DNA Double-Strand Break Repairs and Their Application in Plant DNA Integration
Abstract
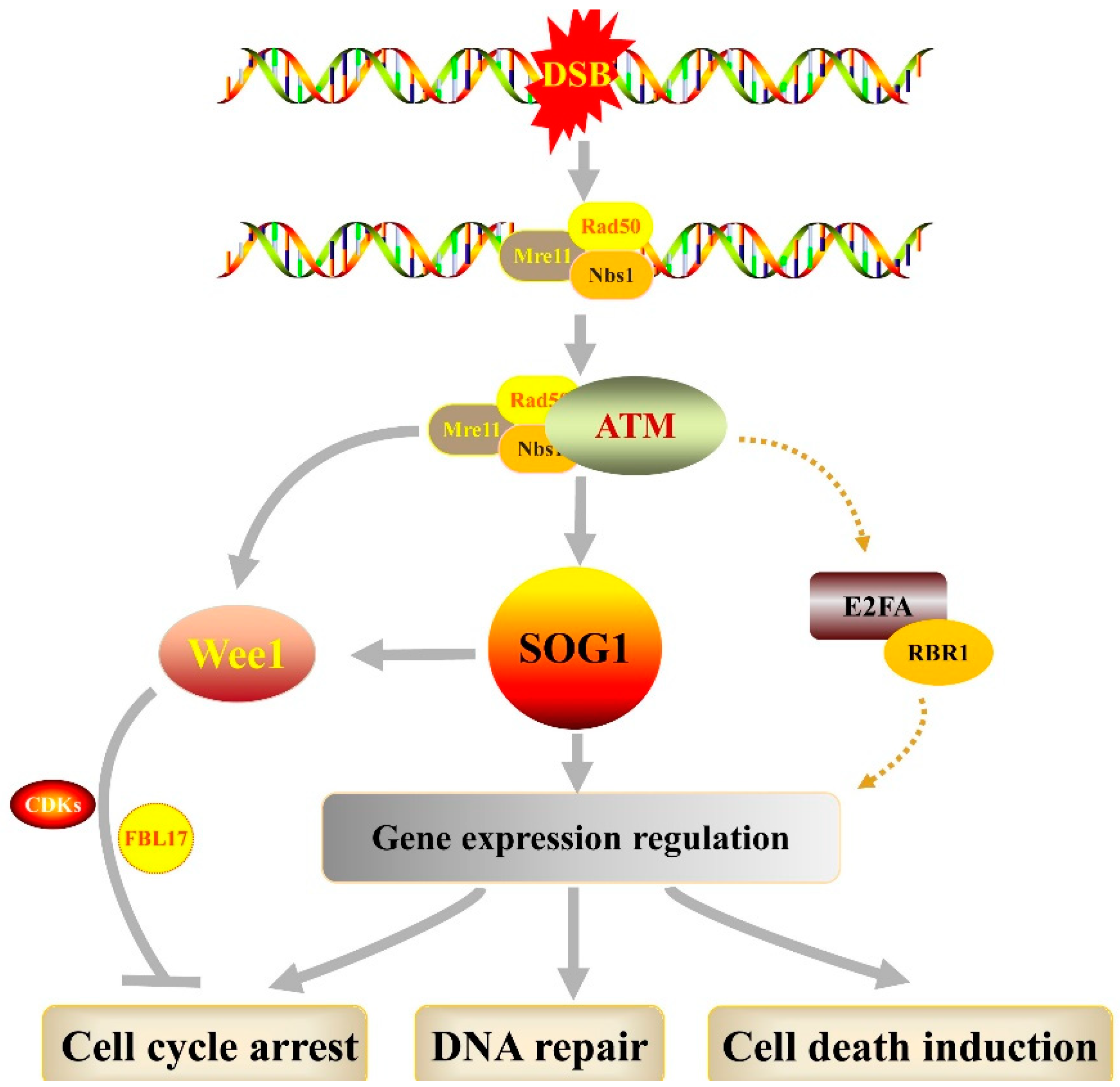
:1. Introduction
2. DNA Damage Response
3. DSB Repair via Homologous Recombination
4. DSB Repair via Non-Homologous End-Joining
5. DSB Repair Pathway and Agrobacterium-Mediated T-DNA Integration
6. DSB Repair Pathway and Gene Targeting
7. Strategies for DSB Induction
8. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khanna, K.K.; Jackson, S.P. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet. 2001, 27, 247–254. [Google Scholar] [CrossRef]
- Manova, V.; Gruszka, D. DNA damage and repair in plants—From models to crops. Front. Plant Sci. 2015, 6, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Hayut, S.F.; Bessudo, C.M.; Levy, A.A. Targeted recombination between homologous chromosomes for precise breeding in tomato. Nat. Commun. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- Siebert, R.; Puchta, H. Efficient repair of genomic double-strand breaks by homologous recombination between directly repeated sequences in the plant genome. Plant Cell 2002, 14, 1121–1131. [Google Scholar] [CrossRef] [Green Version]
- Stinson, B.M.; Loparo, J.J. Repair of DNA Double-Strand Breaks by the Nonhomologous End Joining Pathway. Annu. Rev. Biochem. 2021, 90, 137–164. [Google Scholar] [CrossRef]
- Puchta, H. The repair of double-strand breaks in plants: Mechanisms and consequences for genome evolution. J. Exp. Bot. 2005, 56, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Spampinato, C.P. Protecting DNA from errors and damage: An overview of DNA repair mechanisms in plants compared to mammals. Cell. Mol. Life Sci. 2017, 74, 1693–1709. [Google Scholar] [CrossRef]
- Block, W.D.; Yu, Y.; Lees-Miller, S.P. Phosphatidyl inositol 3-kinase-like serine/threonine protein kinases (PIKKs) are required for DNA damage-induced phosphorylation of the 32 kDa subunit of replication protein A at threonine 21. Nucleic Acids Res. 2004, 32, 997–1005. [Google Scholar] [CrossRef] [Green Version]
- Falck, J.; Coates, J.; Jackson, S.P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 2005, 434, 605–611. [Google Scholar] [CrossRef]
- Shiloh, Y. The ATM-mediated DNA-damage response: Taking shape. Trends Biochem. Sci. 2006, 31, 402–410. [Google Scholar] [CrossRef]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R.; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef] [Green Version]
- Tomimatsu, N.; Mukherjee, B.; Burma, S. Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient cells. EMBO Rep. 2009, 10, 629–635. [Google Scholar] [CrossRef] [Green Version]
- Meyer, B.; Voss, K.O.; Tobias, F.; Jakob, B.; Durante, M.; Taucher-Scholz, G. Clustered DNA damage induces pan-nuclear H2AX phosphorylation mediated by ATM and DNA-PK. Nucleic Acids Res. 2013, 41, 6109–6118. [Google Scholar] [CrossRef] [Green Version]
- White, D.E.; Negorev, D.; Peng, H.; Ivanov, A.V.; Maul, G.G.; Rauscher, F.J. KAP1, a novel substrate for PIKK family members, colocalizes with numerous damage response factors at DNA lesions. Cancer Res. 2006, 66, 11594–11599. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Saha, J.; Beckmann, P.J.; Hendrickson, E.A.; Davis, A.J. DNA-PKcs promotes chromatin decondensation to facilitate initiation of the DNA damage response. Nucleic Acids Res. 2019, 47, 9467–9479. [Google Scholar] [CrossRef] [Green Version]
- Garcia, V.; Bruchet, H.; Camescasse, D.; Granier, F.; Bouchez, D.; Tissier, A. AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 2003, 15, 119–132. [Google Scholar] [CrossRef] [Green Version]
- Culligan, K.M.; Robertson, C.E.; Foreman, J.; Doerner, P.; Britt, A.B. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 2006, 48, 947–961. [Google Scholar] [CrossRef]
- Cools, T.; De Veylder, L. DNA stress checkpoint control and plant development. Curr. Opin. Plant Biol. 2009, 12, 23–28. [Google Scholar] [CrossRef]
- Hu, Z.; Cools, T.; De Veylder, L. Mechanisms Used by Plants to Cope with DNA Damage. Annu. Rev. Plant Biol. 2016, 67, 439–462. [Google Scholar] [CrossRef]
