1. Introduction
Parkinson’s disease (PD) is a neurodegenerative disease caused by the death of neurons (called substantia nigra) that generate dopamine [
1]. Dopamine is an organic chemical of the catecholamine and phenethylamine families that controls physical movement by transmitting messages between the brain and the substantia nigra, thereby enabling coordinated movement [
2]. When 60–80% of the cells that produce dopamine are lost, the amount of dopamine is not enough to control a person’s movements and, thus, symptoms of PD appear [
3]. The lack of dopamine neuron production leads to losing control over the body’s motor functions [
4]. Four symptoms of PD are specific to the motor system, including tremors (shaking of jaws, hands, legs, and arms), rigidity (inflexibility of limbs and trunk), poor balance, and slow movement [
5]. Nonmotor features of PD include dementia, depression, restless legs, temperature sensitivity, and digestion problems [
6]. Although PD is still incurable, some treatment options for patients with motor and nonmotor symptoms have been developed. These options include noninvasive (drugs) and invasive (surgical) detection and treatment methods. The medications are used to block nerve impulses to control the motor system [
7]. All drugs and surgical procedures have side effects. Voice disorder testing is a useful and noninvasive option for the early detection of PD since approximately 90% of PD patients have dysphonia or vocal impairment, differentiating them from healthy people [
8]. Therefore, diagnosing PD through voice disorders is one of the promising and effective methods. Affecting approximately 10 million people worldwide, PD is the second type of neurodegenerative disorder after Alzheimer’s Disease [
9] After age 65, people are more susceptible to the disease, and men are more susceptible than women. Common symptoms of this disease, such as loss of smell, constipation, and sleep disturbances, appear years before motor symptoms do. Thereafter, new symptoms such as tremors, imbalance, and voice impairments also appear. In the early stages, appropriate treatments that help slow the progression of the disease or stop it from developing are essential. Nevertheless, diagnosing PD according to clinical symptoms is still difficult and complex [
10]. Since 90% of people with Parkinson’s have a voice disorder, detecting PD using voice data is one of the most important approaches recently [
11]. In this approach, acoustic signals play an important role in the early diagnosis of PD [
12]. In the early stages of PD, voice abnormalities are indistinguishable for listeners but can be discerned by analysing voice cues [
13]. The PD and movement disorders are divided into two phases: the preclinical phase, where the patient suffers from neurodegeneration but there are no clinical indications, and the prodromal phase, where the patient suffers from clinical symptoms but there are not enough indications for a diagnosis [
14]. Therefore, early diagnosis in both phases is essential to allow doctors to discover the disease and provide medical intervention on time. To date, there are no confirmed biomarkers to provide early detection of PD efficiently. Hence, there is an urgent need to use artificial intelligence (AI) techniques to help the healthcare sector diagnose PD early and effectively [
15]. Thus, developing a computer-aided diagnostic system is necessary to analyse voice data to distinguish between PWP and healthy voices. Some researchers have recently proposed noninvasive methods for diagnosing PD using acoustic-signal analysis [
16].
Although there are several efforts from researchers to provide satisfactory results for diagnosing PD, achieving better accuracy is still yet to be realized. Hence, this paper proposes novel techniques to optimize the techniques of early diagnosis of PD by differentiating patients through acoustic-data analysis to help neurologists make appropriate diagnostic decisions.
Parkinson’s disease is a neurodegenerative condition that affects a person’s movement due to a lack of dopamine production in the brain. Early diagnosis of Parkinson’s disease is crucial for timely treatment and management of the condition. However, the current diagnostic process involves multiple tests and specialist examinations, which can be time-consuming and costly. This study proposes a novel approach to early diagnosis by analyzing voice disorders associated with Parkinson’s disease. The researchers extract a set of features from recordings of a person’s voice and apply machine-learning methods to analyze and distinguish between Parkinson’s cases and healthy individuals.
However, it is inferred to determine whether the proposed techniques, which involve feature selection, hyperparameter tuning, dataset balancing, and dimension reduction, improve the accuracy of Parkinson’s disease diagnosis based on voice analysis. Addressing a knowledge gap, the study addresses the need for improved techniques in the early detection of Parkinson’s disease. By exploring the potential of analyzing voice disorders and employing machine-learning algorithms, the study offers a novel approach to enhance diagnostic accuracy. Guiding the methodology, the purpose is to guide the selection of appropriate methods and techniques to achieve the study’s objectives. They employ feature selection, hyperparameter tuning, dataset balancing, and dimension reduction techniques to optimize the diagnostic process.
