Single Nucleotide Polymorphisms in XMN1-HBG2, HBS1L-MYB, and BCL11A and Their Relation to High Fetal Hemoglobin Levels That Alleviate Anemia
Abstract
:1. Introduction
2. Fetal Hemoglobin
3. Polymorphisms Regulate the Expression of Fetal Hemoglobin
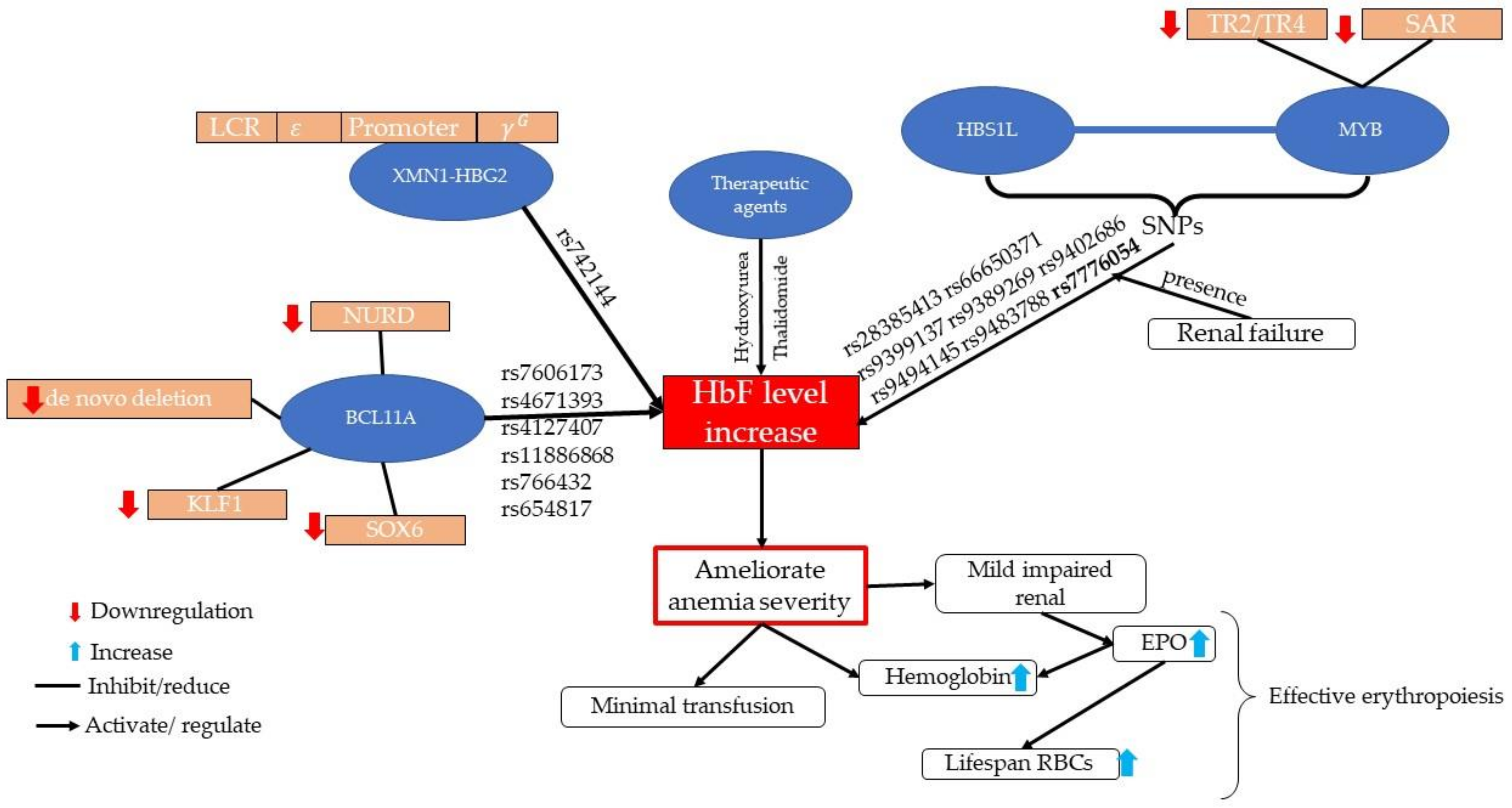
3.1. Epidemiology and Molecular Aspects of XMN1-HBG2
3.2. Epidemiology and Molecular Aspects of HBS1L-MYB
3.3. Epidemiology and Molecular Aspect of BCL11A
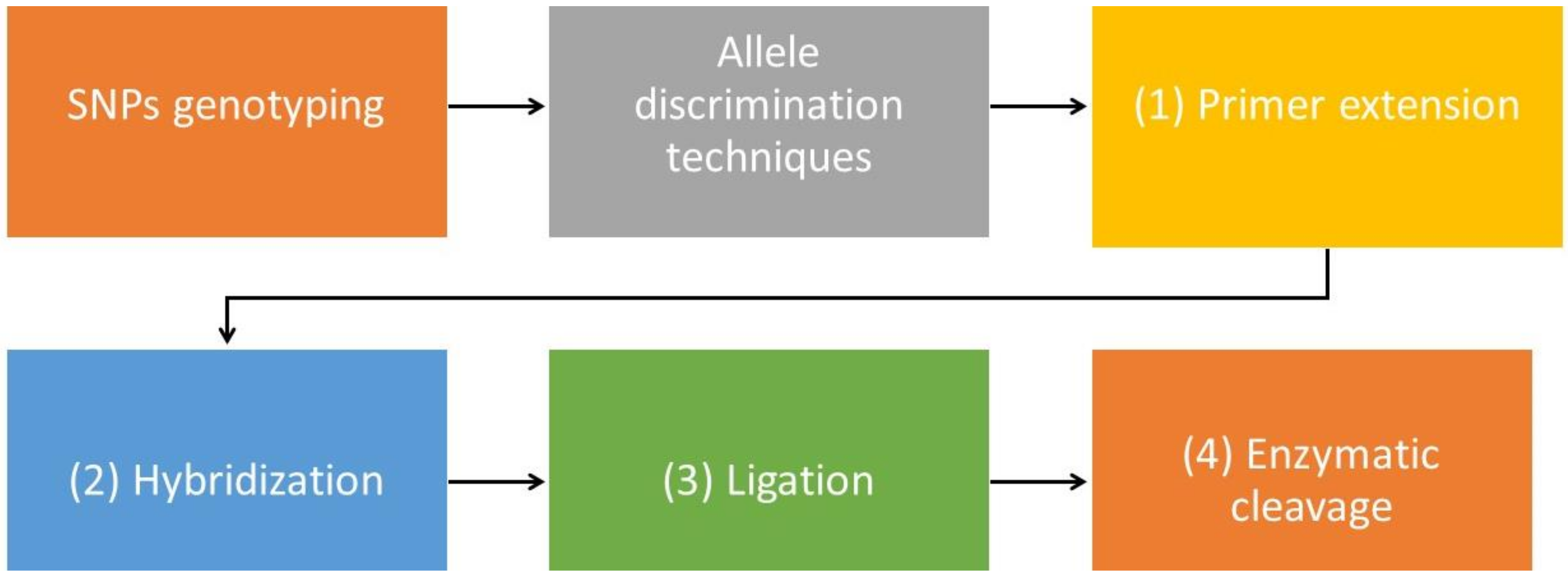
4. Genotyping Technologies in Single Nucleotide Polymorphism Recognition
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weiss, G.; Goodnough, L.T. Anemia of Chronic Disease. N. Engl. J. Med. 2005, 352, 1011–1023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madu, A.J.; Ughasoro, M.D. Anaemia of Chronic Disease: An In-Depth Review. Med Princ. Pract. 2017, 26, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chaparro, C.M.; Suchdev, P.S. Anemia epidemiology, pathophysiology, and etiology in low- and middle-income countries. Ann. N. Y. Acad. Sci. 2019, 1450, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Domenica Cappellini, M.; Motta, I. Anemia in Clinical Practice-Definition and Classification: Does Hemoglobin Change with Aging? Semin. Hematol. 2015, 52, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Chulilla, J.A.M.; Colás, M.S.R.; Martín, M.G. Classification of anemia for gastroenterologists. World J. Gastroenterol. 2009, 15, 4627–4637. [Google Scholar] [CrossRef]
- Akinsheye, I.; Alsultan, A.; Solovieff, N.; Ngo, D.; Baldwin, C.T.; Sebastiani, P.; Chui, D.H.K.; Steinberg, M.H. Fetal hemoglobin in sickle cell anemia. Blood 2011, 118, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Thein, S.L.; Menzel, S. Discovering the genetics underlying foetal haemoglobin production in adults. Br. J. Haematol. 2009, 145, 455–467. [Google Scholar] [CrossRef]