- Yoshiyama, K.O.; Kobayashi, J.; Ogita, N.; Ueda, M.; Kimura, S.; Maki, H.; Umeda, M. ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep. 2013, 14, 817–822. [Google Scholar] [CrossRef] [Green Version]
- Bourbousse, C.; Vegesna, N.; Law, J.A. SOG1 activator and MYB3R repressors regulate a complex DNA damage network in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E12453–E12462. [Google Scholar] [CrossRef] [Green Version]
- Ogita, N.; Okushima, Y.; Tokizawa, M.; Yamamoto, Y.Y.; Tanaka, M.; Seki, M.; Makita, Y.; Matsui, M.; Okamoto-Yoshiyama, K.; Sakamoto, T.; et al. Identifying the target genes of SUPPRESSOR OF γ RESPONSE 1, a master transcription factor controlling DNA damage response in Arabidopsis. Plant J. 2018, 94, 439–453. [Google Scholar] [CrossRef] [Green Version]
- De Schutter, K.; Joubes, J.; Cools, T.; Verkest, A.; Corellou, F.; Babiychuk, E.; Van Der Schueren, E.; Beeckman, T.; Kushnir, S.; Inze, D.; et al. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 2007, 19, 211–225. [Google Scholar] [CrossRef] [Green Version]
- Pan, T.; Qin, Q.; Nong, C.; Gao, S.; Wang, L.; Cai, B.; Zhang, M.; Wu, C.; Chen, H.; Li, T.; et al. A novel WEE1 pathway for replication stress responses. Nat. Plants 2021, 7, 209–218. [Google Scholar] [CrossRef]
- Horvath, B.M.; Kourova, H.; Nagy, S.; Nemeth, E.; Magyar, Z.; Papdi, C.; Ahmad, Z.; Sanchez-Perez, G.F.; Perilli, S.; Blilou, I.; et al. Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO J. 2017, 36, 1261–1278. [Google Scholar] [CrossRef]
- Biedermann, S.; Harashima, H.; Chen, P.; Heese, M.; Bouyer, D.; Sofroni, K.; Schnittger, A. The retinoblastoma homolog RBR 1 mediates localization of the repair protein RAD 51 to DNA lesions in Arabidopsis. EMBO J. 2017, 36, 1279–1297. [Google Scholar] [CrossRef]
- Nisa, M.U.; Huang, Y.; Benhamed, M.; Raynaud, C. The plant DNA damage response: Signaling pathways leading to growth inhibition and putative role in response to stress conditions. Front. Plant Sci. 2019, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Heyer, W.-D.; Ehmsen, K.T.; Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010, 44, 113–139. [Google Scholar] [CrossRef] [Green Version]
- Serra, H.; Da Ines, O.; Degroote, F.; Gallego, M.E.; White, C.I. Roles of XRCC2, RAD51B and RAD51D in RAD51-Independent SSA Recombination. PLoS Genet. 2013, 9, e1003971. [Google Scholar] [CrossRef] [Green Version]
- Schuermann, D.; Molinier, J.; Fritsch, O.; Hohn, B. The dual nature of homologous recombination in plants. Trends Genet. 2005, 21, 172–181. [Google Scholar] [CrossRef]
- Mimitou, E.P.; Symington, L.S. DNA end resection—Unraveling the tail. DNA Repair 2011, 10, 344–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owalczykowski, S.T.C.K. DNA annealing by Rad52 Protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. USA 1998, 95, 6049–6054. [Google Scholar]
- Sugiyama, T.; Zaitseva, E.M.; Kowalczykowski, S.C. A Single-stranded DNA-binding Protein Is Needed for Efficient Presynaptic Complex Formation by the Saccharomyces cerevisiae Rad51 Protein. J. Biol. Chem. 1997, 272, 7940–7945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edlinger, B.; Schlögelhofer, P. Have a break: Determinants of meiotic DNA double strand break (DSB) formation and processing in plants. J. Exp. Bot. 2011, 62, 1545–1563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mannuss, A.; Trapp, O.; Puchta, H. Gene regulation in response to DNA damage. Biochim. Biophys. Acta-Gene Regul. Mech. 2012, 1819, 154–165. [Google Scholar] [CrossRef]
- Mazin, A.V.; Alexeev, A.A.; Kowalczykowski, S.C. A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 2003, 278, 14029–14036. [Google Scholar] [CrossRef] [Green Version]
- Wolner, B.; Peterson, C.L. ATP-dependent and ATP-independent roles for the Rad54 chromatin remodeling enzyme during recombinational repair of a DNA double strand break. J. Biol. Chem. 2005, 280, 10855–10860. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Sugiyama, T.; Kowalczykowski, S.C. DNA annealing mediated by Rad52 and Rad59 proteins. J. Biol. Chem. 2006, 281, 15441–15449. [Google Scholar] [CrossRef] [Green Version]
- Baudat, F.; de Massy, B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 2007, 15, 565–577. [Google Scholar] [CrossRef]
