Abstract
Nitrogen removal through heterotrophic nitrification–aerobic denitrification (HN–AD) has been acknowledged as one of the most efficient and cost-effective nitrogen removal processes. This study involved the isolation of a novel HN–AD bacterium (Rhodococcus sp. S2) from landfill leachate. Rhodococcus sp. S2 exhibited high nitrogen removal performance under aerobic conditions without the accumulation of nitrite as an intermediate. The maximum removal efficiencies for NH4+-N, NO2−-N, and NO3−-N were found to be 99.97 ± 0.3%, 99.79 ± 0.14%, and 83.53 ± 0.31%, respectively. Additionally, Rhodococcus sp. S2 demonstrated simultaneous nitrifying and denitrifying capabilities and showed a preference for utilizing NH4+-N in mixed nitrogen sources. The optimal nitrogen removal conditions for Rhodococcus sp. S2 were as follows: sodium acetate as a carbon source, a C/N ratio of 16, a shaking speed of 200 rpm, a pH of 9, and a temperature of 35 °C. Genome sequencing results revealed the presence of nitrate reductases (NarG), nitrate oxidoreductase (NxrA), and nitrite reductase (NirBD) in Rhodococcus sp. S2, providing further evidence of its HN–AD capability. In treating raw wastewater under the aforementioned experimental conditions, S2 achieved a maximum TN removal efficiency of 57.16 ± 0.52% with the addition of sodium acetate as a carbon source. These results suggest that Rhodococcus sp. S2 might be a promising candidate for wastewater nitrogen removal.
1. Introduction
Nitrogen removal is essential for averting adverse environmental and human health impacts stemming from improper wastewater disposal. Biological nitrogen removal is widely employed in sewage treatment due to its advantages over physical and chemical treatment technologies, such as its efficient removal capacity, cost-effectiveness, and the absence of secondary pollution [1]. The conventional process for biological nitrogen removal involves aerobic nitrification and anoxic denitrification. Nitrifiers oxidize ammonium to nitrite or nitrate under aerobic conditions, while anoxic denitrifiers convert nitrate and nitrite into N2 under anoxic conditions [2,3,4]. Performing these two processes requires different facilities and operating conditions, leading to increased facility costs, energy consumption, and time requirements. Heterotrophic nitrification–aerobic denitrification (HN–AD) bacteria have the ability to simultaneously carry out nitrification and denitrification under aerobic conditions [5]. They offer significant advantages owing to their faster growth rate, environmental adaptability, and capacity to utilize organic compounds. Consequently, research on HN–AD bacteria has garnered increasing attention in recent years.
At present, numerous HN–AD bacteria, such as Klebsiella sp. [6], Pseudomonas sp. [7], Acinetobacter sp. [8], Chryseobacterium sp. [9], and Thauera sp. [10], have been isolated and studied. However, a study found that there was a significant build-up of nitrite in the biological processes involved in HN–AD when both ammonia and nitrate were used as nitrogen sources, ultimately resulting in the retention of total nitrogen (TN) [11,12,13]. The accumulation of nitrite in the reported HN–AD bacteria may be attributed to two factors: (a) the lack of specific genes encoding nitrite reductase (Nir), nitric oxide reductase (Nor), and nitrous oxide reductase (Nos) or reductase damage [14] and (b) the impact of environmental factors, such as high dissolved oxygen (DO) (3 mg L−1) or low-carbon sources (C/N = 2.5), on gene expression and enzyme activity, leading to incomplete metabolic processes [15]. Moreover, in other studies, even a low concentration of nitrite accumulation, such as 2.04 mg L−1, was found to be toxic to certain environmental bacteria (such as Paracoccus versutus LYM and Pseudomonas stutzeri D6), with it inhibiting their ability to remove nitrogen and limiting their application in wastewater treatment [4,16]. Consequently, it is of great significance to explore new HN–AD bacteria that can address multiple nitrogen contaminants without accumulating nitrite.
It has been reported that some HN–AD bacteria exhibit effective nitrogen removal performance even under extreme conditions, including low pH (3–6), low temperature (5–15 ℃), or high salinity (30–50 g L−1) [17]. Landfill leachate, one of the most complex forms of urban waste, has garnered attention due to its high concentrations of ammonium (50–2200 mg/L), high salinity, and high chemical oxygen demand (COD) concentration (commonly ≤4000 mg L−1) and the presence of heavy metals (0.01 to 1.0 × 104 mg/kg) [18,19,20]. Landfill leachate may serve as an ideal source for salt/heavy metal-resistant HN–AD bacteria. Furthermore, HN–AD bacteria existing in landfill leachate may be able to adjust to high ammonium concentrations. In previous studies, HN–AD bacteria such as Stenotrophomonas maltophilia DQ01 [21] and Alcaligenes faecalis TF-1 [22] were isolated from landfill leachates and exhibited good performance in carrying out nitrogen removal. However, although these strains showed considerable nitrification ability, they only showed low denitrification ability. Furthermore, it remains unclear as to how a high initial NH4+-N concentration affects HN–AD bacteria. On the other hand, functional genes in the nitrogen metabolism pathways of HN–AD are worth investigating, with such investigations being useful in understanding the HN–AD mechanism of different strains. Hence, the separation and screening of microbes with efficient nitrogen removal capacity from landfill leachate offer significant potential for application.
In this study, our aim is to investigate a high-performance HN–AD bacterium that can remove nitrogen without nitrite accumulation under aerobic conditions. In this experiment, Rhodococcus sp. S2 was isolated from landfill leachate collected from a wastewater treatment plant in Shenzhen, China. Complete genome sequencing was conducted to explore the key genes related to the HN–AD process and elucidate its nitrogen metabolism mechanism. Additionally, the nitrogen removal capacity of S2 was analyzed through the use of different nitrogen sources. The optimal growth conditions of S2 were investigated under various carbon sources, carbon-to-nitrogen ratio (C/N), dissolved oxygen (DO), pH, temperature, and initial NH4+-N concentrations. Finally, S2 was introduced to actual sewage to verify its nitrogen removal capacity.
2. Materials and Methods
2.1. Media
During the bacterial screening process, Luria–Bertani (LB) media were utilized, and bromothymol blue (BTB) agar plates were employed to screen aerobic denitrifiers. The specific formulation of these two media matched that of Wei et al. [18]. The heterotrophic nitrification medium (HNM) was employed to measure nitrification capacity and consisted of the following composition per liter: 4.052 g sodium succinate, 0.47 g (NH4)2SO4, 0.25 g K2HPO4, 0.125 g NaCl, 0.125 g MgSO4·7 H2O, 0.0025 g MnSO4·4 H2O, 0.0025 g FeSO4·7 H2O, and pH 7.5
2.2. Isolation and Identification
Landfill leachate samples were collected from a wastewater treatment plant (Laohukeng) in Shenzhen, China, in November 2019. The initial NH4+-N and COD concentrations were 1200 and 40,000 mg/L, respectively. A 10 mL sample was mixed with 90 mL sterilized 0.9% NaCl in a 250 mL flask and cultured at 30 °C at 150 rpm. After shaking for 8 h, a 10 mL suspension was added to 90 mL LB medium and incubated for 7 days. For subsequent selection, a 2% portion of the bacterial suspension was transferred to 150 mL sterilized HNM and incubated for 3 days. This procedure was repeated three times. The obtained supernatant (100 µL) was diluted 10−4-, 10−5-, 10−6-, and 10−7-fold by distilled water, and then, 200 µL diluent was applied to the HNM plate reproduction for 3 days at 30 °C. The selected isolates were cultured in HNM and then on BTB plates at 30 °C for 24 h. Colonies with blue circles were separated and kept in 25% glycerol solution at −80 °C.
The genomic DNA of strain S2 was extracted using a TIANamp Bacteria DNA Kit (TIANGEN Biotech, Beijing, China). The PCR product was obtained using a TaKaRa 16S rRNA Genetic Identification PCR Kit (TaKaRa, Japan) with universal primers 27F and 1492R. Subsequently, it was sent to BGI Company (Shenzhen, China) for sequencing. The corresponding phylogenetic tree was constructed using MEGA 7 software based on BLAST analysis.
2.3. Complete Genome Sequencing and Functional Annotation
S2 was cultured in LB media at 30 °C and shaken at 150 rpm for 16 h. The DNA was extracted as described previously. The concentration of extracts was detected using NanoDrop 2000 (Thermo Scientific, Waltham, MA, USA), and the purity and integrity were assessed using a Qubit fluorometer (Thermo Scientific, USA). The sequencing library was constructed by BGI Company (Shenzhen, China) using a Nanopore sequencing platform (Oxford Nanopore Technologies, Oxford, UK). The fragment size and library concentration were determined using an Agilent 2100 Bioanalyzer. Functional genes in the nitrogen metabolism pathways of S2 were investigated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
2.4. Factors Affecting N Removal Capacity
After being enriched with LB media for 15 h, S2 was centrifuged at 8000 rpm for 10 min to obtain bacterial pellets. The pellets were resuspended in sterilized NaCl (0.9%) to obtain the experimental bacterial suspensions (OD600 = 0.8). A 3 mL bacterial suspension was added to 150 mL HNM and cultured for up to 72 h at 30 °C and 150 rpm to detect the nitrification capacity. Samples were taken at different times (0 h, 4 h, 8 h, 12 h, 1 day, 2 days, and 3 days) to measure the OD600 value and nitrogen concentration (NH4+-N, NO2−-N, and NO3−-N). Next, to determine the aerobic denitrification capacity of S2, experiments were conducted through S2 incubation in HNM media with (NH4)2SO4 replaced by NaNO2 or KNO3. S2 was also cultured in HNM with mixed nitrogen sources (with the addition of 100 mg L−1 of (NH4)2SO4, NaNO2, and KNO3) to detect the simultaneous nitrification and denitrification capacity. Additionally, experiments were performed to identify the factors affecting ammonium removal efficiency. Sodium succinate in HNM was substituted by glucose, sucrose, sodium acetate, and sodium citrate to explore the impact of the carbon sources on S2. Based on the HNM medium, the C/N ratio was set to 4, 8, 12, 16, and 20, while the shaking speeds were adjusted from 50 to 250 rpm to explore the effects of the C/N ratio and shaking speed. Furthermore, the initial pH was set at 5, 6, 7, 8, 9, and 10 to examine the effect of pH. The temperatures were varied from 20 to 40 °C to assess the effect of temperature. Lastly, the initial NH4+-N concentrations were set to 5, 10, 20, 100, and 300 mg L−1 to explore the tolerance of S2 to high ammonium concentrations. The variables, other than those mentioned, were kept the same throughout the experiment.