- Amato, A.; Cappabianca, M.P.; Perri, M.; Zaghis, I.; Grisanti, P.; Ponzini, D.; Di Biagio, P. Interpreting elevated fetal hemoglobin in pathology and health at the basic laboratory level: New and known γ- gene mutations associated with hereditary persistence of fetal hemoglobin. Int. J. Lab. Hematol. 2014, 36, 13–19. [Google Scholar] [CrossRef]
- Thein, S.L.; Menzel, S.; Lathrop, M.; Garner, C. Control of fetal hemoglobin: New insights emerging from genomics and clinical implications. Hum. Mol. Genet. 2009, 18, 216–223. [Google Scholar] [CrossRef] [Green Version]
- Fanis, P.; Kousiappa, I.; Phylactides, M.; Kleanthous, M. Genotyping of BCL11A and HBS1L-MYB SNPs associated with fetal haemoglobin levels: A SNaPshot minisequencing approach. BMC Genom. 2014, 15, 108. [Google Scholar] [CrossRef] [Green Version]
- Zakaria, N.; Islam, A.; Abdullah, W.; Bahar, R.; Yusoff, A.M.; Wahab, R.A.; Shamsuddin, S.; Johan, M. Epigenetic Insights and Potential Modifiers as Therapeutic Targets in β–Thalassemia. Biomolecules 2021, 11, 755. [Google Scholar] [CrossRef] [PubMed]
- Shastry, B.S. SNPs: Impact on gene function and phenotype. Methods Mol. Biol. 2009, 578, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Ismail, S.; Essawi, M. Genetic polymorphism studies in humans. Middle East J. Med. Genet. 2012, 1, 57–63. [Google Scholar] [CrossRef]
- Sukhumsirichart, W. Polymorphisms. In Genetic Diversity and Disease Susceptibility; BoD: Norderstedt, Germany, 2018; Available online: http://www.intechopen.com/books/genetic-diversity-and-disease-susceptibility/polymorphisms (accessed on 7 February 2022).
