genet. 2016, 12, e1006277. [Google Scholar] [CrossRef] [PubMed]
- Fei, L.; Xu, H. Role of MCM2–7 protein phosphorylation in human cancer cells. Cell Biosci. 2018, 8, 43. [Google Scholar] [CrossRef]
- Sheu, Y.-J.; Kinney, J.B.; Lengronne, A.; Pasero, P.; Stillman, B. Domain within the helicase subunit Mcm4 integrates multiple kinase signals to control DNA replication initiation and fork progression. Proc. Natl. Acad. Sci. 2014, 111, E1899–E1908. [Google Scholar] [CrossRef]
- Wallace, M.D.; Southard, T.L.; Schimenti, K.J.; Schimenti, J.C. Role of DNA damage response pathways in preventing carcinogenesis caused by intrinsic replication stress. Oncogene 2013, 33, 3688–3695. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Komata, M.; Bando, M.; Araki, H.; Shirahige, K. The Direct Binding of Mrc1, a Checkpoint Mediator, to Mcm6, a Replication Helicase, Is Essential for the Replication Checkpoint against Methyl Methanesulfonate-Induced Stress. Mol. Cell. Biol. 2009, 29, 5008–5019. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, H.; Liu, W.; Yan, H.; Zhang, Y.; Cheung, A.H.K.; Zhang, J.; Chen, B.; Liang, L.; Zhou, Z.; et al. MCM6 is a critical transcriptional target of YAP to promote gastric tumorigenesis and serves as a therapeutic target. Theranostics 2022, 12, 6509–6526. [Google Scholar] [CrossRef]
- Wagner, S.A.; Oehler, H.; Voigt, A.; Dalic, D.; Freiwald, A.; Serve, H.; Beli, P. ATR inhibition rewires cellular signaling networks induced by replication stress. Proteomics 2015, 16, 402–416. [Google Scholar] [CrossRef] [PubMed]
- Yun, H.J.; Hyun, S.K.; Park, J.H.; Kim, B.W.; Kwon, H.J. Widdrol activates DNA damage checkpoint through the signaling Chk2-p53-Cdc25A-p21-MCM4 pathway in HT29 cells. Mol Cell Biochem. 2012, 363, 281–289. [Google Scholar] [CrossRef]
- Cortez, D.; Guntuku, S.; Qin, J.; Elledge, S.J. ATR and ATRIP: Partners in Checkpoint Signaling. Science 2001, 294, 1713–1716. [Google Scholar] [CrossRef] [PubMed]
- Tsao, C.-C.; Geisen, C.; Abraham, R.T. Interaction between human MCM7 and Rad17 proteins is required for replication checkpoint signaling. EMBO J. 2004, 23, 4660–4669. [Google Scholar] [CrossRef]
- Wei, Q.; Li, J.; Liu, T.; Tong, X.; Ye, X. Phosphorylation of Minichromosome Maintenance Protein 7 (MCM7) by Cyclin/Cyclin-dependent Kinase Affects Its Function in Cell Cycle Regulation. J. Biol. Chem. 2013, 288, 19715–19725. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, R.; Wu, J.; Dang, Z.; Zhang, Q.; Li, B. Knockdown of minichromosome maintenance proteins inhibits foci forming of mediator of DNA-damage checkpoint 1 in response to DNA damage in human esophageal squamous cell carcinoma TE-1 cells. Biochemistry 2016, 81, 1221–1228. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Jones, M.J.; Yin, Y.; Crist, S.B.; Colnaghi, L.; Sims, R.J.; Rothenberg, E.; Jallepalli, P.V.; Huang, T.T. ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress. Mol. Cell 2015, 58, 323–338. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Weng, C.; Zhang, H.; Sun, J.; Yuan, Y. A Direct Interaction Between P53-Binding Protein 1 and Minichromosome Maintenance Complex in Hepg2 Cells. Cell. Physiol. Biochem. 2018, 47, 2350–2359. [Google Scholar] [CrossRef] [PubMed]