2.5. Nitrogen Balance Experiment
During this experiment, 0.6 mL of bacterial suspension was inoculated into 30 mL HNM in 60 mL vials to prepare the samples for N2 determination and nitrogen balance during the ammonium removal process. Sodium acetate was used as the sole carbon source, and the initial C/N ratio, pH, and DO concentration were 16, 9, and 6.2 mg L−1, respectively. The initial NH4+-N concentration was set at 100 mg L−1. The vial was sealed and incubated for 24 h at 35 °C and 200 rpm. Approximately 1 mL of ZnCl2 (50%, w/v) was added to the vial to end the reaction at 0 and 24 h, respectively. The N2 production in the vials was determined using a gas chromatography (GC) system (Agilent 7890B, Agilent, Technologies, Lexington, MA, USA). Some parallel vials were prepared to determine the nitrogen balance in this reaction system, and cultures were collected at 0 h and 24 h to determine the TN concentration. Biomass nitrogen was calculated by deducting the TN of cultures following the filter (0.22 μm, Jinteng, China) from the TN of unfiltered cultures.
2.6. Nitrogen Removal Performance of S2 on Sewage
The sewage sample was taken from the Guanlan Sewage Treatment Plant in Shenzhen, China. To examine the nitrogen removal performance of S2 on sewage, the bacterium was cultured in a 1 L flask with a 500 mL sewage sample at a 2% inoculation ratio. The initial pH of the sewage was 7.44. Sodium acetate was added to achieve an initial C/N ratio of 16. The flasks were shaken at 200 rpm at 35 °C for up to 18 days. Samples were collected at different time points and centrifuged, and the supernatants were analyzed for NH4+-N, NO2−-N, NO3−-N, and TN. Additionally, sewage samples (500 mL) without S2 inoculation, as well as sterilized sewage samples (500 mL) with S2 inoculation incubated under the same conditions, served as controls.
2.7. Analytical Methods
The growth trend of S2 was measured using a multifunctional microplate reader (Lx, BioTek, VT, USA) at 600 nm. The concentrations of NH4+-N, NO2−-N, NO3−-N, and TN were calculated with reference to APHA’s methods by using Berthelot colorimetry, N-(1-naphthalene)-diaminoethane dihydrochloride, sulfamic acid, and alkaline potassium persulphate methods, respectively [19]. Biomass nitrogen was tested using Roun et al.’s method [20], deducting the TN of inoculated media following the filter (0.22 μm, Jinteng, China) from the TN of the unfiltered samples.
3. Results and Discussion
3.1. Isolation and Identification of Strain S2
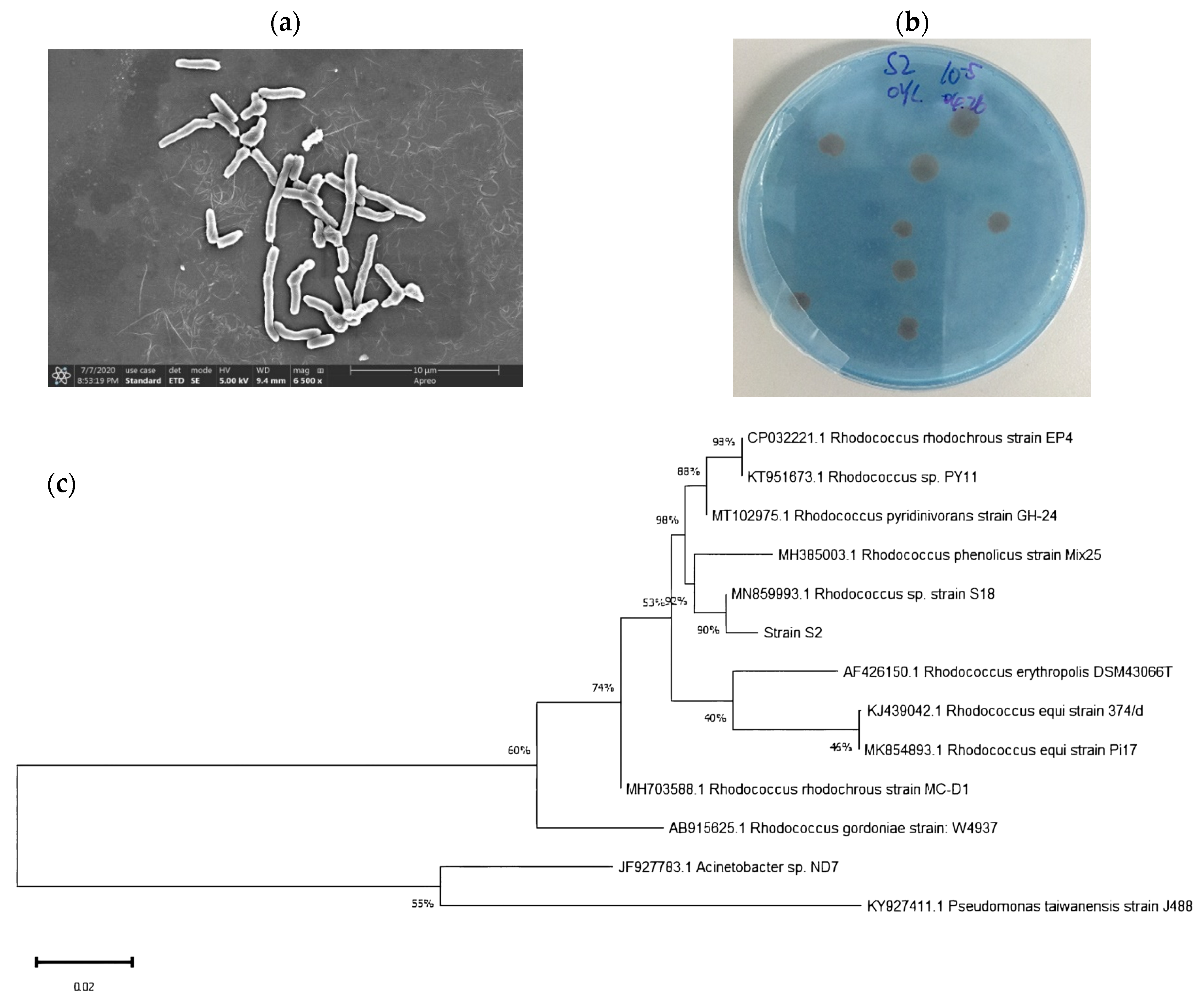
Strain S2 appeared as a rod-shaped bacterium (Figure 1a) with reddish brown, circular-shaped colonies, a rough surface, and complete edges (Figure 1b). Moreover, a 1322 bp 16S rRNA gene sequence was amplified and obtained by means of PCR. The BLAST results indicated 100% homology between the strain S2 and Rhodococcus sp. S18 (MN859993.1), and a phylogenetic tree constructed using the neighbor-joining method confirmed the identification of strain S2 as Rhodococcus sp. (Figure 1c).
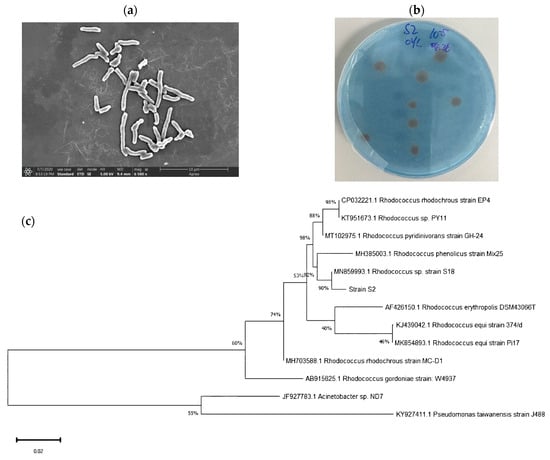
Figure 1.
Scanning electron micrograph (6500×) (a), the colony on the BTB plate (b), and the phylogenetic characteristics of Rhodococcus sp. S2 (c).
Some researchers have reported that Rhodococcus sp., a Gram-positive bacterium, could be used in bioremediate environmental pollutants [21,22,23]. It has been found to exhibit impressive tolerance to various environments such as coking wastewater [24], Antarctic sea ice [25], and polluted soil [26]. However, only a few studies have confirmed the HN–AD capability of Rhodococcus sp. Previously, Rhodococcus erythropolis strain Y10 isolated from surface flow constructed wetlands (SFCWs) was proven to have heterotrophic ammonium assimilation and aerobic denitrification capacity [27]. Rhodococcus sp. CPZ 24 isolated from swine wastewater also performed simultaneous nitrification and denitrification [28]. The isolation of Rhodococcus sp. S2 in this study extends the important role of Rhodococcus sp. in the nitrogen removal process.