- Lambert, G.; Tsinajinnie, D.; Duggan, D. Single Nucleotide Polymorphism Genotyping Using BeadChip Microarrays. Curr. Protoc. Hum. Genet. 2013, 78, 2–9. [Google Scholar] [CrossRef]
- Butler, J.M.; Coble, M.; Vallone, P. STRs vs. SNPs: Thoughts on the future of forensic DNA testing. Forensic Sci. Med. Pathol. 2007, 3, 200–205. [Google Scholar] [CrossRef]
- Teama, S. DNA Polymorphisms: DNA-Based Molecular Markers and Their Application in Medicine. In Genetic Diversity and Disease Susceptibility; BoD: Norderstedt, Germany, 2018; Available online: https://www.intechopen.com/chapters/62578 (accessed on 9 May 2022).
- Vallejos-Vidal, E.; Reyes-Cerpa, S.; Rivas-Pardo, J.A.; Maisey, K.; Yáñez, J.M.; Valenzuela, H.; Cea, P.A.; Castro-Fernandez, V.; Tort, L.; Sandino, A.M.; et al. Single-Nucleotide Polymorphisms (SNP) Mining and Their Effect on the Tridimensional Protein Structure Prediction in a Set of Immunity-Related Expressed Sequence Tags (EST) in Atlantic Salmon (Salmo salar). Front. Genet. 2020, 10, 1406. [Google Scholar] [CrossRef] [Green Version]
- Edoh, D.; Antwi-Boasiako, C.; Amuzu, D. Fetal hemoglobin during infancy and in sickle cell adults. Afr. Health Sci. 2006, 6, 51–54. [Google Scholar] [CrossRef]
- Dadheech, S.; Jain, S.; Madhulatha, D.; Sharma, V.; Joseph, J.; Jyothy, A.; Munshi, A. Association of Xmn1 −158 γG variant with severity and HbF levels in β-thalassemia major and sickle cell anaemia. Mol. Biol. Rep. 2014, 41, 3331–3337. [Google Scholar] [CrossRef]
- Thein, S.L. Molecular basis of β thalassemia and potential therapeutic targets. Blood Cells Mol. Dis. 2018, 70, 54–65. [Google Scholar] [CrossRef]
- Gardner, K.; Fulford, T.; Silver, N.; Rooks, H.; Angelis, N.; Allman, M.; Nkya, S.; Makani, J.; Howard, J.; Kesse-Adu, R.; et al. G(HbF): A genetic model of fetal hemoglobin in sickle cell disease. Blood Adv. 2018, 2, 235–239. [Google Scholar] [CrossRef]
- Lim, W.F.; Muniandi, L.; George, E.; Sathar, J.; Teh, L.; Lai, M.I. HbF in HbE/β-thalassemia: A clinical and laboratory correlation. Hematology 2014, 20, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Farrell, J.J.; Sherva, R.; Chen, Z.-Y.; Luo, H.; Chu, B.F.; Ha, S.Y.; Li, C.K.; Lee, A.; Li, R.C.H.; Yuen, H.L.; et al. A 3-bp deletion in the HBS1L-MYB intergenic region on chromosome 6q23 is associated with HbF expression. Blood 2011, 117, 4935–4945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, M.; Noor, F.; Bhuyan, G.S.; Qadri, S.S.; Mannoor, K. Role of XmnI polymorphism in HbF induction in HbE/β and β-thalassaemia patients. Bangladesh Med. Res. Counc. Bull. 2019, 45, 133–142. [Google Scholar] [CrossRef]
- Sokolova, A.; Mararenko, A.; Rozin, A.; Podrumar, A.; Gotlieb, V. Hereditary persistence of hemoglobin F is protective against red cell sickling. A case report and brief review. Hematol. Oncol. Stem Cell Ther. 2019, 12, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Wood, W.G. 7 Increased HbF in adult life. Baillière’s Clin. Haematol. 1993, 6, 177–213. [Google Scholar] [CrossRef]
- Fitzhugh, C.D.; Hsieh, M.M.; Allen, D.; Coles, W.A.; Seamon, C.; Ring, M.; Zhao, X.; Minniti, C.P.; Rodgers, G.P.; Schechter, A.N.; et al. Hydroxyurea-Increased Fetal Hemoglobin Is Associated with Less Organ Damage and Longer Survival in Adults with Sickle Cell Anemia. PLoS ONE 2015, 10, e0141706. [Google Scholar] [CrossRef] [PubMed]
- Lübbert, M.; Ihorst, G.; Sander, P.; Bogatyreva, L.; Becker, H.; Wijermans, P.W.; Suciu, S.; Bissé, E.; Claus, R. Elevated fetal haemoglobin is a predictor of better outcome in MDS/AML patients receiving 5-aza-2′-deoxycytidine (Decitabine). Br. J. Haematol. 2017, 176, 609–617. [Google Scholar] [CrossRef]
- Pule, G.D.; Mowla, S.; Novitzky, N.; Wiysonge, C.S.; Wonkam, A. A Systematic Review of Known Mechanisms of Hydroxyurea-induced Foetal Haemoglobin for Treatment of Sickle Cell Disease. Expert Rev. Hematol. 2015, 8, 669–679. [Google Scholar] [CrossRef] [Green Version]
- Mosca, A.; Paleari, R.; Leone, D.; Ivaldi, G. The relevance of hemoglobin F measurement in the diagnosis of thalassemias and related hemoglobinopathies. Clin. Biochem. 2009, 42, 1797–1801. [Google Scholar] [CrossRef]
- Karimi, M.; Zarei, T.; Haghpanah, S.; Moghadam, M.; Ebrahimi, A.; Rezaei, N.; Heidari, G.; Vazin, A.; Khavari, M.; Miri, H. Relationship between Some Single-nucleotide Polymorphism and Response to Hydroxyurea Therapy in Iranian Patients with β-Thalassemia Intermedia. J. Pediatr. Hematol. 2017, 39, e171–e176. [Google Scholar] [CrossRef]
- Ware, R.E. How I use hydroxyurea to treat young patients with sickle cell anemia. Blood 2010, 115, 5300–5311. [Google Scholar] [CrossRef]
- Panja, A.; Basu, A. Pharmacogenomics of the Drugs used for the Treatment of Thalassemia. J. Cytol. Histol. 2015, 6, 5. Available online: https://www.readcube.com/articles/10.4172%2F2157-7099.1000360 (accessed on 25 January 2022).
