Automated Detection of Cervical Carotid Artery Calcifications in Cone Beam Computed Tomographic Images Using Deep Convolutional Neural Networks
Abstract
:1. Introduction
2. Materials and Methods
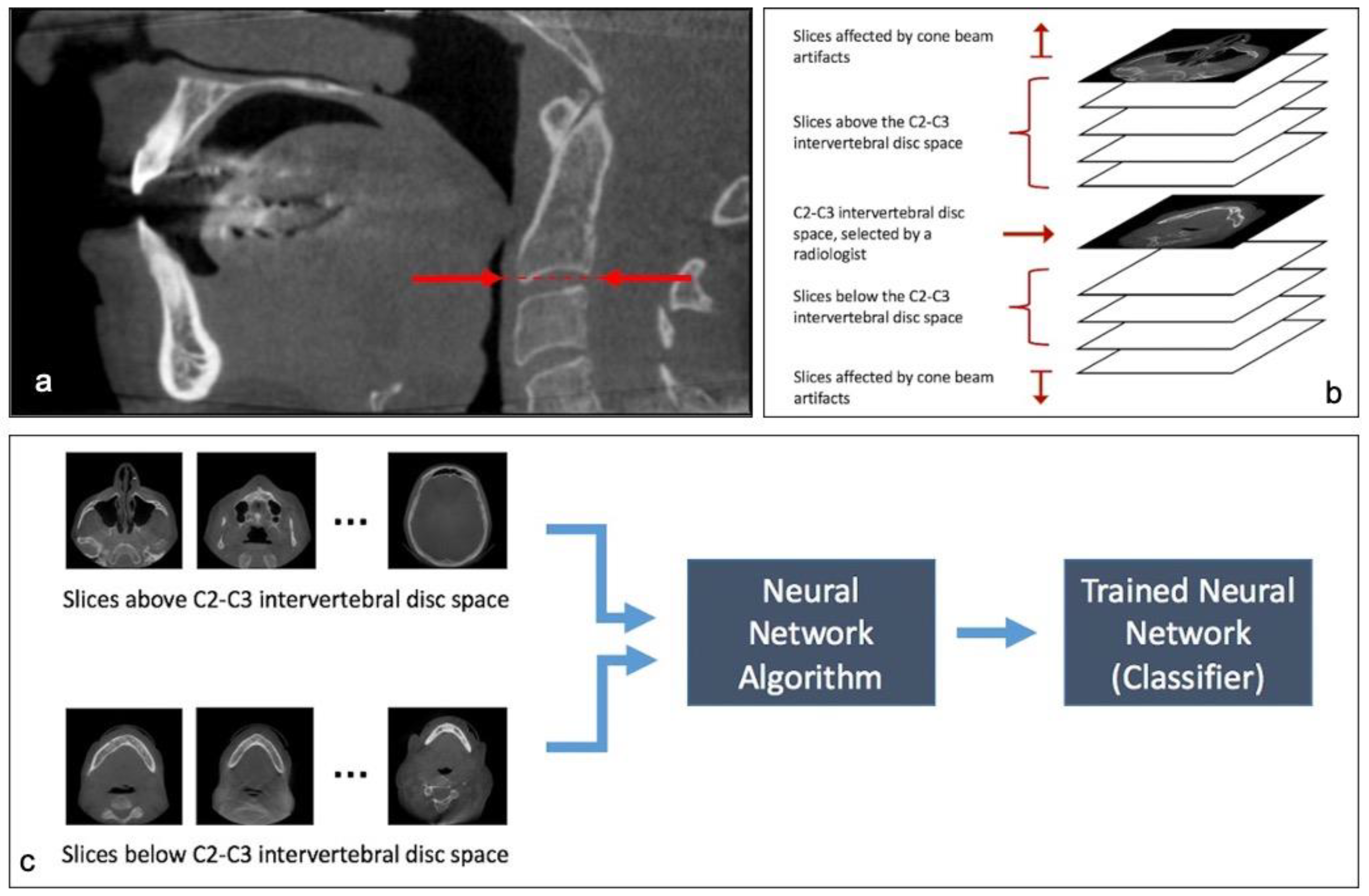
- Finding axial slices that are below the C2–C3 disc space (Classification of slices)
- Finding carotid artery calcifications in slices from step 1.