- Lossaint, G.; Larroque, M.; Ribeyre, C.; Bec, N.; Larroque, C.; Décaillet, C.; Gari, K.; Constantinou, A. FANCD2 Binds MCM Proteins and Controls Replisome Function upon Activation of S Phase Checkpoint Signaling. Mol. Cell 2013, 51, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.; Navadgi, V.M.; Mallikarjuna, K.; Rao, B.J. Interaction of hRad51 and hRad52 with MCM complex: A cross-talk between recombination and replication proteins. Biochem. Biophys. Res. Commun. 2005, 329, 1240–1245. [Google Scholar] [CrossRef] [PubMed]
- Stead, B.E.; Brandl, C.J.; Davey, M.J. Phosphorylation of Mcm2 modulates Mcm2–7 activity and affects the cell’s response to DNA damage. Nucleic Acids Res. 2011, 39, 6998–7008. [Google Scholar] [CrossRef] [PubMed]
- McKinzey, D.R.; Gomathinayagam, S.; Griffin, W.C.; Klinzing, K.N.; Jeffries, E.P.; Rajkovic, A.; Trakselis, M.A. Motifs of the C-terminal domain of MCM9 direct localization to sites of mitomycin-C damage for RAD51 recruitment. J. Biol. Chem. 2021, 296, 100355. [Google Scholar] [CrossRef]
- Lutzmann, M.; Grey, C.; Traver, S.; Ganier, O.; Maya-Mendoza, A.; Ranisavljevic, N.; Bernex, F.; Nishiyama, A.; Montel, N.; Gavois, E.; et al. MCM8- and MCM9-Deficient Mice Reveal Gametogenesis Defects and Genome Instability Due to Impaired Homologous Recombination. Mol. Cell 2012, 47, 523–534. [Google Scholar] [CrossRef]
- Nishimura, K.; Ishiai, M.; Horikawa, K.; Fukagawa, T.; Takata, M.; Takisawa, H.; Kanemaki, M.T. Mcm8 and Mcm9 Form a Complex that Functions in Homologous Recombination Repair Induced by DNA Interstrand Crosslinks. Mol. Cell 2012, 47, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Im, J.-S.; Shibata, E.; Park, J.; Handa, N.; Kowalczykowski, S.C.; Dutta, A. MCM8-9 complex promotes resection of double-strand break ends by MRE11-RAD50-NBS1 complex. Nat. Commun. 2015, 6, 7744. [Google Scholar] [CrossRef] [PubMed]
- Hustedt, N.; Saito, Y.; Zimmermann, M.; Alvarez-Quilon, A.; Setiaputra, D.; Adam, S.; McEwan, A.; Yuan, J.Y.; Olivieri, M.; Zhao, Y.; et al. Control of homologous recombination by the HROB-MCM8-MCM9 pathway. Genes Dev. 2019, 33, 1397–1415. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.-W.; Acharya, A.; Taglialatela, A.; Nambiar, T.S.; Cuella-Martin, R.; Leuzzi, G.; Hayward, S.B.; Joseph, S.A.; Brunette, G.J.; Anand, R.; et al. MCM8IP activates the MCM8-9 helicase to promote DNA synthesis and homologous recombination upon DNA damage. Nat. Commun. 2020, 11, 2948. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Long, D.T.; Lee, K.Y.; Abbas, T.; Shibata, E.; Negishi, M.; Luo, Y.; Schimenti, J.C.; Gambus, A.; Walter, J.C.; et al. The MCM8-MCM9 Complex Promotes RAD51 Recruitment at DNA Damage Sites To Facilitate Homologous Recombination. Mol. Cell. Biol. 2013, 33, 1632–1644. [Google Scholar] [CrossRef] [PubMed]