3.2. Heterotrophic Nitrification and Aerobic Denitrification Capacity of S2
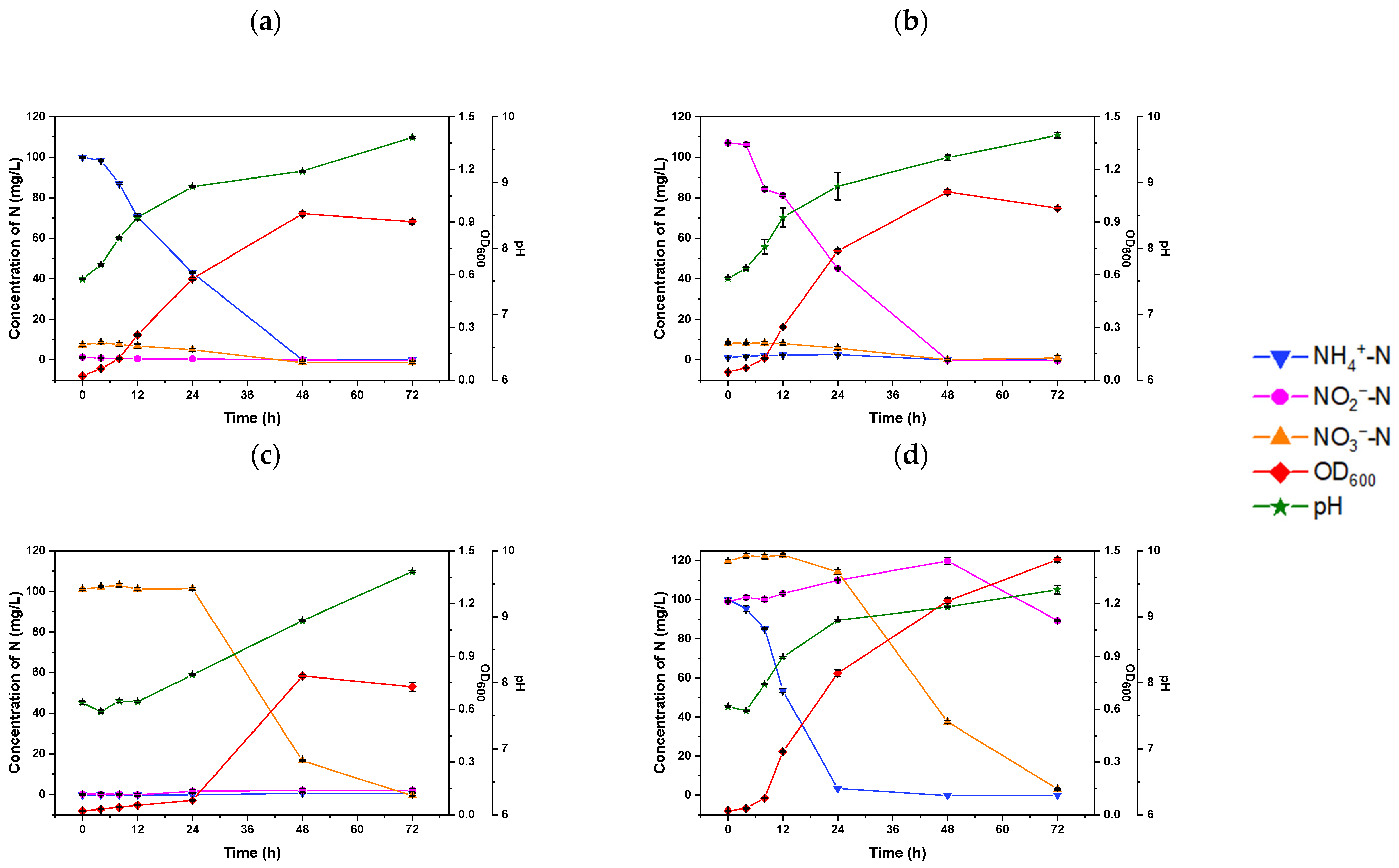
The heterotrophic nitrification capacity of strain S2 was studied using (NH4)2SO4 as the sole nitrogen source. As depicted in Figure 2a, the ammonium-N removal capacity of S2 exhibited a close relationship with cell growth.
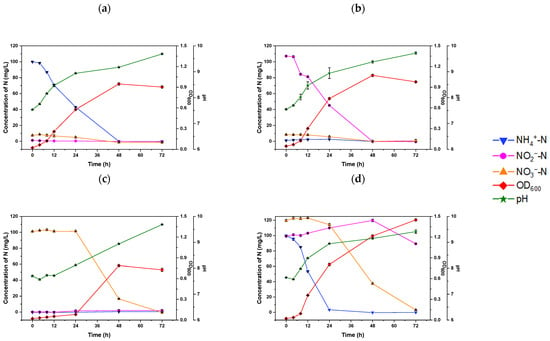
Figure 2.
The changes in nitrogen concentration in HNM-cultured Rhodococcus sp. S2 with the addition of (a) 100 mg/L NH4+-N, (b) 100 mg/L NO2−-N, (c) 100 mg/L NO3−-N, and (d) a mixture of N sources (100 mg/L NH4+-N, 100 mg/L NO2−-N, and 100 mg/L NO3−-N).
In the first 8 h, S2 exhibited a low ammonium-N removal rate of 1.62 ± 0.07 mg L−1h−1. However, its removal rate increased significantly to a maximum of 2.08 ± 0.03 mg L−1h−1 after 48 h of incubation, surpassing that of Bacillus sp. LY (0.43 mg L−1h−1) [14], Pesudomonas sp. ADN-42 (1.38 mg L−1h−1) [29], and Pseudomonas tolaasii Y-11 (2.04 mg L−1h−1) [30]. Although the ammonium-N removal rate of S2 was lower than Rhodococcus sp. CPZ 24 (3.10 mg L−1h−1) [28], notably, there was no significant accumulation of nitrite or nitrate during the NH4+-N removal process, indicating the effective removal of nitrogen and demonstrating the potential denitrification capacity of S2. Similarly, Ochrobactrum anthropic LJ81 [12] and Acinetobacter junii YB [31] showed no intermediate product accumulation during the ammonium removal process, with the pH gradually rising from 7.68 to 9.69 at 72 h due to alkali production in the denitrification process. This shift in pH is supported by the findings of Zhu et al., who demonstrated that minimal pH fluctuations could create a favorable environment for bacterial growth [32]. These results indicate that the S2 strain possesses excellent heterotrophic nitrification ability; a high pH value (pH > 8) could lead to the conversion of ammonium to ammonia.
To evaluate aerobic denitrification ability, the experiment was conducted using HNM, wherein (NH4)2SO4 was replaced with NaNO2 or NaNO3. As illustrated in Figure 2b,c, the bacteria in the two groups exhibited a similar growth phenomenon that was closely related to the nitrogen removal rates, albeit with significant differences between the two substrates. When S2 was cultured with nitrite as the sole nitrogen source, it displayed an OD600 of 1.11 ± 0.01 and 99.95 ± 0.14% NO2−-N removal efficiency (Figure 2b). Conversely, S2 cultured with NaNO3 experienced a 24-h lag phase, reaching its maximum biomass (OD600 = 0.79 ± 0.008) and NO3–-N removal efficiency of 83.53 ± 0.3% after 48 h (Figure 2c). These results suggest that strain S2 can utilize nitrate as a nitrogen source, although it requires more time to adapt to this condition, resulting in an extended lag phase (Figure 2c). This phenomenon has been well-documented in various HN–AD strains [33,34,35].
Moreover, the removal capacity of S2 was tested in a mixed nitrogen medium containing (NH4)2SO, NaNO2, and NaNO3. In Figure 2d, the growth rate of S2 in the mixture media surpassed that of the medium with a singular nitrogen source, with it entering the logarithmic growth phase with a maximum OD600 value of 1.45 ± 0.01 at 72 h, indicating the higher removal efficiency of ammonium-N at 24 h (96.60 ± 0.43%) (Table 1).

Table 1.
Maximum nitrogen removal efficiencies and rates of Rhodococcus sp. S2 using different nitrogen sources (C/N = 12, shaking speed of 150 rpm, initial pH of 7.5, sodium acetate as the carbon source, and incubated at 30 °C).
The NH4+-N removal rate of 4.17 ± 0.69 mg L−1 h−1 in the relevant condition was found to be higher than that in the medium containing ammonium (2.46 ± 0.13 mg L−1 h−1). Subsequently, S2 initiated the removal of NO3–-N and achieved 97.40 ± 0.37% removal efficiency at 72 h. The decrease in NH4+-N and NO3–-N resulted in an increase in NO2−-N from 99.10 ± 0.28 mg L−1 to 119.72 ± 1.72 mg L−1 at 48 h, followed by a gradual reduction to 89.30 ± 0.25 mg L−1 at 72 h. These results underline the efficient HN–AD capacity of S2 in utilizing the three nitrogen sources. An additional experiment using sealed vials was carried out to confirm the production of N2 gas and the nitrogen balance during the NH4+-N removal process. During this experiment, the TN concentration dropped from 88.77 ± 2.69 mg L−1 to 71.44 ± 0.10 mg L−1 between 0 and 24 h; however, only 2.21 ± 0.41 mg L−1 of biomass nitrogen accumulated (Table S1). These findings suggest that the majority of the N was removed as N2 gas. The proportion of N2 in the vial increased from 79.70 ± 0.26% to 89.30 ± 0.64%, further confirming S2’s aerobic denitrifying ability (Fig.S1). Overall, the results provide evidence that bacterial denitrification primarily drove ammonium removal, demonstrating S2’s high nitrogen removal capacity.