2.1. Step 1: Classification of Slices
2.2. Step 2: Detection and Localization of Calcifications
2.2.1. Method A
2.2.2. Method B
- Classifier + U-Net + Multi-patch: this is the model presented as Method A and acts as the baseline model.
- Classifier + U-Net + 2-patch: this is the model presented as first improvement from Method-A. In this case we have added broad supervision for localizing the calcification areas on patch-level.
- Classifier + U-Net + 2-patch + class weight balancing loss: this is the final improvement compared to the previous models, as in this case we have changed the loss function used to train the U-Net architecture, thereby making the architecture more robust to class-imbalance.
- Data preparation: For Method A the input to U-Net is a cropped region of 192 × 192 of the original scanned image. These patches are randomly generated and overlap between patches is allowed. However, for Method B, we crop the image from specific regions. The goal is to use human level supervision to broadly localize the calcification regions, and feed cropped regions from these localized areas as input to the network.
- Loss function: One of the major challenges of segmenting out the calcification region is that the calcification region is extremely small compared to the full size of the CBCT scan. Thus, implying that, most pixels in the input are labelled as background or non-calcification regions. This creates a class-imbalance problem while training the network.To overcome this, in Method-B we used class-weighted loss function. Essentially, we increased the penalty that is put on the network parameters for predicting the calcification region incorrectly compared to incorrectly predicting the non-calcification region. To compare the results, we used U-Nets trained in Method-A with general cross-entropy loss as baselines.
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Dental Association Council on Scientific Affairs. The use of cone-beam computed tomography in dentistry: An advisory statement from the American Dental Association Council on Scientific Affairs. Am. J. Dent. Assoc. 2012, 143, 899–902. [Google Scholar] [CrossRef] [PubMed]
- Dief, S.; Veitz-Keenan, A.; Amintavakoli, N.; McGowan, R. A systematic review on incidental findings in cone beam computed tomography (CBCT) scans. Dentomaxillofac. Radiol. 2019, 48, 20180396. [Google Scholar] [CrossRef] [PubMed]
- Mupparapu, M.; Kim, I.H. Calcified carotid artery atheroma and stroke: A systematic review. J. Am. Dent. Assoc. 2007, 138, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Nandalur, K.R.; Baskurt, E.; Hagspiel, K.D. Carotid artery calcification on CT may independently predict stroke risk. AJR Am. J. Roentgenol. 2006, 186, 547–552. [Google Scholar] [CrossRef] [Green Version]
- Schulze, R.; Friedlander, A.H. Cone beam CT incidental findings: Intracranial carotid artery calcification—A cause for concern. Dentomaxillofac. Radiol. 2013, 42, 20130347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, L. Discrimination between calcified triticeous cartilage and calcified carotid atheroma on panoramic radiography. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2000, 90, 108–110. [Google Scholar] [CrossRef] [PubMed]
- Allareddy, V.; Vincent, S.D.; Hellstein, J.W.; Qian, F.; Smoker, W.R.; Ruprecht, A. Incidental findings on cone beam computed tomography images. Int. J. Dent. 2012, 2012, 871532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, L.C. Soft tissue calcifications and ossifications. In White and Pharoah’s Oral Radiology: Principles and Interpretation, 8th ed.; Mallya, S.M., Lam, E.W.N., Eds.; Elsevier Inc.: Maryland Heights, MO, USA, 2019; pp. 607–621. [Google Scholar]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Hinton, G.E.; Osindero, S.; Teh, Y.W. A fast learning algorithm for deep belief nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.W. Adapting mask-rcnn for automatic nucleus segmentation. arXiv 2018, arXiv:1805.00500. [Google Scholar]