- Traver, S.; Coulombe, P.; Peiffer, I.; Hutchins, J.R.; Kitzmann, M.; Latreille, D.; Mechali, M. MCM9 Is Required for Mammalian DNA Mismatch Repair. Mol Cell 2015, 59, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Wang, Y.; Zhu, Q.; Li, P.; Chen, J.; Tang, Z.; Shen, Y.; Cheng, X.; Lu, L.-Y.; Liu, Y. Aberrantly expressed HORMAD1 disrupts nuclear localization of MCM8–MCM9 complex and compromises DNA mismatch repair in cancer cells. Cell Death Dis. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Griffin, W.C.; McKinzey, D.R.; Klinzing, K.N.; Baratam, R.; Eliyapura, A.; Trakselis, M.A. A multi-functional role for the MCM8/9 helicase complex in maintaining fork integrity during replication stress. Nat Commun. 2022, 13, 5090. [Google Scholar] [CrossRef]
- Weng, Z.; Zheng, J.; Zhou, Y.; Lu, Z.; Wu, Y.; Xu, D.; Li, H.; Liang, H.; Liu, Y. Structural and mechanistic insights into the MCM8/9 helicase complex. eLife 2023, 12, RP87468. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Bang, S.W.; Kim, S.H.; Hwang, D.S. Knockdown of human MCM10 activates G2 checkpoint pathway. Biochem. Biophys. Res. Commun. 2007, 365, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kaur, M.; Kar, A.; Ranade, S.M.; Saxena, S. Ultraviolet Radiation Stress Triggers the Down-regulation of Essential Replication Factor Mcm10. J. Biol. Chem. 2010, 285, 8352–8362. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Fu, P.; Alcivar, A.L.; Fu, H.; Redon, C.; Foo, T.K.; Zuo, Y.; Ye, C.; Baxley, R.; Madireddy, A.; et al. BRCA2 associates with MCM10 to suppress PRIMPOL-mediated repriming and single-stranded gap formation after DNA damage. Nat. Commun. 2021, 12, 5966. [Google Scholar] [CrossRef] [PubMed]
- Sollier, J.; Cimprich, K.A. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 2015, 25, 514–522. [Google Scholar] [CrossRef] [PubMed]
- García-Muse, T.; Aguilera, A. Transcription–replication conflicts: How they occur and how they are resolved. Nat. Rev. Mol. Cell Biol. 2016, 17, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Kumar, C.; Remus, D. Looping out of control: R-loops in transcription-replication conflict. Chromosoma 2023, 133, 37–56. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.T.; Coulson, R.L.; LaSalle, J.M. R-loop formation at immunoglobulin class switch regions is regulated by the MCM helicase complex. Nat. Commun. 2013, 4, 1334. [Google Scholar] [CrossRef]
- Cortez, D. Preventing replication fork collapse to maintain genome integrity. DNA Repair 2015, 32, 149–157. [Google Scholar] [CrossRef]
- Fatoba, S.T.; Tognetti, S.; Berto, M.; Leo, E.; Mulvey, C.M.; Godovac-Zimmermann, J.; Pommier, Y.; Okorokov, A.L. Human SIRT1 regulates DNA binding and stability of the Mcm10 DNA replication factor via deacetylation. Nucleic Acids Res. 2013, 41, 4065–4079. [Google Scholar] [CrossRef]