3.3. Factors Affecting the Ammonium Removal Efficiency of the S2 Strain
3.3.1. Influence of Carbon Sources
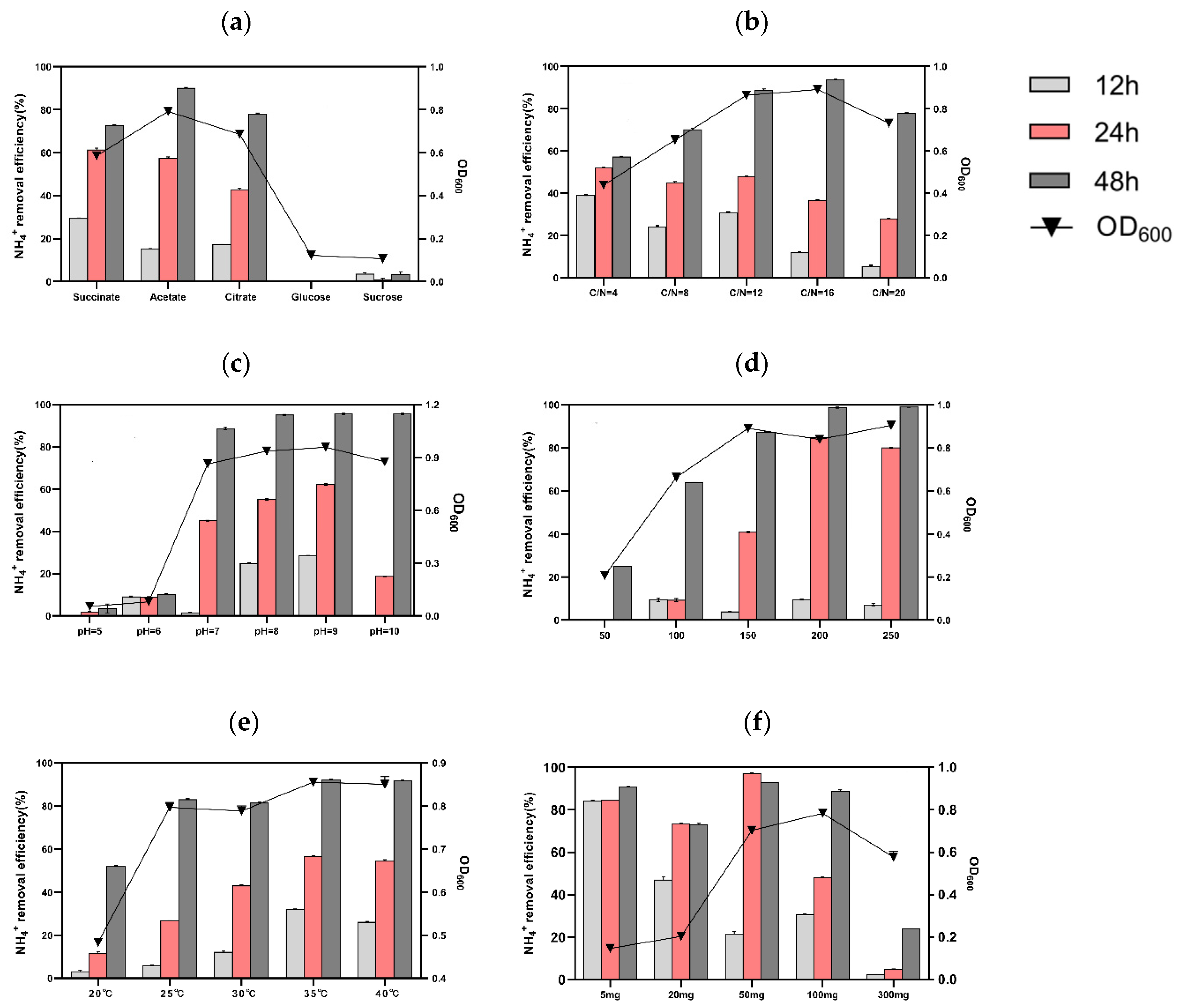
Organic carbon is consistently considered an energy resource for microorganisms to proliferate, yet different species exhibit a distinct preference for organic substrates [36]. To identify the optimal carbon source for S2, experiments were conducted with various carbon sources at a C/N ratio of 12:1 (Figure 3a). Among these, the medium containing sodium acetate exhibited the highest NH4+-N removal rate (90.05 ± 0.34%) and supported the most notable cell growth (with an OD600 peak of 0.79± 0.01) within 48 h.
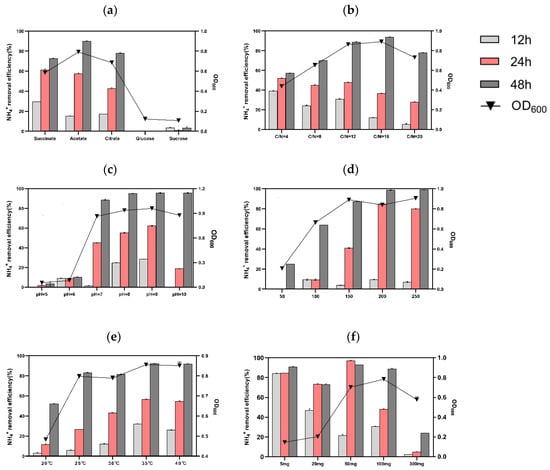
Figure 3.
Influencing factors of (a) carbon source, (b) C/N ratio, (c) pH, (d) shaking speed, (e) temperature, and (f) initial NH4+-N concentration on the growth and nitrogen removal capacity of Rhodococcus sp. S2 incubated for 12, 24, and 48 h.
It has been documented that carbon sources with a simple structure and low molecular mass may be more suitable for bacterial growth and as electron donors in the nitrification process due to their ability to readily participate in the straightforward biochemical pathway of carbon source utilization [4,17,31]. Acetate can be utilized by the majority of the known HN–AD bacteria. Considering the growth and NH4+-N removal rate, sodium acetate was selected for the subsequent experiment.
3.3.2. Carbon–Nitrogen Ratio
It is generally reported that most HN–AD bacteria require a higher carbon–nitrogen ratio (C/N) to achieve high nitrogen removal efficiency, and a low C/N ratio results in poor HN–AD performance [36]. Therefore, the NH4+-N removal efficiency of S2 was tested under different C/N ratios (Figure 3b). The results demonstrated an improvement in removal efficiency from 57.22 ± 0.28% to 93.85 ± 0.2% when the C/N ratio increased from 4 to 16. Additionally, the OD600 value rose to 0.89 ± 0.01 at a C/N ratio of 16. However, when the C/N ratio was adjusted to 20, the NH4+-N removal efficiency slightly decreased. Therefore, the optimal C/N ratio for strain S2, considering energy savings, was 16. These findings align with the high NH4+-N removal capacity exhibited by Rhodococcus erythropolis Y10 [27] and Acinetobacter indicus ZJB20129 [3] with a C/N ratio of 15.
3.3.3. Initial pH
The impact of pH on strain S2’s growth and nitrogen removal capacity is illustrated in Figure 3c. The growth of strain S2 was poor at pH 5 and 6 (OD600 values of 0.05 and 0.08, respectively), resulting in removal efficiencies of only 3.45 ± 1.31% and 10.23 ± 0.28% within 48 h. In contrast, at pH levels between 7 and 10, the OD600 values were 0.86, 0.94, 0.96, and 0.86, with respective maximum removal efficiencies reaching up to 88.67 ± 0.18%, 95.03 ± 0.29%, 95.66 ± 0.16%, and 95.65 ± 0.56% within 48 h. These results indicate that strain S2 is an alkaline HN–AD bacteria, consistent with Marinobacter NNA5 [37]. However, a high pH value (>8) might result in the conversion of ammonium to ammonia.
3.3.4. Dissolved Oxygen
The influence of dissolved oxygen content on ammonium removal efficiency by S2 was investigated by varying the shaking speed, as depicted in Figure 3d. When the rotational speed was adjusted from 50 to 200 rpm, the nitrogen removal efficiency improved from 24.95 ± 0.05% to 98.76 ± 0.37%. Furthermore, the OD600 peak at 200 rpm reached 0.84 ± 0.01. Notably, at a speed of 250 rpm, the NH4+-N removal efficiency was consistent with that at 200 rpm, indicating that S2 could tolerate a high oxygen concentration, in line with findings on Acinetobacter tandoii MZ-5 [38] and Pseudomonas putida NP5 [39]. It has been reported that a higher or lower DO concentration has a negative impact on the nitrogen removal performance of HN–AD bacteria [40]. Hence, the shaking speed of 200 rpm was conducive to faster growth and a more efficient ammonium removal rate.
3.3.5. Temperature
The susceptibility of HN–AD bacteria to temperature variation, affecting cell membrane fluidity and enzyme activity, was reported in one example study [11]. In this study, we investigated the influence of temperature on strain S2, and the experimental results are presented in Figure 3e. It is evident that strain S2 exhibited a temperature-dependent response within the nitrogen removal range of 20–35 °C. At 20 °C, the maximum removal efficiency and OD600 value were approximately 52.73% ± 0.18 and 0.482 ± 0.01 within 48 h, demonstrating that the low temperature primarily slowed down microorganisms’ propagation, leading to a reduction in ammonium removal efficiency. Conversely, at 35 °C, the OD600 and maximum removal efficiency reached 0.86 ± 0.01 and 92.5% ± 0.18, respectively. Interestingly, at 40 °C, the NH4+-N removal capacity of S2 was lower than at 35 °C (27.47% ± 0.31 vs. 35.49% ± 0.18 at 12 h). These results indicate that the optimal culture temperature of S2 was 35 °C, consistent with the mesophilic nature of microorganisms [17].