- de Vos, D.B.; Wolterink, J.M.; de Jong, P.A.; Leiner, T.; Viergever, M.A.; Isgum, I. ConvNet-Based Localization of Anatomical Structures in 3-D Medical Images. IEEE Trans. Med. Imaging 2017, 36, 1470–1481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lévy, D.; Arzav, J. Breast mass classification from mammograms using deep convolutional neural networks. arXiv 2016, arXiv:1612.00542. [Google Scholar]
- Zhu, W.; Liu, C.; Fan, W.; Xie, X. Deeplung: 3d deep convolutional nets for automated pulmonary nodule detection and classification. arXiv 2017, arXiv:1709.05538. [Google Scholar]
- Guo, Y.; Hao, Z.; Zhao, S.; Gong, J.; Yang, F. Artificial Intelligence in Health Care: Bibliometric Analysis. J. Med. Internet Res. 2020, 22, e18228. [Google Scholar] [CrossRef]
- Agrawal, P.; Nikhade, P. Artificial Intelligence in Dentistry: Past, Present, and Future. Cureus 2022, 14, e27405. [Google Scholar] [CrossRef]
- Ossowska, A.; Kusiak, A.; Swietlik, D. Artificial Intelligence in Dentistry-Narrative Review. Int. J. Environ. Res. Public Health 2022, 19, 3449. [Google Scholar] [CrossRef]
- Thurzo, A.; Urbanova, W.; Novak, B.; Czako, L.; Siebert, T.; Stano, P.; Marekova, S.; Fountoulaki, G.; Kosnacova, H.; Varga, I. Where Is the Artificial Intelligence Applied in Dentistry? Systematic Review and Literature Analysis. Healthcare 2022, 10, 1269. [Google Scholar] [CrossRef]
- Sawagashira, T.; Hayashi, T.; Hara, T.; Katsumata, A.; Muramatsu, C.; Zhou, X.; Iida, Y.; Katagi, K.; Fujita, H. An automatic detection method for carotid artery calcifications using top-hat filter on dental panoramic radiographs. IEICE Trans. Inf. Syst. 2013, 96, 1878–1881. [Google Scholar] [CrossRef] [Green Version]
- Bortsova, G.; Tulder, G.V.; Dubost, F.; Peng, T.; Navab, N.; Lugt, A.V.; Bos, D.; Bruijne, M.D. Segmentation of intracranial arterial calcification with deeply supervised residual dropout networks. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 11–13 September 2017; Springer: Cham, Switzerland, 2017; pp. 356–364. [Google Scholar]
- Bortsova, G.; Bos, D.; Dubost, F.; Wernooij, M.W.; Ikram, K.; Van Tulder, G.; de Bruijne, M. Automated Segmentation and Volume Measurement of Intracranial Internal Carotid Artery Calcification at Noncontrast CT. Radiol. Artif. Intell. 2021, 3, e200226. [Google Scholar] [CrossRef] [PubMed]
- Kats, L.; Vered, M.; Zlotogorski-Hurvitz, A.; Harpaz, I. Atherosclerotic carotid plaque on panoramic radiographs: Neural network detection. Int. J. Comput. Dent. 2019, 22, 163–169. [Google Scholar] [PubMed]
- Lindsey, T.; Garami, Z. Automated Stenosis Classification of Carotid Artery Sonography using Deep Neural Networks. In Proceedings of the 18th IEEE International Conference on Machine Learning and Applications (ICMLA), Boca Raton, FL, USA, 16–19 December 2019; pp. 1880–1884. [Google Scholar]
- Hyde, D.E.; Naik, S.; Habets, D.F.; Holdsworth, D.W. Cone-beam CT of the internal carotid artery. In Proceedings of the Medical Imaging 2002: Visualization, Image-Guided Procedures, and Display, SPIE 4681, San Diego, CA, USA, 23–28 February 2002. [Google Scholar]
- Yadav, S.S.; Jadhav, S.M. Deep convolutional neural network based medical image classification for disease diagnosis. J. Big Data 2019, 6, 113. [Google Scholar] [CrossRef]