- Takei, Y.; Swietlik, M.; Tanoue, A.; Tsujimoto, G.; Kouzarides, T.; Laskey, R. MCM3AP, a novel acetyltransferase that acetylates replication protein MCM3. Embo Rep. 2001, 2, 119–123. [Google Scholar] [CrossRef]
- Li, Z.; Xu, X. Post-Translational Modifications of the Mini-Chromosome Maintenance Proteins in DNA Replication. Genes 2019, 10, 331. [Google Scholar] [CrossRef]
- Giaginis, C.; Vgenopoulou, S.; Vielh, P.; Theocharis, S. MCM proteins as diagnostic and prognostic tumor markers in the clinical setting. Histol. Histopathol. 2010, 25, 351–370. [Google Scholar] [CrossRef]
- Wang, L.; Liu, X. Pan-Cancer Multi-Omics Analysis of Minichromosome Maintenance Proteins (MCMs) Expression in Human Cancers. Front. Biosci. 2023, 28, 230. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.S.; Kang, Y.H. The Human Replicative Helicase, the CMG Complex, as a Target for Anti-cancer Therapy. Front. Mol. Biosci. 2018, 5, 26. [Google Scholar] [CrossRef]
- Majid, S.; Dar, A.A.; Saini, S.; Chen, Y.; Shahryari, V.; Liu, J.; Zaman, M.S.; Hirata, H.; Yamamura, S.; Ueno, K.; et al. Regulation of Minichromosome Maintenance Gene Family by MicroRNA-1296 and Genistein in Prostate Cancer. Cancer Res. 2010, 70, 2809–2818. [Google Scholar] [CrossRef]
- Yadav, A.K.; Polasek-Sedlackova, H. Quantity and quality of minichromosome maintenance protein complexes couple replication licensing to genome integrity. Commun. Biol. 2024, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Liu, X.; Yang, C.; Wang, X.; Yu, T.; Han, C.; Huang, K.; Zhu, G.; Su, H.; Qin, W.; et al. Distinct Diagnostic and Prognostic Values of Minichromosome Maintenance Gene Expression in Patients with Hepatocellular Carcinoma. J. Cancer 2018, 9, 2357–2373. [Google Scholar] [CrossRef]
- Yu, S.; Wang, G.; Shi, Y.; Xu, H.; Zheng, Y.; Chen, Y. MCMs in Cancer: Prognostic Potential and Mechanisms. Anal. Cell. Pathol. 2020, 2020, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, H.; Zhang, J.; Cheng, A.S.; Yu, J.; To, K.F.; Kang, W. MCM family in gastrointestinal cancer and other malignancies: From functional characterization to clinical implication. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2020, 1874, 188415. [Google Scholar] [CrossRef]
- Winther, T.L.; Torp, S.H. MCM7 expression is a promising predictor of recurrence in patients surgically resected for meningiomas. J. Neuro-Oncol. 2016, 131, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Mughal, M.J.; Chan, K.I.; Mahadevappa, R.; Wong, S.W.; Wai, K.C.; Kwok, H.F. Over-Activation of Minichromosome Maintenance Protein 10 Promotes Genomic Instability in Early Stages of Breast Cancer. Int. J. Biol. Sci. 2022, 18, 3827–3844. [Google Scholar] [CrossRef]


| MCMs | Notably Effects in DDR | Model | Reference |
|---|---|---|---|
| MCM2 | MCM2 mutant was sensitive to MMS and caffeine. MCM2 regulates DNA replication in response to DNA damage | Saccharomyces cerevisiae | [57] |
| MCM2 | DNA damage requires MCM2DENQ mutants to progress through G2/M | Saccharomyces cerevisiae | [59] |