- Tayebi, B.; Abrishami, F.; Alizadeh, S.; Minayi, N.; Mohammadian, M.; Soleimani, M.; Dehghanifard, A.; Atwan, H.; Ajami, M.; Ajami, M. Modulation of microRNAs expression in hematopoietic stem cells treated with sodium butyrate in inducing fetal hemoglobin expression. Artif. Cells Nanomed. Biotechnol. 2016, 45, 146–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aerbajinai, W.; Zhu, J.; Gao, Z.; Chin, K.; Rodgers, G.P. Thalidomide induces γ-globin gene expression through increased reactive oxygen species–mediated p38 MAPK signaling and histone H4 acetylation in adult erythropoiesis. Blood 2007, 110, 2864–2871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Wei, Z.; Yang, G.; Lai, Y.; Liu, R. Investigating the Efficacy and Safety of Thalidomide for Treating Patients With ß-Thalassemia: A Meta-Analysis. Front. Pharmacol. 2022, 12, 3959. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Morikawa, M.; Yamada, T.; Nishida, R.; Takeda, M.; Kawaguchi, S.; Minakami, H. Changes in hemoglobin F levels in pregnant women unaffected by clinical fetomaternal hemorrhage. Clin. Chim. Acta 2013, 415, 124–127. [Google Scholar] [CrossRef] [PubMed]
- Kahhaleh, F.; Sulaiman, M.A.; Alquobaili, F. Association of Xmn1 polymorphism and consanguineous marriage with fetal hemoglobin in Syrian patients with sickle cell disease. Cogent Med. 2019, 6, 1639243. Available online: https://www.tandfonline.com/action/journalInformation?journalCode=oamd20 (accessed on 6 January 2021).
- Prasing, W.; Odawara, T.; Traisathit, P.; Yamashiro, Y.; Hattori, Y.; Pornprasert, S. Analysis of theXmn1-Gγ polymorphism in β-thalassemia/hemoglobin E (HbE) and homozygous HbE patients with low and high levels of HbF. Int. J. Lab. Hematol. 2015, 37, e25–e28. [Google Scholar] [CrossRef]
- Rujito, L.; Basalamah, M.; Siswandari, W.; Setyono, J.; Wulandari, G.; Mulatsih, S.; Sofro, A.S.M.; Sadewa, A.H.; Sutaryo, S. Modifying effect of XmnI, BCL11A, and HBS1L-MYB on clinical appearances: A study on β-thalassemia and hemoglobin E/β-thalassemia patients in Indonesia. Hematol. Oncol. Stem Cell Ther. 2016, 9, 55–63. [Google Scholar] [CrossRef] [Green Version]
- Yusof, W.; Zulkifli, M.M.; Azman, N.F.; Ab Hamid, S.A.; Othman, A.; Draman, N.; Zilfalil, B.A.; Hassan, R.; Abdullah, W.Z. Factors affecting health-related quality of life and its association with the Xmn1-Gγ polymorphism among adolescents with transfusion-dependent beta thalassemia and HbE/β-thalassemia in East Coast Malaysia. Pediatr. Hematol. Oncol. J. 2020, 5, 30–36. [Google Scholar] [CrossRef]
- Sivalingam, M.; Looi, M.L.; Zakaria, S.Z.S.; Hamidah, N.H.; Alias, H.; Latiff, Z.A.; Ibrahim, H.; Jamal, R. Molecular study and genotype/phenotype correlation of β thalassemia in Malaysia. Int. J. Lab. Hematol. 2012, 34, 377–382. [Google Scholar] [CrossRef]
- Green, N.S.; Barral, S. Genetic modifiers of HbF and response to hydroxyurea in sickle cell disease. Pediatr. Blood Cancer 2011, 56, 177–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lettre, G.; Sankaran, V.G.; André Bezerra, M.C.; Araú Jo, A.S.; Uda, M.; Sanna, S. DNA polymorphisms at the BCL11A, HBS1L-MYB, and-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc. Natl. Acad. Sci. USA 2008, 105, 11869–11874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuinoon, M.; Kruachan, K.; Sengking, W.; Horpet, D.; Sungyuan, U. Thalassemia and Hemoglobin E in Southern Thai Blood Donors. Adv. Hematol. 2014, 2014, 932306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badens, C.; Joly, P.; Agouti, I.; Thuret, I.; Gonnet, K.; Fattoum, S.; Francina, A.; Simeoni, M.C.; Loundou, A.; Pissard, S. Variants in genetic modifiers of β-Thalassemia can help to predict the major or intermedia type of the disease. Haematologica 2011, 96, 1712–1714. [Google Scholar] [CrossRef] [Green Version]