- Moynahan, M.E.; Jasin, M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 2010, 11, 196–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prakash, R.; Satory, D.; Dray, E.; Papusha, A.; Scheller, J.; Kramer, W.; Krejci, L.; Klein, H.; Haber, J.E.; Sung, P.; et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: Implications for crossover control in mitotic recombination. Genes Dev. 2009, 23, 67–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Kantake, N.; Sugiyama, T.; Kowalczykowski, S.C. Rad51 protein controls Rad52-mediated DNA annealing. J. Biol. Chem. 2008, 283, 14883–14892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lydeard, J.R.; Jain, S.; Yamaguchi, M.; Haber, J.E. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 2007, 448, 820–823. [Google Scholar] [CrossRef] [PubMed]
- Schubert, I.; Schubert, V.; Fuchs, J. No evidence for “break-induced replication” in a higher plant—But break-induced conversion may occur. Front. Plant Sci. 2011, 2, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef] [Green Version]
- McVey, M.; Lee, S.E. MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. 2008, 24, 529–538. [Google Scholar] [CrossRef] [Green Version]
- Ramsden, D.A.; Carvajal-Garcia, J.; Gupta, G.P. Mechanism, cellular functions and cancer roles of polymerase-theta-mediated DNA end joining. Nat. Rev. Mol. Cell Biol. 2022, 23, 125–140. [Google Scholar] [CrossRef]
- Van Gent, D.C.; van der Burg, M. Non-homologous end-joining, a sticky affair. Oncogene 2007, 26, 7731–7740. [Google Scholar] [CrossRef] [Green Version]
- Shen, H.; Strunks, G.D.; Klemann, B.J.P.M.; Hooykaas, P.J.J.; de Pater, S. CRISPR/Cas9-Induced Double-Strand Break Repair in Arabidopsis Nonhomologous End-Joining Mutants. G3 Genes Genomes Genet. 2017, 7, 193–202. [Google Scholar] [CrossRef] [Green Version]
- Waterworth, W.M.; Drury, G.E.; Bray, C.M.; West, C.E. Repairing breaks in the plant genome: The importance of keeping it together. New Phytol. 2011, 192, 805–822. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.R.; Corpina, R.A.; Goldberg, J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 2001, 412, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Valuchova, S.; Fulnecek, J.; Prokop, Z.; Stolt-Bergner, P.; Janouskova, E.; Hofr, C.; Riha, K. Protection of arabidopsis blunt-ended telomeres is mediated by a physical association with the ku heterodimer. Plant Cell 2017, 29, 1533–1545. [Google Scholar] [CrossRef]
- Roy, S.; Choudhury, S.R.; Sengupta, D.N.; Das, K.P. Involvement of AtPolλ in the repair of high salt- and DNA cross-linking agent-induced double strand breaks in Arabidopsis. Plant Physiol. 2013, 162, 1195–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West, C.E.; Waterworth, W.M.; Jiang, Q.; Bray, C.M. Arabidopsis DNA ligase IV is induced by γ-irradiation and interacts with an Arabidopsis homologue of the double strand break repair protein XRCC4. Plant J. 2000, 24, 67–78. [Google Scholar] [CrossRef]
- Deriano, L.; Roth, D.B. Modernizing the nonhomologous end-joining repertoire: Alternative and classical NHEJ share the stage. Annu. Rev. Genet. 2013, 47, 433–455. [Google Scholar] [CrossRef] [PubMed]
- Caldecott, K.W. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008, 9, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Haince, J.F.; McDonald, D.; Rodrigue, A.; Dery, U.; Masson, J.Y.; Hendzel, M.J.; Poirier, G.G. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008, 283, 1197–1208. [Google Scholar] [CrossRef] [Green Version]
- Aguilar-quesada, R.; Muñoz-gámez, J.A.; Martín-Oliva, D.; Peralta, A.; Valenzuela, M.T.; Matínez-Romero, R.; Quiles-pérez, R.; Menissier-de Murcia, J.; de Murcia, G.; Ruiz de Almodóvar, M.; et al. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol. Biol. 2007, 8, 29. [Google Scholar] [CrossRef] [Green Version]
- Jia, Q.; Den Dulk-Ras, A.; Shen, H.; Hooykaas, P.J.J.; de Pater, S. Poly(ADP-ribose)polymerases are involved in microhomology mediated back-up non-homologous end joining in Arabidopsis thaliana. Plant Mol. Biol. 2013, 82, 339–351. [Google Scholar] [CrossRef]