| MCM2 | DDK phosphorylates MCM2 to respond to replicative stress but not to induce checkpoint | Saccharomyces pombe | [75] |
| MCM2 | ATM and ATR phosphorylate MCM2 on Serine 92 | Xenopus laevis | [55] |
| MCM2 | ATR phosphorylates MCM2 on Serine 108 | HeLa and human dermal fibroblasts | [56] |
| MCM2 | ATR phosphorylates MCM2 in response to pyrrole–imidazole polyamides | LNCaP, LNAR and DU145 | [48] |
| MCM2 | Cdc7/Dbf4 mediates phosphorylation on Serine 108 and Serine 40 of human MCM2 in the absence of DNA damage | A549 and HCT116 | [49] |
| MCM3 | CyclinE/Cdk2 phosphorylate MCM3 Threonine 722 | HEK293-T and HeLa | [54] |
| MCM3 | Chk1 phosphorylates MCM3 on Serine 205 | HEK293-T, HeLa, U2OS, A549 | [53] |
| MCM4 | MCM4 interacts with Cds1 in response to HU MCM4 interacts with overexpressed Rad22 in response to HU MCM4 interacts with Rhp51 in response to HU | Saccharomyces pombe | [15] |
| MCM4 | Early treatment of Widdrol leads to MCM4 downregulation only | HT-29 and SC-1 | [66] |
| MCM4 | Double mutant MCM4 and ATM led to decreased tumor latency and increased tumor susceptibility | Murine models | [62] |
| MCM4 | N-terminal serine/threonine domain (NSD) on MCM4 mutants have elevated levels of Rad53 and increased γH2AX Deletion of NSD leads to weakened checkpoint response | Saccharomyces cerevisiae | [61] |
| MCM6 | Mrc1 interacts with MCM6 and acts as checkpoint sensor for methanesulfonate (MMS)-induced DNA damage | Saccharomyces cerevisiae | [63] |
| MCM6 | ATR phosphorylates MCM6 on Serine 13 | U2OS | [65] |
| MCM7 | Rad17 interacts with MCM7 | U2OS and A549 | [68] |
| MCM7 | MCM7 is phosphorylated on Serine 121 by cyclin E/Cdk2 and cyclin B/Cdk1 MCM7 overexpression leads to S phase block Phosphorylation of MCM7 is necessary for proper mitotic exit | HEK293-T, HeLa and HCT116 | [69] |
| MCM2 MCM3 | MCM2 and MCM3 interact with Rad51 and Rad52. MCM2/3 and /Rad51 promotes non-HR repair | Saccharomyces cerevisiae | [47] |
| MCM2 MCM3 MCM7 | ATR phosphorylates MCM2 on Serine 108 in response to ionizing radiation (IR), UV and Hydroxyurea (HU) ATM phosphorylates MCM3 on Serine 535 in response to IR ATRIP interacts with MCM7 | HEK293, U2OS, HeLa | [30] |
| MCM2 MCM3 | MCM3 siRNA led to G2 block and Chk1 and Chk2 activation MCM2 siRNA led to the activation of Chk2 | HeLa | [19] |
| MCM2 MCM3 | MCM2 and MCM3 knockdown causes reduction in Chk1 and Chk2 signaling in response to etoposide MCM2 and MCM3 knockdown causes reduction in HRR and NHEJ | HEK293 and U2OS | [31] |
| MCM2 MCM3 | MCM3 interacts with Rad51 and Rad52 MCM2 interacts with Rad52 | HeLa | [74] |
| MCM2 MCM5 | MCM2 interacts with ASF1 in response to etoposide | U2OS | [32] |
| MCM2/3/5/6 | 53BP1 interacts with MCM 2/3/5/6. Knockdown of MCM2/6 reduces 53BP1 interaction with chromatin and reduces 53BP1 foci formation | HepG2 | [72] |
| MCM2/3/MCM5/6 | MDC1 interacts with MCM2/3/5/6. MDC1 interacts with MCM2/6 on chromatin | TE-1 | [70] |