- Nuntakarn, L.; Fucharoen, S.; Fucharoen, G.; Sanchaisuriya, K.; Jetsrisuparb, A.; Wiangnon, S. Molecular, hematological and clinical aspects of thalassemia major and thalassemia intermedia associated with Hb E-β-thalassemia in Northeast Thailand. Blood Cells Mol. Dis. 2009, 42, 32–35. [Google Scholar] [CrossRef]
- Allard, P.; Alhaj, N.; Lobitz, S.; Cario, H.; Jarisch, A.; Grosse, R.; Oevermann, L.; Hakimeh, D.; Tagliaferri, L.; Kohne, E.; et al. Genetic modifiers of fetal hemoglobin affect the course of sickle cell disease in patients treated with hydroxyurea. Haematologica 2021. Available online: https://pubmed.ncbi.nlm.nih.gov/34706496/ (accessed on 26 January 2022).
- Lolis, D.; Georgiou, I.; Loizou, P.; Makrydimas, G. High HbF in pregnancy is associated with the Xmn I polymorphism at the-158bp of the Gq,-globin gene. Eur. J. Obstet. Gynecol. Reprod. Biol. 1995, 60, 153–156. [Google Scholar] [CrossRef]
- Prasing, W.; Mekki, C.; Traisathit, P.; Pissard, S.; Pornprasert, S. Genotyping of BCL11A and HBS1L-MYB Single Nucleotide Polymorphisms in β-thalassemia/HbE and Homozygous HbE Subjects with Low and High Levels of HbF. Walailak J. Sci. Technol. 2018, 15, 627–636. [Google Scholar] [CrossRef]
- Chaouch, L.; Moumni, I.; Ouragini, H.; Darragi, I.; Kalai, M.; Chaouachi, D.; Boudrigua, I.; Hafsia, R.; Abbes, S. rs11886868 and rs4671393 of BCL11A associated with HbF level variation and modulate clinical events among sickle cell anemia patients. Hematology 2016, 21, 425–429. [Google Scholar] [CrossRef]
- Nicolau, M.; Vargas, S.; Silva, M.; Coelho, A.; Ferreira, E.; Mendonça, J.; Vieira, L.; Kjöllerström, P.; Maia, R.; Silva, R.; et al. Genetic modulators of fetal hemoglobin expression and ischemic stroke occurrence in African descendant children with sickle cell anemia. Ann. Hematol. 2019, 98, 2673–2681. [Google Scholar] [CrossRef]
- Dadheech, S.; Madhulatha, D.; Jain, S.; Joseph, J.; Jyothy, A.; Munshi, A. Association of BCL11A genetic variant (Rs11886868) with severityin β-thalassaemia major & sickle cell anaemia. Indian J. Med. Res. 2016, 143, 449–454. [Google Scholar]
- Xu, J.; Sankaran, V.G.; Ni, M.; Menne, T.F.; Puram, R.V.; Kim, W.; Orkin, S.H. Transcriptional silencing of γ-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev. 2010, 24, 783–789. [Google Scholar] [CrossRef] [Green Version]
- Wilber, A.; Nienhuis, A.W.; Persons, D.A. Transcriptional regulation of fetal to adult hemoglobin switching: New therapeutic opportunities. Blood 2011, 117, 3945–3953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Ghamrawy, M.; Yassa, M.E.; Tousson, A.M.S.; El-Hady, M.A.; Mikhaeil, E.; Mohamed, N.B.; Khorshied, M.M. Association between BCL11A, HSB1L-MYB, and XmnI γG-158 (C/T) gene polymorphism and hemoglobin F level in Egyptian sickle cell disease patients. Ann. Hematol. 2020, 99, 2279–2288. [Google Scholar] [CrossRef] [PubMed]
- Friedrisch, J.R.; Sheehan, V.; Flanagan, J.M.; Baldan, A.; Summarell, C.C.G.; Bittar, C.M.; Friedrisch, B.K.; Wilke, I.I.; Ribeiro, C.B.; Daudt, L.E.; et al. The role of BCL11A and HMIP-2 polymorphisms on endogenous and hydroxyurea induced levels of fetal hemoglobin in sickle cell anemia patients from southern Brazil. Blood Cells Mol. Dis. 2016, 62, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Uda, M.; Galanello, R.; Sanna, S.; Lettre, G.; Sankaran, V.G.; Chen, W.; Usala, G.; Busonero, F.; Maschio, A.; Albai, G.; et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of-thalassemia. Proc. Natl. Acad. Sci. USA 2008, 105, 1620–1625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menzel, S.; Jiang, J.; Silver, N.; Gallagher, J.; Cunningham, J.; Surdulescu, G.; Lathrop, M.; Farrall, M.; Spector, T.D.; Thein, S.L. The HBS1L-MYB intergenic region on chromosome 6q23.3 influences erythrocyte, platelet, and monocyte counts in humans. Blood J. Am. Soc. Hematol. 2007, 105, 3624–3626. [Google Scholar] [CrossRef] [Green Version]