- Uanschou, C.; Siwiec, T.; Pedrosa-Harand, A.; Kerzendorfer, C.; Sanchez-Moran, E.; Novatchkova, M.; Akimcheva, S.; Woglar, A.; Klein, F.; Schlögelhofer, P. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 2007, 26, 5061–5070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, J.; Jiang, G.; Willers, H.; Xia, F. Exonuclease function of human Mre11 promotes deletional nonhomologous end joining. J. Biol. Chem. 2009, 284, 30565–30573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee-Theilen, M.; Matthews, A.J.; Kelly, D.; Zheng, S.; Chaudhuri, J. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat. Struct. Mol. Biol. 2011, 18, 75–79. [Google Scholar] [CrossRef]
- Zhang, Y.; Jasin, M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat. Struct. Mol. Biol. 2011, 18, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Crespan, E.; Czabany, T.; Maga, G.; Hübscher, U. Microhomology-mediated DNA strand annealing and elongation by human DNA polymerases λ and β on normal and repetitive DNA sequences. Nucleic Acids Res. 2012, 40, 5577–5590. [Google Scholar] [CrossRef] [Green Version]
- Weinfeld, M.; Mani, R.S.; Abdou, I.; Aceytuno, R.D.; Glover, J.N.M. Tidying up loose ends: The role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem. Sci. 2011, 36, 262–271. [Google Scholar] [CrossRef] [Green Version]
- Chan, S.H.; Yu, A.M.; McVey, M. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010, 6, e1001005. [Google Scholar] [CrossRef] [Green Version]
- Kent, T.; Chandramouly, G.; McDevitt, S.M.; Ozdemir, A.Y.; Pomerantz, R.T. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat. Struct. Mol. Biol. 2015, 22, 230–237. [Google Scholar] [CrossRef] [Green Version]
- He, P.; Yang, W. Template and primer requirements for DNA Pol θ-mediated end joining. Proc. Natl. Acad. Sci. USA 2018, 115, 7747–7752. [Google Scholar] [CrossRef] [Green Version]
- Mateos-Gomez, P.A.; Kent, T.; Deng, S.K.; Mcdevitt, S.; Kashkina, E.; Hoang, T.M.; Pomerantz, R.T.; Sfeir, A. The helicase domain of Polθ counteracts RPA to promote alt-NHEJ. Nat. Struct. Mol. Biol. 2017, 24, 1116–1123. [Google Scholar] [CrossRef]
- Schrempf, A.; Slyskova, J.; Loizou, J.I. Targeting the DNA Repair Enzyme Polymerase θ in Cancer Therapy. Trends Cancer 2021, 7, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Koole, W.; van Schendel, R.; Karambelas, A.E.; van Heteren, J.T.; Okihara, K.L.; Tijsterman, M. A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat. Commun. 2014, 5, 3216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mateos-Gomez, P.A.; Gong, F.; Nair, N.; Miller, K.M.; Lazzerini-Denchi, E.; Sfeir, A. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 2015, 518, 254–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Kregten, M.; De Pater, S.; Romeijn, R.; Van Schendel, R.; Hooykaas, P.J.J.; Tijsterman, M. T-DNA integration in plants results from polymerase-θ-mediated DNA repair. Nat. Plants 2016, 2, 16164. [Google Scholar] [CrossRef]
- Wyatt, D.W.; Feng, W.; Conlin, M.P.; Yousefzadeh, M.J.; Roberts, S.A.; Mieczkowski, P.; Wood, R.D.; Gupta, G.P.; Ramsden, D.A. Essential Roles for Polymerase θ-Mediated End Joining in the Repair of Chromosome Breaks. Mol. Cell 2016, 63, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Inagaki, S.; Nakamura, K.; Morikami, A. A link among DNA replication, recombination, and gene expression revealed by genetic and genomic analysis of TEBICHI gene of Arabidopsis thaliana. PLoS Genet. 2009, 5, e1000613. [Google Scholar] [CrossRef] [Green Version]
- Nisa, M.; Bergis, C.; Pedroza-Garcia, J.A.; Drouin-Wahbi, J.; Mazubert, C.; Bergounioux, C.; Benhamed, M.; Raynaud, C. The plant DNA polymerase theta is essential for the repair of replication-associated DNA damage. Plant J. 2021, 106, 1197–1207. [Google Scholar] [CrossRef]
- Audebert, M.; Salles, B.; Calsou, P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004, 279, 55117–55126. [Google Scholar] [CrossRef] [Green Version]
- Simsek, D.; Brunet, E.; Wong, S.Y.W.; Katyal, S.; Gao, Y.; McKinnon, P.J.; Lou, J.; Zhang, L.; Li, J.; Rebar, E.J.; et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011, 7, e1002080. [Google Scholar] [CrossRef] [Green Version]