3.3.6. Initial NH4+-N Concentration
The nitrogen removal capacity of strain S2 was assessed at initial NH4+-N concentrations ranging from 5–300 mg/L, as shown in Figure 3f. The cell growth demonstrated a gradual increase with rising NH4+-N concentration within the range of 5–100 mg L−1. Notably, at 24 h, when the initial NH4+-N concentrations were set to 5, 20, and 50 mg/L, the nitrogen removal efficiencies were 84.47% ± 0.12, 73.36% ± 0.26, and 97.05% ± 0.49, respectively. Conversely, under the conditions of 100 and 300 mg/L NH4+-N, the maximum removal efficiencies for 48 h were 88.67% ± 0.65 and 23.93% ± 0.12, with OD600 values reaching 0.78 ± 0.01 and 0.58 ± 0.02, respectively, suggesting that high concentrations of ammonium nitrogen inhibited cell growth and nitrogen removal performance. These findings align with previous reports indicating that ammonium becomes an inhibitor above a certain threshold, hindering microorganism growth during wastewater treatment [12,41]. Our results also concur with Acinetobacter tandoii MZ-5, which displayed robust growth at a wide range of NH4+-N concentrations [38], suggesting the potential applicability of strain S2 in wastewater treatment.
3.4. Genome Sequencing and KEGG Pathway Analysis
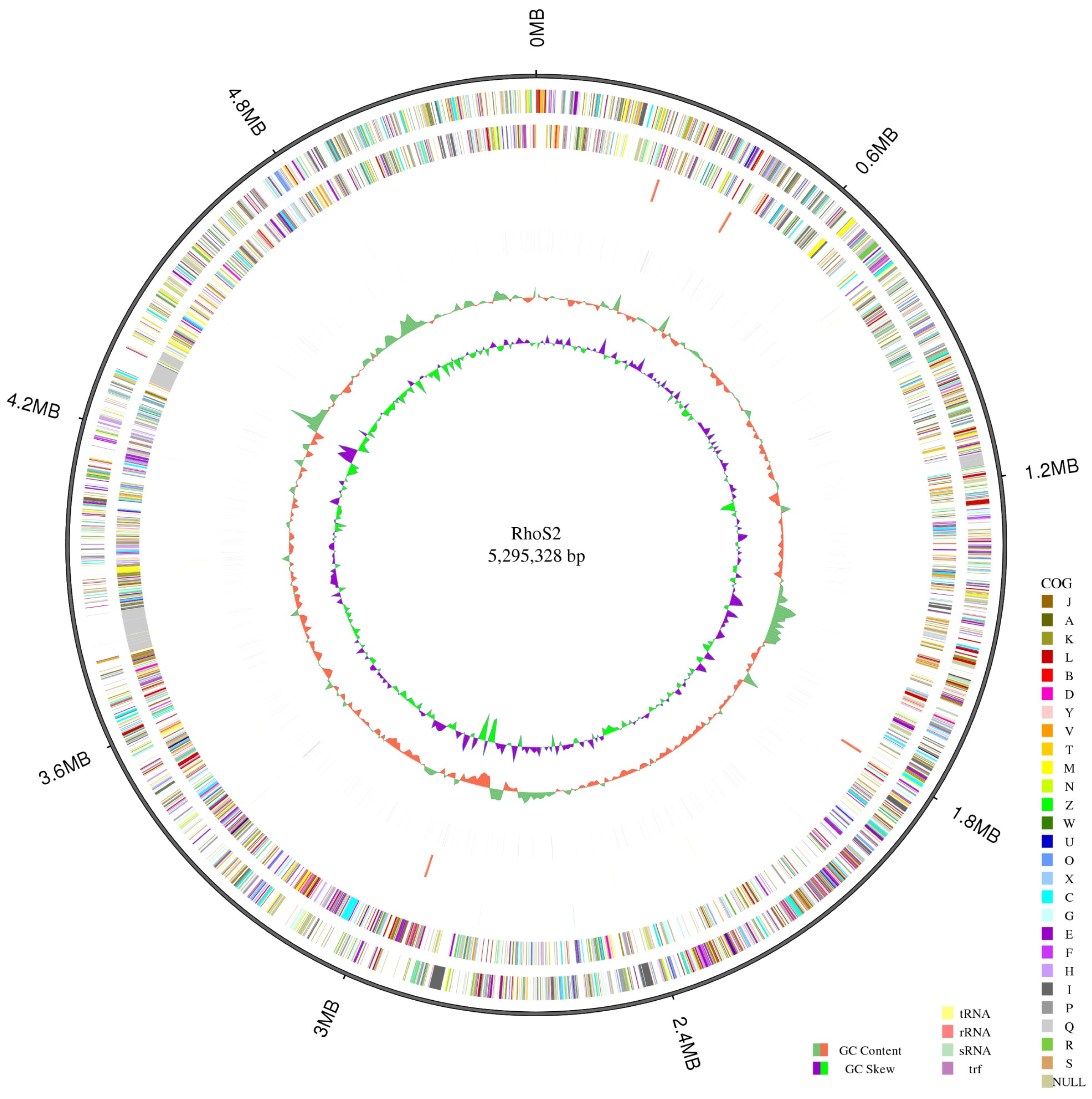
The length of strain S2’s genome is 5,295,328 bp, with a GC content of 68.14%, as shown in Figure 4. Through gene prediction, the total gene length was determined to be 5,495,886 bp, with a GC content of 68.02%. Of the 5822 annotated genes, 53 were tRNA genes, and 12 were rRNA genes. The number of genes annotated in different databases varied, yielding the following results: Swiss-Prot (n = 2061), NR (n = 5516), GO (n = 3204), COG (n = 3947), and KEGG (n = 3092).
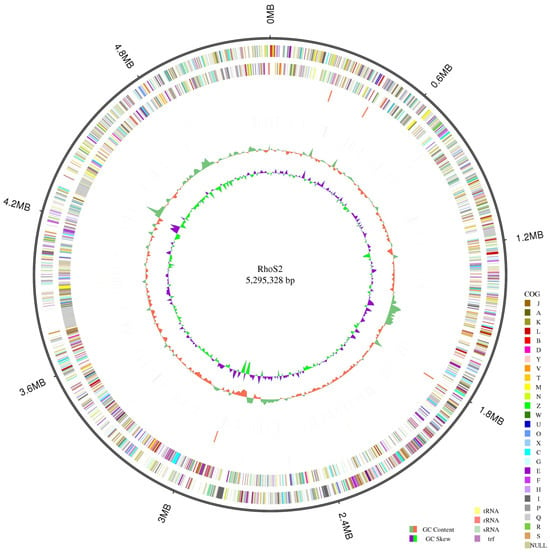
Figure 4.
Schematic genome of the Rhodococcus sp. S2. Figure note: The first circle is the genome from the outside to the inside. The second and third circles of the circle diagram are the CDS on the positive or the negative chain, colored according to a cluster of orthologous groups (COG) classification. Different colors indicate different classifications of COG functions. The fourth circle corresponds to the repetition sequence. The fifth circle is the GC content information, and the last circle is the GC skew.
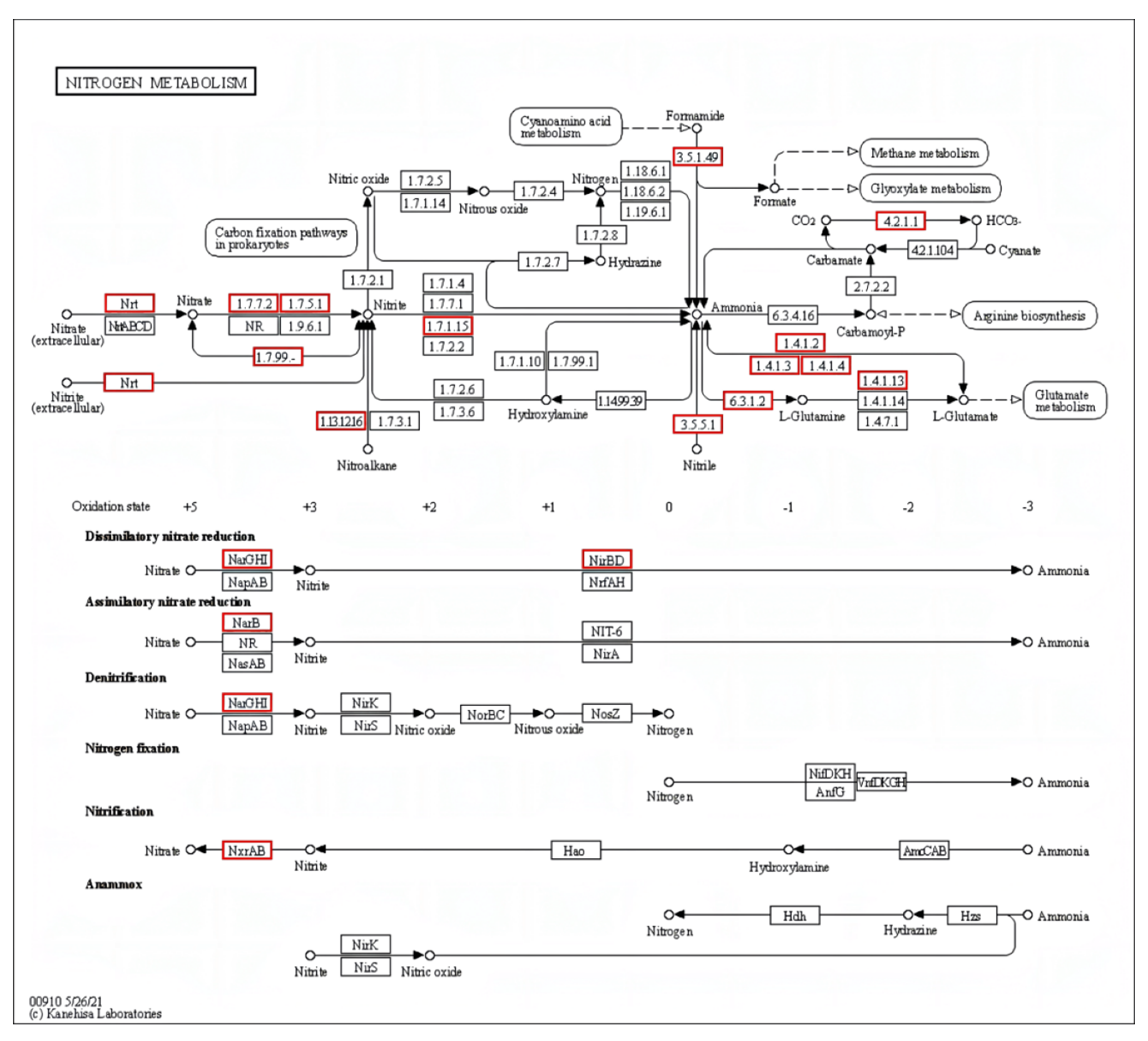
In the conventional nitrogen removal pathway for HN–AD bacteria, inorganic nitrogen compounds such as nitrite, ammonium, hydroxylamine, and nitrate are converted into nitrogen gas. Essential enzymes in the HN–AD process, including hydroxylamine oxidoreductase (Hao), nitrite reductase (NiR), ammonia monooxygenase (Amo), and nitrate reductase (NaR), were identified. Genome annotation revealed that strain S2 contained a total of nine genes related to nitrogen metabolism, including NarB (ferredoxin–nitrate reductase), NarG, NxrA, NirBD, NRT (the major facilitator superfamily transporter), ncd2 (nitronate monooxygenase), GDH2 (glutamate dehydrogenase), and gltD (glutamate synthase (NADPH) small chain) (Figure 5).