- Adeyemo, T.A.; Ojewunmi, O.O.; Oyetunji, I.A.; Rooks, H.; Rees, D.C.; Akinsulie, A.O.; Akanmu, A.S.; Thein, S.L.; Menzel, S. A survey of genetic fetal-haemoglobin modifiers in Nigerian patients with sickle cell anaemia. PLoS ONE 2018, 13, e0197927. [Google Scholar] [CrossRef] [Green Version]
- The, L.K.; Yu, K.S.; Chua, S.M.; George, E.; Lai, M.I.; Wong, L. Genetic Variants in HBS1L-MYB rs9399137 and rs11759553 Associated with Elevated HbF Levels Among Filipino β°-deletion Carriers. J. Biomed. Clin. Sci. 2017, 2, 28–29. [Google Scholar]
- Lim, L.N.; Teh, L.K.; Yu, K.S.; Chua, S.M.; George, E.; Lai, M.I.; Wong, L. Genetic variants of HBS1L-MYB with Hb subtypes level among Filipino β°-deletion carriers co-inherited with −α3.7 deletion thalassaemia. Meta Gene 2020, 26, 100769. [Google Scholar] [CrossRef]
- Pourfarzad, F.; von Lindern, M.; Azarkeivan, A.; Hou, J.; Kia, S.K.L.; Esteghamat, F.; van IJcken, W.; Philipsen, S.; Najmabadi, H.; Grosveld, F. Hydroxyurea responsiveness in β-thalassemic patients is determined by the stress response adaptation of erythroid progenitors and their differentiation propensity. Haematologica 2013, 98, 696–704. [Google Scholar] [CrossRef] [Green Version]
- Wong, H.; Mylrea, K.; Feber, J.; Drukker, A.; Filler, G. Prevalence of complications in children with chronic kidney disease according to KDOQI. Kidney Int. 2006, 70, 585–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bashir, S.; Mahmood, S.; Mohsin, S.; Tabassum, I.; Ghafoor, M.; Sajjad, O. Modulatory effect of single nucleotide polymorphism in Xmn1, BCL11A and HBS1L-MYB loci on foetal haemoglobin levels in β-thalassemia major and Intermedia patients. J. Pak. Med. Assoc. 2021, 71, 1394–1398. [Google Scholar] [CrossRef] [PubMed]
- Stadhouders, R.; Aktuna, S.; Thongjuea, S.; Aghajanirefah, A.; Pourfarzad, F.; van IJcken, W.; Lenhard, B.; Rooks, H.; Best, S.; Menzel, S.; et al. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J. Clin. Investig. 2014, 124, 1699–1710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandit, R.A.; Svasti, S.; Sripichai, O.; Munkongdee, T.; Triwitayakorn, K.; Winichagoon, P.; Fucharoen, S.; Peerapittayamongkol, C. Association of SNP in exon 1 of HBS1L with hemoglobin F level in β0-thalassemia/hemoglobin E. Int. J. Hematol. 2008, 88, 357–361. [Google Scholar] [CrossRef]
- So, C.C.; Song, Y.Q.; Tsang, S.T.; Tang, L.F.; Chan, A.Y.; Ma, E.S.; Chan, L.C. The HBS1L-MYB intergenic region on chromosome 6q23 is a quantitative trait locus controlling fetal haemoglobin level in carriers of beta-thalassaemia. J. Med. Genet. 2008, 45, 745–751. [Google Scholar] [CrossRef]
- Jiang, J.; Best, S.; Menzel, S.; Silver, N.; Lai, M.I.; Surdulescu, G.L.; Spector, T.D.; Thein, S.L. cMYB is involved in the regulation of fetal hemoglobin production in adults. Blood 2006, 108, 1077–1083. [Google Scholar] [CrossRef] [Green Version]
- Thein, S.L.; Menzel, S.; Peng, X.; Best, S.; Jiang, J.; Close, J.; Silver, N.; Gerovasilli, A.; Ping, C.; Yamaguchi, M.; et al. Intergenic variants of HBS1L-MYB are responsible for a major quantitative trait locus on chromosome 6q23 influencing fetal hemoglobin levels in adults. Proc. Natl. Acad. Sci. USA 2007, 104, 11346–11351. [Google Scholar] [CrossRef] [Green Version]
- Menzel, S.; Thein, S.L. Genetic Modifiers of Fetal Haemoglobin in Sickle Cell Disease. Mol. Diagnosis Ther. 2018, 23, 235–244. [Google Scholar] [CrossRef]
- Beverborg, N.G.; Verweij, N.; Klip, I.T.; van der Wal, H.H.; Voors, A.A.; van Veldhuisen, D.J.; Gansevoort, R.T.; Bakker, S.J.L.; van der Harst, P.; van der Meer, P. Erythropoietin in the General Population: Reference Ranges and Clinical, Biochemical and Genetic Correlates. PLoS ONE 2015, 10, e0125215. Available online: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0125215 (accessed on 7 December 2021).