- Waterworth, W.M.; Kozak, J.; Provost, C.M.; Bray, C.M.; Angelis, K.J.; West, C.E. DNA ligase 1 deficient plants display severe growth defects and delayed repair of both DNA single and double strand breaks. BMC Plant Biol. 2009, 9, 79. [Google Scholar] [CrossRef] [Green Version]
- Arakawa, H.; Bednar, T.; Wang, M.; Paul, K.; Mladenov, E.; Bencsik-Theilen, A.A.; Iliakis, G. Functional redundancy between DNA ligases I and III in DNA replication in vertebrate cells. Nucleic Acids Res. 2012, 40, 2599–2610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, K.; Wang, M.; Mladenov, E.; Bencsik-Theilen, A.; Bednar, T.; Wu, W.; Arakawa, H.; Iliakis, G. DNA ligases I and III cooperate in alternative non-homologous end-joining in vertebrates. PLoS ONE 2013, 8, e59505. [Google Scholar] [CrossRef] [PubMed]
- Bundock, P.; den Dulk-Ras, A.; Beijersbergen, A.; Hooykaas, P.J. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995, 14, 3206–3214. [Google Scholar] [CrossRef] [PubMed]
- Offringa, R.; de Groot, M.J.; Haagsman, H.J.; Does, M.P.; van den Elzen, P.J.; Hooykaas, P.J. Extrachromosomal homologous recombination and gene targeting in plant cells after Agrobacterium mediated transformation. EMBO J. 1990, 9, 3077–3084. [Google Scholar] [CrossRef] [PubMed]
- Tzfira, T.; Li, J.; Lacroix, B.; Citovsky, V. Agrobacterium T-DNA integration: Molecules and models. Trends Genet. 2004, 20, 375–383. [Google Scholar] [CrossRef]
- Gelvin, S.B. Plant Proteins Involved in Agrobacterium -Mediated Genetic Transformation. Annu. Rev. Phytopathol. 2010, 48, 45–68. [Google Scholar] [CrossRef] [Green Version]
- Rodenburg, K.W.; de Groot, M.J.; Schilperoort, R.A.; Hooykaas, P.J. Single-stranded DNA used as an efficient new vehicle for transformation of plant protoplasts. Plant Mol. Biol. 1989, 13, 711–719. [Google Scholar] [CrossRef]
- Gheysen, G.; Villarroel, R.; Van Montagu, M. Illegitimate recombination in plants: A model for T-DNA integration. Genes Dev. 1991, 5, 287–297. [Google Scholar] [CrossRef] [Green Version]
- Mayerhofer, R.; Koncz-kalman, Z.; Nawrath, C.; Bakkeren, G.; Crameri, A.; Angelis, K.; Redei, G.P.; Schell, J.; Hohn, B.; Koncz, C. T-DNA integration: In plants mode of illegitimate recombination. EMBO J. 1991, 10, 697–704. [Google Scholar] [CrossRef]
- Salomon, S.; Puchta, H. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 1998, 17, 6086–6095. [Google Scholar] [CrossRef]
- Chilton, M.-D.M.; Que, Q. Targeted integration of T-DNA into the tobacco genome at double-stranded breaks: New insights on the mechanism of T-DNA integration. Plant Physiol. 2003, 133, 956–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzfira, T.; Frankman, L.R.; Vaidya, M.; Citovsky, V. Site-specific integration of Agrobacterium tumefaciens T-DNA via double-stranded intermediates. Plant Physiol. 2003, 133, 1011–1023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Xing, H.L.; Wang, Z.P.; Zhang, H.Y.; Yang, F.; Wang, X.C.; Chen, Q.J. Potential high-frequency off-target mutagenesis induced by CRISPR/Cas9 in Arabidopsis and its prevention. Plant Mol. Biol. 2018, 96, 445–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Bent, A.F. Microbial Pathogens Trigger Host DNA Double-Strand Breaks Whose Abundance Is Reduced by Plant Defense Responses. PLoS Pathog. 2014, 10, e1004226. [Google Scholar] [CrossRef] [PubMed]
- Van Attikum, H.; Bundock, P.; Hooykaas, P.J. Non-homologous end-joining proteins are required for Agrobacterium T-DNA integration. EMBO J. 2001, 20, 6550–6558. [Google Scholar] [CrossRef] [Green Version]
- Van Attikum, H.; Hooykaas, P.J.J. Genetic requirements for the targeted integration of Agrobacterium T-DNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2003, 31, 826–832. [Google Scholar] [CrossRef] [Green Version]
- Friesner, J.; Britt, A.B. Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J. 2003, 34, 427–440. [Google Scholar] [CrossRef] [Green Version]
- Gallego, M.E.; Bleuyard, J.-Y.; Daoudal-Cotterell, S.; Jallut, N.; White, C.I. Ku80 plays a role in non-homologous recombination but is not required for T-DNA integration in Arabidopsis. Plant J. 2003, 35, 557–565. [Google Scholar] [CrossRef]