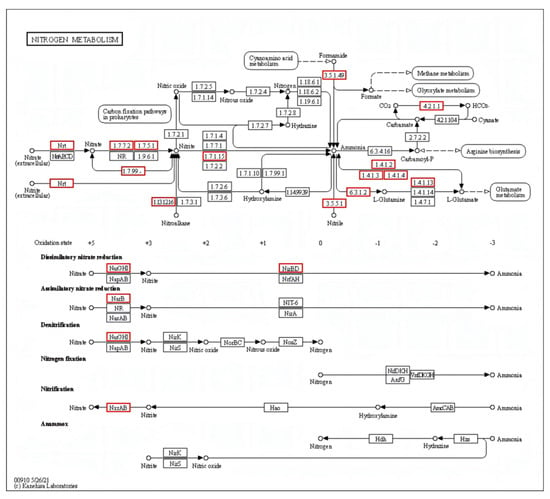
Figure 5.
Key genes of S2 in the nitrogen metabolism pathway annotated in the KEGG database. The red boxes indicate the existence of genes in the nitrogen metabolism pathway.
The major facilitator superfamily transporter, responsible for the transport of nitrate and nitrite in the biofilm, is encoded by the NRT gene [42]. The genes ncd2 and NarB control the enzymes nitronate monooxygenase and ferredoxin–nitrate reductase, respectively. Gene gltD encodes glutamine synthetase (GS), which converts ammonia into glutamine for assimilation into the cell biomass [43]. Additionally, GDH2 encodes Glutamate dehydrogenase (GDH), which plays a crucial role in ammonium detoxification [44]. Nitrate reductases, nitrate oxidoreductase, and nitrite reductase are encoded by NarG, NxrA, and NirBD genes, respectively, and they are essential for the HN–AD nitrogen removal process.
However, genome sequencing did not annotate the Amo and Hao genes related to the HN–AD process or the key genes (NirK and NirS) involved in nitrite reductase synthesis. This result is consistent with other HN–AD bacteria, such as Pseudomonas taiwanensis J488 (with only the NirB gene mapped) [45] and Klebsiella pneumoniae EGD-HP19-C (with only NR genes annotated) [46]. The inability to map some key genes could be attributed to two factors. First, the new genes associated with the HN–AD process in strain S2 may differ from those found in traditional HN–AD bacteria. The second factor may be the incomplete genome draft. Our study showed that strain S2 exhibited efficient ammonium, nitrite, and nitrate removal capacity, producing N2 when using (NH4)2SO4 as the sole nitrogen source, thus confirming its classification as an HN–AD bacterium (results shown in Figure 2 and Figure S1).
3.5. Nitrogen Removal Capacity of Strain S2 in Raw Wastewater
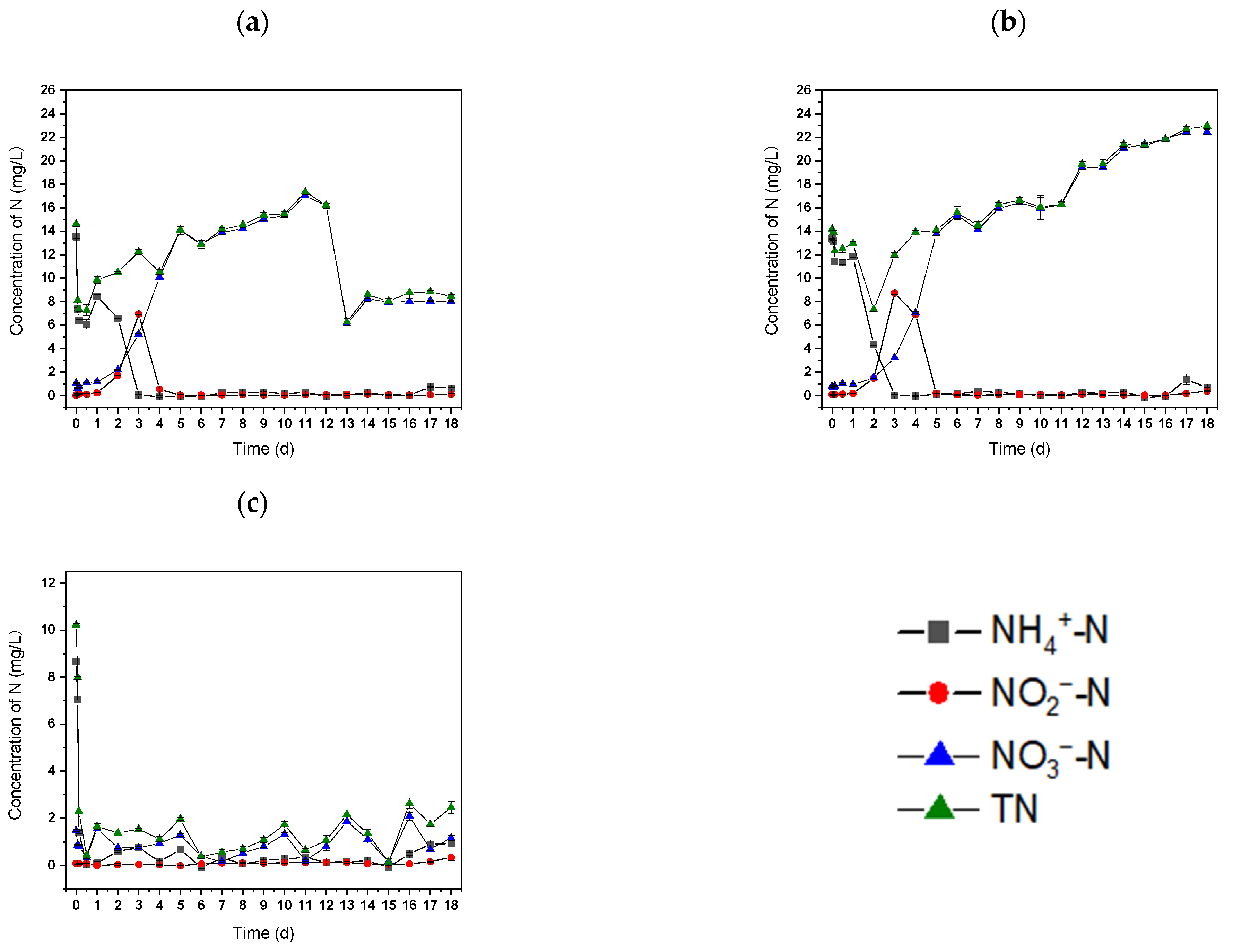
The ability of strain S2 to treat wastewater was evaluated. The NH4+-N content in raw wastewater decreased from 13.51 ± 0.13 to 6.08 ± 0.41 mg L−1 within 12 h when strain S2 and sodium acetate were added, resulting in NH4+-N removal efficiency of 55.29 ± 0.43% (Figure 6a).
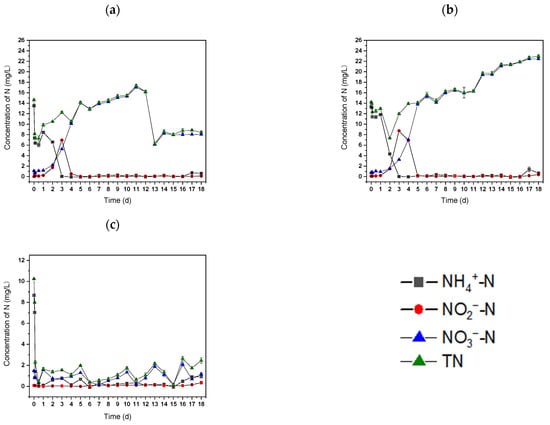
Figure 6.
The N concentrations in (a) raw wastewater with Rhodococcus sp. S2, (b) raw wastewater, and (c) sterilized wastewater with Rhodococcus sp. S2.