| MCM8 MCM9 | Null MCM8 mice have blocked double-strand break repair. MCM8 and MCM9 form a complex and control HR | Murine modes and MEF | [77] |
| MCM8 MCM9 | MCM 8 and MCM9 form a complex and are involved in HR MCM8 and MCM9 are involved in ICL repair and functions downstream of BRCA2/Rad51-Fanconi anemia pathways MCM8 and MCM9 are resistant to DNA interstrand crosslinks (ICL) | Chicken DT40 | [78] |
| MCM8 MCM9 | MCM8/9 are necessary for MRN localization to HR sites MCM8/9 are necessary for ssDNA generation for HR | U2OS, HeLa and HEK293-T | [79] |
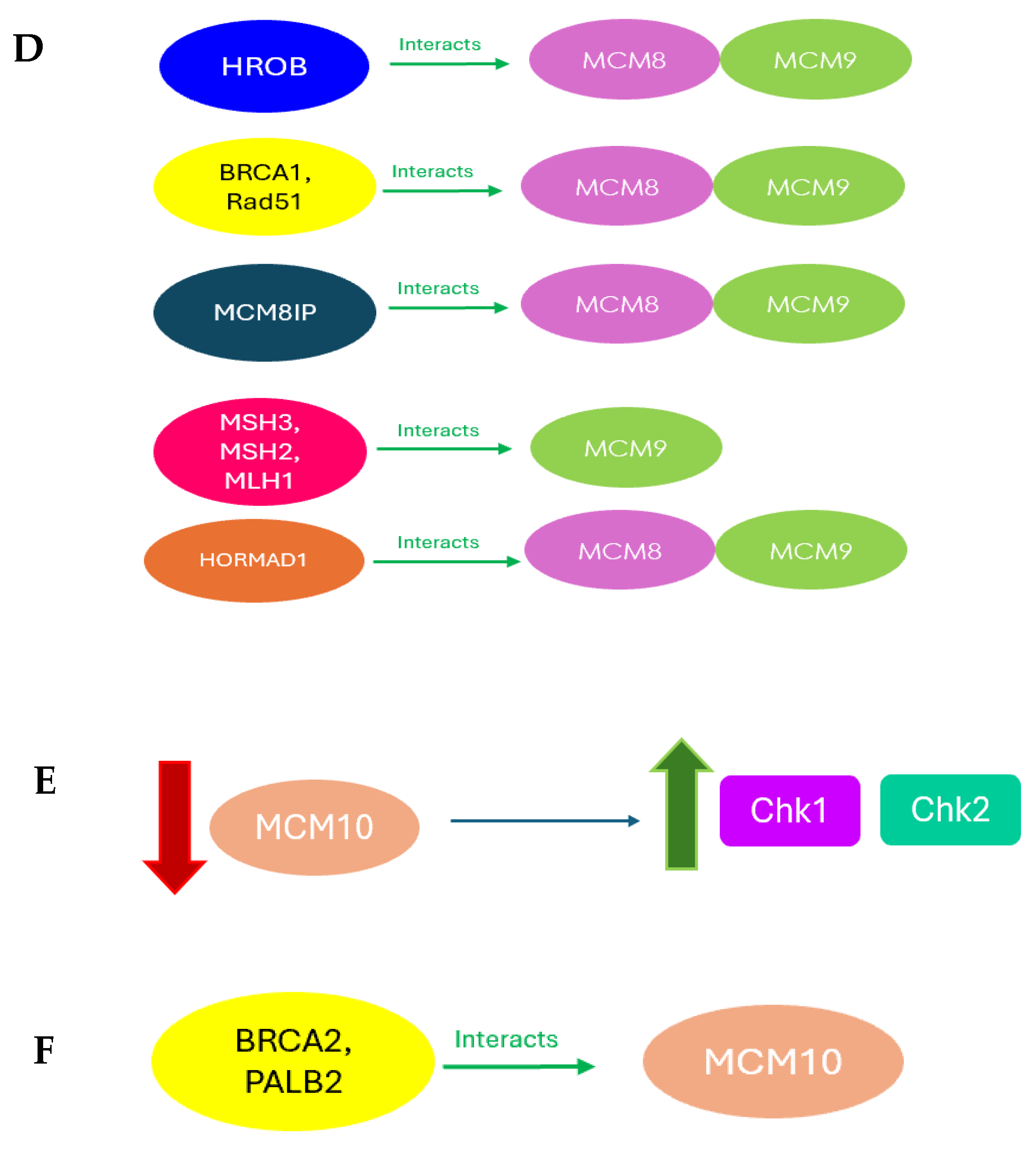
| MCM8 MCM9 | HROB recruits MCM8/9 to DNA damage sites | RPE1-hTERT, HCT116, HEK293-T, murine model | [80] |
| MCM8 MCM9 | MCM8IP interacts with MCM8/9 MCM8IP stimulates the helicase activity of MCM8/9 | HEK293T, HEK293T TREx, U2OS | [81] |
| MCM8 MCM9 | MCM8/9 directs BRCA1 and Rad51 to protect forks from excessive degradation | HEK293 | [85] |
| MCM8 MCM9 | NLS motif is required for MCM8 and MCM9 nuclear localization BRC motif found in MCM9 directly interacts with and recruits Rad51 in response to mitomycin C DNA damage | HEK293-T and U2OS | [76] |
| MCM8 MCM9 | Activated by HROB, the structure of MCM8/9 permits its unwinding ability | HeLa and Chicken DT40 | [86] |
| MCM8 MCM9 | HORMAD1 interacts with MCM8/MCM9 HORMAD1 disrupts MMR through its interaction with MCM8/9 | HEK293-T and U2OS | [84] |
| MCM9 | MCM9 interacts with mismatched repair (MMR) initiator proteins MCM9 is necessary for MMR and has helicase activity MCM9 helicase activity is necessary for MMR | HeLa | [83] |
| MCM10 | Knockdown MCM10 causes an increase in Chk1 and Chk2 expression Prolonged knockdown of MCM10 causes DNA breaks | HeLa, S7, and YZ5 | [87] |
| MCM10 | MCM10 and DNA polymerase alpha interact Knockdown of MCM10 leads to Chk2 activation | HeLa | [39] |
| MCM10 | MCM10 is downregulated in response to UV MCM10 overexpression helps cells recover from UV | U2OS and HeLa | [88] |
| MCM10 | MCM10 interacts with BRCA2 and PALB2 MCM10-BRCA2 prevents fork progression in response to IR or bleomycin | CHO, VC8 and HEK293-T | [89] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malysa, A.; Zhang, X.M.; Bepler, G. Minichromosome Maintenance Proteins: From DNA Replication to the DNA Damage Response. Cells 2025, 14, 12. https://doi.org/10.3390/cells14010012
Malysa A, Zhang XM, Bepler G. Minichromosome Maintenance Proteins: From DNA Replication to the DNA Damage Response. Cells. 2025; 14(1):12. https://doi.org/10.3390/cells14010012
Chicago/Turabian StyleMalysa, Agnes, Xiaohong Mary Zhang, and Gerold Bepler. 2025. "Minichromosome Maintenance Proteins: From DNA Replication to the DNA Damage Response" Cells 14, no. 1: 12. https://doi.org/10.3390/cells14010012
APA StyleMalysa, A., Zhang, X. M., & Bepler, G. (2025). Minichromosome Maintenance Proteins: From DNA Replication to the DNA Damage Response. Cells, 14(1), 12. https://doi.org/10.3390/cells14010012





_Zhang.png)