- Li, J.; Vaidya, M.; White, C.; Vainstein, A.; Citovsky, V.; Tzfira, T. Involvement of KU80 in T-DNA integration in plant cells. Proc. Natl. Acad. Sci. USA 2005, 102, 19231–19236. [Google Scholar] [CrossRef] [Green Version]
- Jia, Q.; Bundock, P.; Hooykaas, P.J.J.; de Pater, S. Agrobacterium tumefaciens T-DNA Integration and Gene Targeting in Arabidopsis thaliana Non-Homologous End-Joining Mutants. J. Bot. 2012, 2012, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Mestiri, I.; Norre, F.; Gallego, M.E.; White, C.I. Multiple host-cell recombination pathways act in Agrobacterium-mediated transformation of plant cells. Plant J. 2014, 77, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Vaghchhipawala, Z.; Vasudevan, B.; Lee, L.-Y.; Shen, Y.; Singer, K.; Waterworth, W.M.; Zhang, Z.J.; West, C.E.; Mysore, K.S.; et al. Agrobacterium T-DNA integration into the plant genome can occur without the activity of key non-homologous end-joining proteins. Plant J. 2015, 81, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Kleinboelting, N.; Huep, G.; Appelhagen, I.; Viehoever, P.; Li, Y.; Weisshaar, B. The Structural Features of Thousands of T-DNA Insertion Sites Are Consistent with a Double-Strand Break Repair-Based Insertion Mechanism. Mol. Plant 2015, 8, 1651–1664. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa-Yokoi, A.; Saika, H.; Hara, N.; Lee, L.Y.; Toki, S.; Gelvin, S.B. Agrobacterium T-DNA integration in somatic cells does not require the activity of DNA polymerase θ. New Phytol. 2021, 229, 2859–2872. [Google Scholar] [CrossRef] [PubMed]
- Kralemann, L.; de Pater, S.; Shen, H.; Kloet, S.; van Schendel, R.; Hooykaas, P.J.J.; Tijsterman, M. T-DNA integration in plants requires MRE11- or TDP2-mediated removal of the 5 ’ bound Agrobacterium protein VirD2. Nat. Plants 2022. submitted. [Google Scholar]
- Knoll, A.; Fauser, F.; Puchta, H. DNA recombination in somatic plant cells: Mechanisms and evolutionary consequences. Chromosom. Res. 2014, 22, 191–201. [Google Scholar] [CrossRef]
- Shukla, V.K.; Doyon, Y.; Miller, J.C.; Dekelver, R.C.; Moehle, E.A.; Worden, S.E.; Mitchell, J.C.; Arnold, N.L.; Gopalan, S.; Meng, X.; et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 2009, 459, 437–441. [Google Scholar] [CrossRef]
- Ayar, A.; Wehrkamp-Richter, S.; Laffaire, J.B.; Le Goff, S.; Levy, J.; Chaignon, S.; Salmi, H.; Lepicard, A.; Sallaud, C.; Gallego, M.E.; et al. Gene targeting in maize by somatic ectopic recombination. Plant Biotechnol. J. 2013, 11, 305–314. [Google Scholar] [CrossRef] [Green Version]
- De Pater, S.; Pinas, J.E.; Hooykaas, P.J.J.; van der Zaal, B.J. ZFN-mediated gene targeting of the Arabidopsis protoporphyrinogen oxidase gene through Agrobacterium-mediated floral dip transformation. Plant Biotechnol. J. 2013, 11, 510–515. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, K.; Breier, U.; Hensel, G.; Kumlehn, J.; Schubert, I.; Reiss, B. Stable gene replacement in barley by targeted double-strand break induction. J. Exp. Bot. 2016, 67, 1433–1445. [Google Scholar] [CrossRef] [Green Version]
- De Pater, S.; Klemann, B.J.P.M.; Hooykaas, P.J.J. True gene-targeting events by CRISPR/Cas-induced DSB repair of the PPO locus with an ectopically integrated repair template. Sci. Rep. 2018, 8, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Reiss, B.; Schubert, I.; Köpchen, K.; Wendeler, E.; Schell, J.; Puchta, H. RecA stimulates sister chromatid exchange and the fidelity of double-strand break repair, but not gene targeting, in plants transformed by Agrobacterium. Proc. Natl. Acad. Sci. USA 2000, 97, 3358–3363. [Google Scholar] [CrossRef] [PubMed]
- Shaked, H.; Melamed-Bessudo, C.; Levy, A.A. High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. Proc. Natl. Acad. Sci. USA 2005, 102, 12265–12269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kooistra, R.; Hooykaas, P.J.J.; Steensma, H.Y. Efficient gene targeting in Kluyveromyces lactis. Yeast 2004, 21, 781–792. [Google Scholar] [CrossRef]
- De Boer, P.; Bastiaans, J.; Touw, H.; Kerkman, R.; Bronkhof, J.; van den Berg, M.; Offringa, R. Highly efficient gene targeting in Penicillium chrysogenum using the bi-partite approach in Δlig4 or Δku70 mutants. Fungal Genet. Biol. 2010, 47, 839–846. [Google Scholar] [CrossRef]