However, only 14.83 ± 0.20% of NH4+-N was eliminated in the control group (raw wastewater) (Figure 6b), indicating that the addition of strain S2 and sodium acetate would enhance NH4+-N removal from the wastewater. These results validate the substantial NH4+-N removal capability of strain S2 in raw wastewater. In comparison, sterilized wastewater, with the inclusion of strain S2 and sodium acetate, exhibited 99.79 ± 0.45% NH4+-N removal efficiency within 12 h (Figure 6c). A relatively lower NH4+-N removal efficiency from wastewater as compared to sterilized wastewater may be attributed to the complex composition of raw wastewater and the presence of indigenous microbial groups, which could interfere with the metabolic activity of strain S2. Afterward, the NH4+-N concentration decreased to 0.05 ± 0.06 mg L−1 on day 3 (Figure 6a). During this period, NH4+-N was oxidized to NO2−-N and finally converted to NO3−-N. This process was mainly caused by the nitrification of autotrophic bacteria in the raw wastewater. The same phenomenon was observed in the control group (Figure 6b) from day 0 to day 5. This further suggests that the nitrification process is caused by indigenous autotrophic nitrifiers in raw wastewater. Furthermore, the introduction of additional sodium acetate on day 12 led to a decrease in NO3--N content down to 6.12 ± 0.14 mg L−1 in the experimental group (Figure 6a). This result further confirms the aerobic denitrification capacity of S2. In the control group, the NO3--N concentration increased gradually afterward, which may have been caused by the transformation of other forms of nitrogen in the culture process, indicating the significant contribution of indigenous microbial populations to nutrient transformation. Slight increases in NH4+-N were observed at the end of the experiment, which was probably mainly due to the release of NH4+-N from the dead bacteria after the exhaustion of organic matter [47]. Another explanation could possibly be the inhibition of the free ammonia produced at elevated pH caused by denitrification [40]. The maximum TN removal efficiency reached 57.16 ± 0.52% in the experimental group (Figure 6a). Overall, these findings suggest that strain S2 may possess significant nitrogen removal capacity when a carbon source is present in wastewater. The successful isolation and characterization of strain S2 from landfill leachate could offer a promising foundation for future in situ biotechnological treatment of landfill leachate. This strain may be readily adapted to the contaminated environment. The enhanced in situ indigenous HN–AD offers potential advantages for the bioremediation of contaminated sites and wastewater. To reduce carbon source demand and further improve nitrogen removal capabilities in practical wastewater treatment, biochar or other carrier-immobilized bacteria [48] or bacterial consortiums with HN–AD capability are therefore worth future exploration.
4. Conclusions
In this study, Rhodococcus sp. S2, isolated from landfill leachate, demonstrated efficient HN–AD capacity. The maximum removal efficiency for NH4+-N, NO2−-N, and NO3−-N was 99.97 ± 0.3%, 99.79 ± 0.14%, and 83.53 ± 0.31%, respectively. Additionally, the preferential removal of NH4+-N in the mixed nitrogen source and the capacity for simultaneous nitrification and denitrification were observed. The critical genes of Rhodococcus sp. S2, namely NarG, NxrA, and NirBD, were ascertained through draft genome sequencing, further confirming its HN–AD capability. Under the experimental conditions, Rhodococcus sp. S2 displayed high nitrogen removal performance when used in the treatment of raw wastewater with the addition of a carbon source. These results indicate the potential candidacy of Rhodococcus sp. S2 for wastewater nitrogen removal. Subsequent studies focusing on the application of S2 in real raw wastewater without a carbon source, with large-scale experiments, or the progressive complexity of the reaction medium could be pursued.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/w16030431/s1, Figure S1. The composition of gas in gas-tight vials cultivated S2 at 0 h and 24 h. Values are means ± se, n = 3. Table S1. The nitrogen balance of strain S2 during the NH4+-N removal process.
Author Contributions
X.C., investigation, methodology, formal analysis, and writing—original draft; S.L. (Shuangfei Li), conceptualization, project administration, and supervision; W.Z., investigation and methodology; S.L. (Shaofeng Li), funding acquisition and writing—review and editing; Y.G., writing—review and editing; L.O., conceptualization, funding acquisition, and writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Scientific Research Startup Fund for Shenzhen High-Caliber Personnel of SZPT (No. 6022310040K), the Post-doctoral Later-stage Foundation Project of Shenzhen Polytechnic (6021271015K1), the Shenzhen Science and Technology Program (KCXFZ20201221173203010), the Shenzhen Science and Technology Program (KCXST20221021111206015 and KCXFZ20201221173404012), the National Natural Science Foundation of China (42107396), and the Shenzhen Science and Technology Program (JCYJ20220531094416037).
Data Availability Statement
Some or all data, models, or codes that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to have influenced the work reported in this paper.
References
- Wang, Y.; Zou, Y.L.; Chen, H.; Lv, Y.K. Nitrate removal performances of a new aerobic denitrifier, Acinetobacter haemolyticus ZYL, isolated from domestic wastewater. Bioprocess. Biosyst. Eng. 2021, 44, 391–401. [Google Scholar] [CrossRef]
- Huang, F.; Pan, L.; Lv, N.; Tang, X. Characterization of novel Bacillus strain N31 from mariculture water capable of halophilic heterotrophic nitrification-aerobic denitrification. J. Biosci. Bioeng. 2017, 124, 564–571. [Google Scholar] [CrossRef]
- Ke, X.; Liu, C.; Tang, S.Q.; Guo, T.T.; Pan, L.; Xue, Y.P.; Zheng, Y.G. Characterization of Acinetobacter indicus ZJB20129 for heterotrophic nitrification and aerobic denitrification isolated from an urban sewage treatment plant. Bioresour. Technol. 2022, 347, 126423. [Google Scholar] [CrossRef]
- Xia, L.; Li, X.; Fan, W.; Wang, J. Heterotrophic nitrification and aerobic denitrification by a novel Acinetobacter sp. ND7 isolated from municipal activated sludge. Bioresour. Technol. 2020, 301, 122749. [Google Scholar] [CrossRef]
- Chen, J.; Gu, S.; Hao, H.; Chen, J. Characteristics and metabolic pathway of Alcaligenes sp. TB for simultaneous heterotrophic nitrification-aerobic denitrification. Appl. Microbiol. Biotechnol. 2016, 100, 9787–9794. [Google Scholar] [CrossRef]
- Li, D.; Liang, X.; Jin, Y.; Wu, C.; Zhou, R. Isolation and Nitrogen Removal Characteristics of an Aerobic Heterotrophic Nitrifying-Denitrifying Bacterium, Klebsiella sp. TN-10. Appl. Biochem. Biotechnol. 2019, 188, 540–554. [Google Scholar] [CrossRef]
- Feng, L.; Yang, J.; Ma, F.; Pi, S.; Xing, L.; Li, A. Characterisation of Pseudomonas stutzeri T13 for aerobic denitrification: Stoichiometry and reaction kinetics. Sci. Total Environ. 2020, 717, 135181. [Google Scholar] [CrossRef]
- Lang, X.; Li, Q.; Ji, M.; Yan, G.; Guo, S. Isolation and niche characteristics in simultaneous nitrification and denitrification application of an aerobic denitrifier, Acinetobacter sp. YS2. Bioresour. Technol. 2020, 302, 122799. [Google Scholar] [CrossRef]
- Kundu, P.; Pramanik, A.; Dasgupta, A.; Mukherjee, S.; Mukherjee, J. Simultaneous heterotrophic nitrification and aerobic denitrification by Chryseobacterium sp. R31 isolated from abattoir wastewater. Biomed. Res. Int. 2014, 2014, 436056. [Google Scholar] [CrossRef]
- Wang, Q.; He, J. Complete nitrogen removal via simultaneous nitrification and denitrification by a novel phosphate accumulating Thauera sp. strain SND5. Water Res. 2020, 185, 116300. [Google Scholar] [CrossRef]
- Zou, S.; Yao, S.; Ni, J. High-efficient nitrogen removal by coupling enriched autotrophic-nitrification and aerobic-denitrification consortiums at cold temperature. Bioresour. Technol. 2014, 161, 288–296. [Google Scholar] [CrossRef]
- Lei, X.; Jia, Y.; Chen, Y.; Hu, Y. Simultaneous nitrification and denitrification without nitrite accumulation by a novel isolated Ochrobactrum anthropic LJ81. Bioresour. Technol. 2019, 272, 442–450. [Google Scholar] [CrossRef]
- He, T.; Xie, D.; Li, Z.; Ni, J.; Sun, Q. Ammonium stimulates nitrate reduction during simultaneous nitrification and denitrification process by Arthrobacter arilaitensis Y-10. Bioresour. Technol. 2017, 239, 66–73. [Google Scholar] [CrossRef]
- Zhao, B.; He, Y.L.; Zhang, X.F. Nitrogen removal capability through simultaneous heterotrophic nitrification and aerobic denitrification by Bacillus sp. LY. Environ. Technol. 2010, 31, 409–416. [Google Scholar] [CrossRef]