- Jiwu, L.; Manna, S.; Lai, M.; Ying, Z.; Yanhui, L. Hyperhaemolysis in a pregnant woman with a homozygous β0-thalassemia mutation and two genetic modifiers. Mol. Genet. Genom. Med. 2021, 9, e1696. Available online: https://pmc/articles/PMC8372088/ (accessed on 17 December 2021).
- Liu, N.; Hargreaves, V.V.; Zhu, Q.; Kurland, J.V.; Hong, J.; Kim, W.; Sher, F.; Trevino, C.M.; Rogers, J.M.; Kurita, R.; et al. Direct Promoter Repression by BCL11A Controls the Fetal to Adult Hemoglobin Switch. Cell 2018, 173, 430–442.e17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sankaran, V.G.; Menne, T.F.; Xu, J.; Akie, T.E.; Lettre, G.; Van Handel, B.; Mikkola, H.K.A.; Hirschhorn, J.N.; Cantor, A.B.; Orkin, S.H. Human Fetal Hemoglobin Expression Is Regulated by the Developmental Stage-Specific Repressor BCL11A. Science 2008, 322, 1839–1842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Li, R.; Teichert, K.; Montbleau, K.E.; Verboon, J.M.; Voit, R.A.; Sankaran, V.G. Pathogenic BCL11A variants provide insights into the mechanisms of human fetal hemoglobin silencing. PLoS Genet. 2021, 17, e1009835. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Liu, K.; Sun, C.W.; Pawlik, K.M.; Townes, T.M. KLF1 regulates BCL11A expression and γ- to β-globin gene switching. Nat. Genet 2010, 42, 742–744. [Google Scholar] [CrossRef]
- Bauer, D.E.; Orkin, S.H. Update on fetal hemoglobin gene regulation in hemoglobinopathies. Curr. Opin. Pediatr. 2011, 23, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Basak, A.; Hancarova, M.; Ulirsch, J.; Balci, T.; Trkova, M.; Pelisek, M.; Vlckova, M.; Muzikova, K.; Cermak, J.; Trka, J.; et al. BCL11A deletions result in fetal hemoglobin persistence and neurodevelopmental alterations. J. Clin. Investig. 2015, 125, 2363–2368. [Google Scholar] [CrossRef] [Green Version]
- Ghedira, E.S.; Lecerf, L.; Faubert, E.; Costes, B.; Moradkhani, K.; Bachir, D.; Galactéros, F.; Pissard, S. Estimation of the difference in HbF expression due to loss of the 5′ δ-globin BCL11A binding region. Haematologica 2013, 98, 305–308. [Google Scholar] [CrossRef] [Green Version]
- Qadah, T.; Noorwali, A.; Alzahrani, F.; Banjar, A.; Filimban, N.; Felimban, R. Detection of BCL11A and HBS1L-MYB Genotypes in Sickle Cell Anemia. Indian J. Hematol. Blood Transfus. 2020, 36, 705–710. [Google Scholar] [CrossRef]
- Nuinoon, M.; Makarasara, W.; Mushiroda, T.; Setianingsih, I.; Wahidiyat, P.A.; Sripichai, O.; Kumasaka, N.; Takahashi, A.; Svasti, S.; Munkongdee, T.; et al. A genome-wide association identified the common genetic variants influence disease severity in β0-thalassemia/hemoglobin e. Qual. Life Res. 2010, 127, 303–314. [Google Scholar] [CrossRef]
- Kim, S.; Misra, A. SNP genotyping: Technologies and biomedical applications. Annu. Rev. Biomed. Eng. 2007, 9, 289–320. [Google Scholar] [CrossRef]
- Chamouine, A.; Saandi, T.; Muszlak, M.; Larmaraud, J.; Lambrecht, L.; Poisson, J.; Balicchi, J.; Pissard, S.; Elenga, N. High fetal hemoglobin level is associated with increased risk of cerebral vasculopathy in children with sickle cell disease in Mayotte. BMC Pediatr. 2020, 20, 302. [Google Scholar] [CrossRef] [PubMed]
- Fedick, A.; Su, J.; Treff, N.R. Development of TaqMan allelic discrimination based genotyping of large DNA deletions. Genomics 2012, 99, 127–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litos, I.K.; Ioannou, P.C.; Christopoulos, T.K.; Traeger-Synodinos, J.; Kanavakis, E. Genotyping of Single-Nucleotide Polymorphisms by Primer Extension Reaction in a Dry-Reagent Dipstick Format. Anal. Chem. 2006, 79, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.; Flower, D.; Rigley, K. Microarrays in hematology. Curr. Opin. Hematol. 2002, 9, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Prince, J.A.; Feuk, L.; Howell, W.M.; Jobs, M.; Emahazion, T.; Blennow, K.; Brookes, A.J. Robust and Accurate Single Nucleotide Polymorphism Genotyping by Dynamic Allele-Specific Hybridization (DASH): Design Criteria and Assay Validation. Genome Res. 2001, 11, 152–162. [Google Scholar] [CrossRef] [Green Version]