- Qi, Y.; Zhang, Y.; Zhang, F.; Baller, J.A.; Cleland, S.C.; Ryu, Y.; Starker, C.G.; Voytas, D.F. Increasing frequencies of site-specific mutagenesis and gene targeting in Arabidopsis by manipulating DNA repair pathways. Genome Res. 2013, 23, 547–554. [Google Scholar] [CrossRef] [Green Version]
- Endo, M.; Mikami, M.; Toki, S. Biallelic gene targeting in rice. Plant Physiol. 2016, 170, 667–677. [Google Scholar] [CrossRef] [Green Version]
- Even-Faitelson, L.; Samach, A.; Melamed-Bessudo, C.; Avivi-Ragolsky, N.; Levy, A.A. Localized egg-cell expression of effector proteins for targeted modification of the Arabidopsis genome. Plant J. 2011, 68, 929–937. [Google Scholar] [CrossRef]
- Saito, S.; Maeda, R.; Adachi, N. Dual loss of human POLQ and LIG4 abolishes random integration. Nat. Commun. 2017, 8, 16112. [Google Scholar] [CrossRef] [Green Version]
- Van Tol, N.; van Schendel, R.; Bos, A.; van Kregten, M.; de Pater, S.; Hooykaas, P.J.J.; Tijsterman, M. Gene targeting in polymerase theta-deficient Arabidopsis thaliana. Plant J. 2022, 109, 112–125. [Google Scholar] [CrossRef]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Durai, S.; Mani, M.; Kandavelou, K.; Wu, J.; Porteus, M.H.; Chandrasegaran, S. Zinc finger nucleases: Custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005, 33, 5978–5990. [Google Scholar] [CrossRef] [PubMed]
- De Pater, S.; Neuteboom, L.W.; Pinas, J.E.; Hooykaas, P.J.J.; Van Der Zaal, B.J. ZFN-induced mutagenesis and gene-targeting in Arabidopsis through Agrobacterium-mediated floral dip transformation. Plant Biotechnol. J. 2009, 7, 821–835. [Google Scholar] [CrossRef]
- Wang, M.; Lu, Y.; Botella, J.R.; Mao, Y.; Hua, K.; Zhu, J. kang Gene Targeting by Homology-Directed Repair in Rice Using a Geminivirus-Based CRISPR/Cas9 System. Mol. Plant 2017, 10, 1007–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fauser, F.; Roth, N.; Pacher, M.; Ilg, G.; Sanchez-Fernandez, R.; Biesgen, C.; Puchta, H. In planta gene targeting. Proc. Natl. Acad. Sci. USA 2012, 109, 7535–7540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolter, F.; Klemm, J.; Puchta, H. Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. Plant J. 2018, 94, 735–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil-Humanes, J.; Wang, Y.; Liang, Z.; Shan, Q.; Ozuna, C.V.; Sánchez-León, S.; Baltes, N.J.; Starker, C.; Barro, F.; Gao, C.; et al. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017, 89, 1251–1262. [Google Scholar] [CrossRef] [Green Version]
- Dahan-Meir, T.; Filler-Hayut, S.; Melamed-Bessudo, C.; Bocobza, S.; Czosnek, H.; Aharoni, A.; Levy, A.A. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 2018, 95, 5–16. [Google Scholar] [CrossRef] [Green Version]
- Miki, D.; Zhang, W.; Zeng, W.; Feng, Z.; Zhu, J.K. CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Begemann, M.B.; Gray, B.N.; January, E.; Gordon, G.C.; He, Y.; Liu, H.; Wu, X.; Brutnell, T.P.; Mockler, T.C.; Oufattole, M. Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Sci. Rep. 2017, 7, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Ma, M.; Zhuang, F.; Hu, X.; Wang, B.; Wen, X.Z.; Ji, J.F.; Xi, J.J. Efficient generation of mice carrying homozygous double-floxp alleles using the Cas9-Avidin/Biotin-donor DNA system. Cell Res. 2017, 27, 578–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aird, E.J.; Lovendahl, K.N.; St Martin, A.; Harris, R.S.; Gordon, W.R. Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun. Biol. 2018, 1, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savic, N.; Ringnalda, F.C.A.S.; Lindsay, H.; Berk, C.; Bargsten, K.; Li, Y.; Neri, D.; Robinson, M.D.; Ciaudo, C.; Hall, J.; et al. Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology-directed repair. eLife 2018, 7, 1–18. [Google Scholar] [CrossRef]