- Chang, J.; Yu, J.; Han, T.; Chang, H.; Park, E. A method for classifying medical images using transfer learning: A pilot study on histopathology of breast cancer. In Proceedings of the 2017 IEEE 19th International Conference on e-Health Networking, Applications and Services (Healthcom), Dalian, China, 12–15 October 2017; pp. 1–4. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional network for biomedical image segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Devito, K.L.; de Souza Barbosa, F.; Felippe Filho, W.N. An artificial multilayer perceptron neural network for diagnosis of proximal dental caries. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2008, 106, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Ekert, T.; Krois, J.; Meinhold, L. Deep learning for the radiographic detection of apical lesions. J. Endod. 2019, 45, 917–922. [Google Scholar] [CrossRef]
- Hiraiwa, T.; Ariji, Y.; Fukuda, M. A deep-learning artificial intelligence system for assessment of root morphology of the mandibular first molar on panoramic radiography. Dentomaxillofac. Radiol. 2019, 48, 20180218. [Google Scholar] [CrossRef]
- Hwang, J.J.; Jung, Y.H.; Cho, B.H.; Heo, M.S. An overview of deep learning in the field of dentistry. Imaging Sci. Dent. 2019, 49, 1–7. [Google Scholar] [CrossRef]
- Karimian, N.; Shahidi Salehi, H.; Mahdian, M.; Alnajjar, H.; Tadinada, A. Deep learning classifier with optical coherence tomography images for early dental caries detection. In Proceedings of the SPIE 10473, Lasers in Dentistry XXIV, San Francisco, CA, USA, 27 January–8 February 2018; Rechmann, P., Fried, D., Eds.; SPIE: San Francisco, CA, USA, 2018. [Google Scholar]
- Orhan, K.; Bayrakdar, I.S.; Ezhov, M.; Kravtsov, A.; Özyürek, T. Evaluation of artificial intelligence for detecting periapical pathosis on cone-beam computed tomography scans. Int. Endod. J. 2020, 53, 680–689. [Google Scholar] [CrossRef]
- Setzer, F.C.; Shi, K.J.; Zhang, Z.; Yan, H.; Yoon, H.; Mupparapu, M.; Li, J. Artificial Intelligence for the Computer-aided Detection of Periapical Lesions in Cone beam Computed Tomographic Images. J. Endod. 2020, 46, 987–993. [Google Scholar] [CrossRef]
- Xu, W.; Yang, X.; Li, Y.; Jiang, G.; Jia, S.; Gong, Z.; Mao, Y.; Zhang, S.; Teng, Y.; Zhu, J.; et al. Deep Learning-Based Automated Detection of Arterial Vessel Wall and Plaque on Magnetic Resonance Vessel Wall Images. Front. Neurosci. 2022, 16, 888814. [Google Scholar] [CrossRef] [PubMed]







| Slices with CAC | Slices without CAC | |
|---|---|---|
| Total number of slices | 189 | 3816 |
| Neural network detected CAC | 178 (TP) | 135 (FP) |
| Neural network did not detect CAC | 11 (FN) | 3681 (TN) |
| Model | Mean IoU |
|---|---|
| Classifier + U-Net + Multi-patch (Method A) | 0.7626 |
| Classifier + U-Net + 2-patch | 0.7975 |
| Classifier + U-Net + 2-patch + class weight balancing loss (Method B) | 0.8251 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ajami, M.; Tripathi, P.; Ling, H.; Mahdian, M. Automated Detection of Cervical Carotid Artery Calcifications in Cone Beam Computed Tomographic Images Using Deep Convolutional Neural Networks. Diagnostics 2022, 12, 2537. https://doi.org/10.3390/diagnostics12102537
Ajami M, Tripathi P, Ling H, Mahdian M. Automated Detection of Cervical Carotid Artery Calcifications in Cone Beam Computed Tomographic Images Using Deep Convolutional Neural Networks. Diagnostics. 2022; 12(10):2537. https://doi.org/10.3390/diagnostics12102537
Chicago/Turabian StyleAjami, Maryam, Pavani Tripathi, Haibin Ling, and Mina Mahdian. 2022. "Automated Detection of Cervical Carotid Artery Calcifications in Cone Beam Computed Tomographic Images Using Deep Convolutional Neural Networks" Diagnostics 12, no. 10: 2537. https://doi.org/10.3390/diagnostics12102537
APA StyleAjami, M., Tripathi, P., Ling, H., & Mahdian, M. (2022). Automated Detection of Cervical Carotid Artery Calcifications in Cone Beam Computed Tomographic Images Using Deep Convolutional Neural Networks. Diagnostics, 12(10), 2537. https://doi.org/10.3390/diagnostics12102537