- Bruse, S.E.; Moreau, M.P.; Azaro, M.A.; Zimmerman, R.; Brzustowicz, L.M.; Moreau, M.P.; Hoffman, A. Improvements to bead-based oligonucleotide ligation SNP genotyping assays. BioTechniques 2008, 45, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Szemes, M.; Bonants, P.; De Weerdt, M.; Baner, J.; Landegren, U.; Schoen, C.D. Diagnostic application of padlock probes--multiplex detection of plant pathogens using universal microarrays. Nucleic Acids Res. 2005, 33, e70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, S.; Long, Y. Genotyping analysis using an RFLP assay. Methods Mol. Biol. 2015, 1245, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.V.N.; Stevens, P.W.; Hall, J.G.; Lyamichev, V.; Neri, B.P.; Kelso, D.M. Genotyping single nucleotide polymorphisms directly from genomic DNA by invasive cleavage reaction on microspheres. Nucleic Acids Res. 2003, 31, e66. [Google Scholar] [CrossRef] [Green Version]
- Olivier, M. The Invader® assay for SNP genotyping. Mutat. Res. 2005, 573, 103. [Google Scholar] [CrossRef] [Green Version]

| Locus | SNPs | Effects of SNPs | References |
|---|---|---|---|
| XMN1-HBG2 | Rs782144 | HbF level highly expressed, with T allele resulting in mild anemia and asymptomatic state in lenient disease. | [40,41,48] |
| The high level of HbF may reduce pain crises, reduce risk of feto-maternal bleeding, reduce blood transfusion requirement, and unusual thalassemia. | [49,50] | ||
| BCL11A | Rs1018987 | Rs1427407, rs10189857, and rs11886868 had no significant difference in <5% and >5% of HbF level. | [51] |
| Rs4127407 | |||
| Rs766432 | The presence of T and G alleles in rs11886868 and rs4671393 is associated with amelioration of SCD phenotype through an increase in HbF level. | [52] | |
| Rs11886868 | |||
| Rs4671393 | |||
| Rs6729815 | The allele C in rs11886868 and allele A in rs4671393 are linked to increased HbF levels (p = 0.026 and 0.028) but have no correlation with stroke. | [53] | |
| Rs1426407 | |||
| Rs6545816 | |||
| Rs7606173 | The downregulation of BCL11A caused by rs11886868 results in high HbF production (11.36–49.40%) in the CC genotype, followed by CT and TT. | [54] | |
| Rs6545817 | |||
| The presence of knockdown of SOX6 and KLF1 may reduce the silencing of the γ-globin gene and enhance HbF levels to more than 28.9%. | [55,56] | ||
| Hydroxyurea treatment is associated with rs11886868, rs4671393, and rs1427407, which may ameliorate anemia. | [57,58] | ||
| The HbF levels may be increased due to Rs11886868, which consists of the C allele; this also ameliorates the clinical phenotype. | [59] | ||
| HBS1L-myb | Rs4895441 | Rs9399137 with C allele associated with increased HbF production and hematologic parameters. | [51] |
| Rs66650371 | |||
| Rs9399137 | The decrease in MYB expression in rs66650371 is associated with high HbF synthesis to produce proper RBC and Hb levels. | [60,61] | |
| Rs11759553 | |||
| Rs28384513 | |||
| Rs9402686 | The rs9402686 at sub-locus is associated with high HbF in the A allele. | [61] | |
| Minor allele frequency of rs9399137 or rs11759553 more than 5% may increase HbF levels and has been proposed as a potent target for therapeutic purposes. | [62,63] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammad, S.N.N.A.; Iberahim, S.; Wan Ab Rahman, W.S.; Hassan, M.N.; Edinur, H.A.; Azlan, M.; Zulkafli, Z. Single Nucleotide Polymorphisms in XMN1-HBG2, HBS1L-MYB, and BCL11A and Their Relation to High Fetal Hemoglobin Levels That Alleviate Anemia. Diagnostics 2022, 12, 1374. https://doi.org/10.3390/diagnostics12061374
Mohammad SNNA, Iberahim S, Wan Ab Rahman WS, Hassan MN, Edinur HA, Azlan M, Zulkafli Z. Single Nucleotide Polymorphisms in XMN1-HBG2, HBS1L-MYB, and BCL11A and Their Relation to High Fetal Hemoglobin Levels That Alleviate Anemia. Diagnostics. 2022; 12(6):1374. https://doi.org/10.3390/diagnostics12061374
Chicago/Turabian StyleMohammad, Siti Nur Nabeela A’ifah, Salfarina Iberahim, Wan Suriana Wan Ab Rahman, Mohd Nazri Hassan, Hisham Atan Edinur, Maryam Azlan, and Zefarina Zulkafli. 2022. "Single Nucleotide Polymorphisms in XMN1-HBG2, HBS1L-MYB, and BCL11A and Their Relation to High Fetal Hemoglobin Levels That Alleviate Anemia" Diagnostics 12, no. 6: 1374. https://doi.org/10.3390/diagnostics12061374
APA StyleMohammad, S. N. N. A., Iberahim, S., Wan Ab Rahman, W. S., Hassan, M. N., Edinur, H. A., Azlan, M., & Zulkafli, Z. (2022). Single Nucleotide Polymorphisms in XMN1-HBG2, HBS1L-MYB, and BCL11A and Their Relation to High Fetal Hemoglobin Levels That Alleviate Anemia. Diagnostics, 12(6), 1374. https://doi.org/10.3390/diagnostics12061374