- Charpentier, M.; Khedher, A.H.Y.; Menoret, S.; Brion, A.; Lamribet, K.; Dardillac, E.; Boix, C.; Perrouault, L.; Tesson, L.; Geny, S.; et al. CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Smih, F.; Rouet, P.; Romanienko, P.J.; Jasin, M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995, 23, 5012–5019. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef] [Green Version]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010, 186, 756–761. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Cong, L.; Lodato, S.; Kosuri, S.; Church, G.M.; Arlotta, P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 2011, 29, 149–153. [Google Scholar] [CrossRef] [Green Version]
- Cermak, T.; Doyle, E.; Christian, M. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82. [Google Scholar] [CrossRef] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; Van Der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, B.; Montoya, G. CRISPR-Cas12a: Functional overview and applications. Biomed. J. 2020, 43, 8–17. [Google Scholar] [CrossRef] [PubMed]


| Homo sapiens | Saccharomyces cerevisiae | Arabidopsis thaliana | Arabidopsis Gene Number | Function |
|---|---|---|---|---|
| Rad51 | Rad51 | Rad51 | At5g20850 | RecA homologue Strand invasion |
| MRN complex: Mre11-Rad50-Nbs1 | MRX complex: Mre11-Rad50-Xrs2 | MRN complex: Mre11-Rad50-Nbs1 | At5g54260 At2g31970 At3g02680 | DNA binding Nuclease activities DSB end processing DNA-damage checkpoints |
| CtIP | Sae2 | Com1 | At3g52115 | DSB end processing DNA strand transition |
| Exo1 | Exo1 | Exo1A Exo1B | At1g29630 At1g18090 | DSB end processing |
| BLM | Sgs1 | RecQ4A RecQ4B | At1g10930 At1g60930 | DSB end processing RecQ helicases |
| RPA1 RPA2 RPA3 | RPA1 RPA2 RPA3 | RPA1 RPA2 RPA3 | At2g06510 At4g19130 At5g45400 At5g08020 At5g61000 | ssDNA binding |
| Rad51B-Rad51C Rad51C-XRCC3 Rad51D-XRCC2 | Rad55-Rad57 | Rad51B-Rad51C Rad51C-XRCC3 Rad51D-XRCC2 | At2g28560 At2g45280 At5g57450 At1g07745 At5g64520 | ssDNA binding Recombination mediator |
| Rad52 | Rad52 | Rad52 | At1g71310 At5g47870 | ssDNA binding and annealing Recombination mediator Interacts with Rad51 and RPA |
| BRCA1 | − 1 | BRCA1 | At4g21070 | Checkpoint mediator Recombination mediator |
| BRCA2 | − 1 | BRCA2-1 BRCA2-2 | At5g01630 At4g00020 | Recombination mediator |
| Rad54 | Rad54 | Rad54 | At3g19210 | ATP-dependent dsDNA translocase Stimulates the D-loop reaction |
| FancM | Mph1 | FancM | At1g35530 | Helicase activity Dissociates D-loop formation and facilitates single-strand annealing |
| Homo sapiens | Saccharomyces cerevisiae | Arabidopsis thaliana | Arabidopsis Gene Number | Function |
|---|---|---|---|---|
| Ku70/Ku80 | Ku70/Ku80 | Ku70/Ku80 | At1g16970 At1g48050 | DSB end binding and protection |
| DNA-PKcs | − 1 | − 1 | protein kinase | |
| Artemis | Snm1/PSO2 | Snm1 | At3g26680 | DNA end processing |
| MRN complex: Mre11-Rad50-Nbs1 | MRX complex: Mre11-Rad50-Xrs2 | MRN complex: Mre11-Rad50-Nbs1 | At5g54260 At2g31970 At3g02680 | DNA binding Nuclease activities DSB ends processing DNA-damage checkpoints |
| PNKP | Tpp1 | ZDP | At3g14890 | DNA end processing |
| Pol λ | − 1 | Pol λ | At1g10520 | DNA polymerase DNA end processing |
| 53BP1 | Rad9 | Rad9 | At3g05480 | DNA end processing |
| DNA ligase IV | Dnl4 | lig4 | At5g57160 | ATP-dependent DNA ligase |
| XRCC4 | Lif1 | XRCC4 | At3g23100 | complex with lig4 |
| XLF/Cernunnos | Nej1 | − 1 | complex with lig4/XRCC4 | |
| Parp1 | − 1 | Parp1 | At2g31320 | DNA end binding NAD + ADP-ribosyltransferase |
| Parp2 | − 1 | Parp2 | At4g02390 | DNA end binding NAD + ADP-ribosyltransferase |
| Parp3 | − 1 | Parp3 | At5g22470 | DNA end binding NAD + ADP-ribosyltransferase |
| CtIP | Sae2 | Com1 | At3g52115 | DNA end processing |
| DNA ligase III | − 1 | − 1 | ATP-dependent DNA ligase | |
| XRCC1 | − 1 | XRCC1 | At1g80420 | complex with lig3 |
| Pol Q | − 1 | Pol θ (Tebichi) | At4g32700 | DNA polymerase DNA end processing |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, H.; Li, Z. DNA Double-Strand Break Repairs and Their Application in Plant DNA Integration. Genes 2022, 13, 322. https://doi.org/10.3390/genes13020322
Shen H, Li Z. DNA Double-Strand Break Repairs and Their Application in Plant DNA Integration. Genes. 2022; 13(2):322. https://doi.org/10.3390/genes13020322
Chicago/Turabian StyleShen, Hexi, and Zhao Li. 2022. "DNA Double-Strand Break Repairs and Their Application in Plant DNA Integration" Genes 13, no. 2: 322. https://doi.org/10.3390/genes13020322