- Sun, Y.; Li, A.; Zhang, X.; Ma, F. Regulation of dissolved oxygen from accumulated nitrite during the heterotrophic nitrification and aerobic denitrification of Pseudomonas stutzeri T13. Appl. Microbiol. Biotechnol. 2015, 99, 3243–3248. [Google Scholar] [CrossRef]
- Soto, O.; Aspé, E.; Roeckel, M. Kinetics of cross-inhibited denitrification of a high load wastewater. Enzym. Microb. Technol. 2007, 40, 1627–1634. [Google Scholar] [CrossRef]
- Song, T.; Zhang, X.; Li, J.; Wu, X.; Feng, H.; Dong, W. A review of research progress of heterotrophic nitrification and aerobic denitrification microorganisms (HNADMs). Sci. Total Environ. 2021, 801, 149319. [Google Scholar] [CrossRef]
- Wei, R.; Hui, C.; Zhang, Y.; Jiang, H.; Zhao, Y.; Du, L. Nitrogen removal characteristics and predicted conversion pathways of a heterotrophic nitrification-aerobic denitrification bacterium, Pseudomonas aeruginosa P-1. Environ. Sci. Pollut. Res. Int. 2021, 28, 7503–7514. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Wastewater, 21st ed.; American Public Health Association: Washington, DC, USA, 2005. [Google Scholar] [CrossRef]
- Rout, P.R.; Bhunia, P.; Dash, R.R. Simultaneous removal of nitrogen and phosphorous from domestic wastewater using Bacillus cereus GS-5 strain exhibiting heterotrophic nitrification, aerobic denitrification and denitrifying phosphorous removal. Bioresour. Technol. 2017, 244, 484–495. [Google Scholar] [CrossRef]
- Yao, N.H.; Du, Y.N.; Xiong, J.X.; Xiao, Y.; He, H.H.; Xie, Z.F.; Huang, D.; Song, Q.; Chen, J.; Yan, D.; et al. Microbial detoxification of 3,5-xylenol via a novel process with sequential methyl oxidation by Rhodococcus sp. CHJ602. Environ. Res. 2023, 220, 115258. [Google Scholar] [CrossRef]
- Ye, X.; Peng, T.; Feng, J.; Yang, Q.; Pratush, A.; Xiong, G.; Huang, T.; Hu, Z. A novel dehydrogenase 17beta-HSDx from Rhodococcus sp. P14 with potential application in bioremediation of steroids contaminated environment. J. Hazard. Mater. 2019, 362, 170–177. [Google Scholar] [CrossRef]
- Mirzakhani, E.; Nejad, F.M. Grasses and Rhodococcus erythropolis Bacteria for Bioremediation of Naturally Polluted Soils with Hydrocarbons. Chem. Eng. Technol. 2016, 39, 1731–1737. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Xiong, J.; Zhao, Z.; Chai, T. The enhancement of pyridine degradation by Rhodococcus KDPy1 in coking wastewater. FEMS Microbiol. Lett. 2019, 366, fny271. [Google Scholar] [CrossRef]
- Qu, C.; Wang, W.; Dong, J.; Wang, X.; Gao, X.; Zhang, H.; Zheng, Z.; Yin, H.; Miao, J. Complete genome sequence of Rhodococcus sp. NJ-530, a DMSP-degrading actinobacterium isolated from Antarctic sea ice. 3 Biotech. 2019, 9, 363. [Google Scholar] [CrossRef]
- Kis, A.E.; Laczi, K.; Zsiros, S.; Kos, P.; Tengolics, R.; Bounedjoum, N.; Kovacs, T.; Rakhely, G.; Perei, K. Characterization of the Rhodococcus sp. MK1 strain and its pilot application for bioremediation of diesel oil-contaminated soil. Acta Microbiol. Immunol. Hung. 2017, 64, 463–482. [Google Scholar] [CrossRef]
- Ma, S.; Huang, S.; Tian, Y.; Lu, X. Heterotrophic ammonium assimilation: An important driving force for aerobic denitrification of Rhodococcus erythropolis strain Y10. Chemosphere 2022, 291, 132910. [Google Scholar] [CrossRef]
- Chen, P.; Li, J.; Li, Q.X.; Wang, Y.; Li, S.; Ren, T.; Wang, L. Simultaneous heterotrophic nitrification and aerobic denitrification by bacterium Rhodococcus sp CPZ24. Bioresour. Technol. 2012, 116, 266–270. [Google Scholar] [CrossRef]
- Jin, R.; Liu, T.; Liu, G.; Zhou, J.; Huang, J.; Wang, A. Simultaneous heterotrophic nitrification and aerobic denitrification by the marine origin bacterium Pseudomonas sp. ADN-42. Appl. Biochem. Biotechnol. 2015, 175, 2000–2011. [Google Scholar] [CrossRef]
- He, T.; Li, Z.; Sun, Q.; Xu, Y.; Ye, Q. Heterotrophic nitrification and aerobic denitrification by Pseudomonas tolaasii Y-11 without nitrite accumulation during nitrogen conversion. Bioresour. Technol. 2016, 200, 493–499. [Google Scholar] [CrossRef]
- Ren, Y.X.; Yang, L.; Liang, X. The characteristics of a novel heterotrophic nitrifying and aerobic denitrifying bacterium, Acinetobacter junii YB. Bioresour. Technol. 2014, 171, 1–9. [Google Scholar] [CrossRef]
- Zhu, L.; Ding, W.; Feng, L.J.; Kong, Y.; Xu, J.; Xu, X.Y. Isolation of aerobic denitrifiers and characterization for their potential application in the bioremediation of oligotrophic ecosystem. Bioresour. Technol. 2012, 108, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Thomas, K.L.; Lloyd, D.; Boddy, L. Effects of oxygen, pH and nitrate concentration on denitrification by Pseudomonas species. FEMS Microbiol. Lett. 1994, 118, 181–186. [Google Scholar] [CrossRef]
- Idi, A.; Ibrahim, Z.; Mohamad, S.E.; Majid, Z.A. Biokinetics of nitrogen removal at high concentrations by Rhodobacter sphaeroides ADZ101. Int. Biodeterior. Biodegrad. 2015, 105, 245–251. [Google Scholar] [CrossRef]
- Xu, Y.; He, T.; Li, Z.; Ye, Q.; Chen, Y.; Xie, E.; Zhang, X. Nitrogen Removal Characteristics of Pseudomonas putida Y-9 Capable of Heterotrophic Nitrification and Aerobic Denitrification at Low Temperature. Biomed. Res. Int. 2017, 2017, 1429018. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Leng, J.; Zhu, J.; Zhang, K.; Zhao, J.; Wu, P.; Xing, Q.; Tang, K.; Li, X.; Hu, B. Influence mechanism of C/N ratio on heterotrophic nitrification- aerobic denitrification process. Bioresour. Technol. 2022, 343, 126116. [Google Scholar] [CrossRef]
- Liu, Y.; Ai, G.M.; Miao, L.L.; Liu, Z.P. Marinobacter strain NNA5, a newly isolated and highly efficient aerobic denitrifier with zero N2O emission. Bioresour. Technol. 2016, 206, 9–15. [Google Scholar] [CrossRef]
- Ouyang, L.; Wang, K.; Liu, X.; Wong, M.H.; Hu, Z.; Chen, H.; Yang, X.; Li, S. A study on the nitrogen removal efficacy of bacterium Acinetobacter tandoii MZ-5 from a contaminated river of Shenzhen, Guangdong Province, China. Bioresour. Technol. 2020, 315, 123888. [Google Scholar] [CrossRef]
- Yang, L.; Wang, X.H.; Cui, S.; Ren, Y.X.; Yu, J.; Chen, N.; Xiao, Q.; Guo, L.K.; Wang, R.H. Simultaneous removal of nitrogen and phosphorous by heterotrophic nitrification-aerobic denitrification of a metal resistant bacterium Pseudomonas putida strain NP5. Bioresour. Technol. 2019, 285, 121360. [Google Scholar] [CrossRef]
- Chen, L.F.; Chen, L.X.; Pan, D.; Ren, Y.L.; Zhang, J.; Zhou, B.; Lin, J.Q. Ammonium removal characteristics of Delftia tsuruhatensis SDU2 with potential application in ammonium-rich wastewater treatment. Int. J. Environ. Sci. Technol. 2023, 20, 3911–3926. [Google Scholar] [CrossRef]
- Kouba, V.; Catrysse, M.; Stryjova, H.; Jonatova, I.; Volcke, E.I.P.; Svehla, P.; Bartacek, J. The impact of influent total ammonium nitrogen concentration on nitrite-oxidizing bacteria inhibition in moving bed biofilm reactor. Water Sci. Technol. 2014, 69, 1227–1233. [Google Scholar] [CrossRef]
- Alvarez, L.; Sanchez-Hevia, D.; Sanchez, M.; Berenguer, J. A new family of nitrate/nitrite transporters involved in denitrification. Int. Microbiol. 2019, 22, 19–28. [Google Scholar] [CrossRef]
- Zhao, Y.; Gao, J.; Su, S.; Shan, X.; Li, S.; Liu, H.; Yuan, Y.; Li, H. Regulation of the activity of maize glutamate dehydrogenase by ammonium and potassium. Biosci. Biotechnol. Biochem. 2021, 85, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Xian, L.; Zhang, Y.; Cao, Y.; Wan, T.; Gong, Y.; Dai, C.; Ochieng, W.A.; Nasimiyu, A.T.; Li, W.; Liu, F. Glutamate dehydrogenase plays an important role in ammonium detoxification by submerged macrophytes. Sci. Total Environ. 2020, 722, 137859. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Chen, M.; Ding, C.; Wu, Q.; Zhang, M. Hypothermia Pseudomonas taiwanensis J488 exhibited strong tolerance capacity to high dosages of divalent metal ions during nitrogen removal process. Bioresour. Technol. 2021, 341, 125785. [Google Scholar] [CrossRef] [PubMed]
- Pal, R.R.; Khardenavis, A.A.; Purohit, H.J. Identification and monitoring of nitrification and denitrification genes in Klebsiella pneumoniae EGD-HP19-C for its ability to perform heterotrophic nitrification and aerobic denitrification. Funct. Integr. Genom. 2015, 15, 63–76. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.; Zhang, Y.; Sun, Z.; Zhang, J.; Chen, G.; Li, J. Simultaneous nitrification and denitrification by a novel isolated Pseudomonas sp. JQ-H3 using polycaprolactone as carbon source. Bioresour. Technol. 2019, 288, 121506. [Google Scholar] [CrossRef]
- Liu, T.F.; Wang, B.J.; Liu, M.; Jiang, K.Y.; Wang, L. Stutzerimonas frequens strain TF18 with superior heterotrophic nitrification-aerobic denitrification ability for the treatment of aquaculture effluent. Process Biochem. 2023, 130, 156–165. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).