Investigating machine-learning methods that utilize feature selection and reduction techniques is of great significance in various fields, including healthcare, finance, and image recognition, among others [
17]. Feature selection and reduction methods aim to identify the most relevant and informative features from a given dataset, which can improve the performance and interpretability of machine-learning models. These techniques play a crucial role in the accurate diagnosis of Parkinson’s disease [
18]. One major advantage of feature selection and reduction methods is the ability to handle high-dimensional data. In many real-world applications, datasets often contain a large number of features, some of which may be redundant or irrelevant. Analyzing such datasets directly can lead to computational inefficiency, increased model complexity, and overfitting. Feature selection methods help identify a subset of features that have the most discriminatory power and contribute the most to the prediction task [
19]. By reducing the dimensionality of the data, these methods can improve the computational efficiency and generalization capability of machine-learning models. Furthermore, feature selection and reduction techniques can enhance model interpretability [
20]. In many domains, understanding the factors or features that contribute to a particular prediction is crucial for gaining insights into the underlying process or making informed decisions. By selecting a smaller set of relevant features, the resulting model becomes more transparent, and the relationship between the features and the target variable becomes easier to comprehend [
21]. In conclusion, investigating machine-learning methods that employ feature selection and reduction techniques has significant implications across various domains. In the context of the paper on Parkinson’s disease detection, these methods can help identify the most informative acoustic features for early diagnosis. However, careful consideration of the challenges and critical issues associated with feature selection is necessary to ensure reliable and robust results.
The contributions of this paper can be summarized as follows:
Proposing a new approach for early detection and diagnosis of PD based on acoustic signals to help doctors with early diagnosis and timely medical interventions;
Proposing and implementing SMOTE technique for a balanced dataset;
Apply Pearson’s coefficient to analyze the correlation between all features and remove attributes with a very high correlation;
Apply the RFE algorithm to give each feature a percentage of its contribution to diagnosing PD;
Apply the t-SNE and PCA algorithms to reduce the number of features in the dataset and select the features correlated with the target characteristic.
The rest of this paper is organised as follows.
Section 2 provides related work.
Section 3 describes the process of PD detection by dysphonia.
Section 3 analyses the materials and methods applied in the study, and presents subsections on processing the features, finding correlations between them and removing the outliers.
Section 4 describes the experiment setup.
Section 5 presents the results of the analysis and compares the results with those of the literature.
Section 6 provides the conclusion.
2. Related Work
This paper study is distinguished from current studies by developing diagnostic systems with various methodologies and tools that can effectively analyze audio data and distinguish between Parkinson’s and healthy people with high precision.
Hui et al. [
22] proposed a CNN for analyzing the EEG recordings of 16 healthy subjects and 15 Parkinson’s patients. Gabor transform converted the EEG signals into spectral diagrams to train a CNN. Majid et al. [
23] developed an approach based on spatial patterns of PD diagnosis for patients taking medication and those not taking medication. The EEG signals were processed to remove noise by a common spatial pattern. Features were extracted from the optimized signals and fed into machine-learning classifiers. The classifier achieved the best results with features extracted from beta and alpha ranges with an accuracy of 95%. Luigi et al. [
24] The frequency features of the velocity and angle signals were extracted, the features were selected, and the machine-learning classifiers were optimized for FOG capture and before. The FOG detection network achieved good results and validated the patients. The network achieved a sensitivity of 84.1%, a specificity of 85.9%, and an accuracy of 86.1%. Thus, the network can predict FOG before it happens. Nalini et al. [
25] utilized three deep-learning networks, RNN, LSTM, and MLP, for diagnosing the voice characteristics of PD patients. They provided loss-function curves for PD detection; the LSTM network achieved better performance than other networks. Arti et al. [
26] utilized three ML and ANN algorithms to diagnose a speech dataset. Data collection and feature selection have been improved based on the wrapper and filtering method. SVM and KNN achieved an accuracy of 87.17%, while naïve Bayes had an accuracy of 74.11%. Hajer et al. [
27] also used three ML algorithms for diagnosing a PD dataset to distinguish PD patients from healthy controls. The data were analyzed by linear discriminant analysis (LDA) and PCA algorithms. K-means and DBSCAN models are built based on feature-reduction algorithms. LDA performs better than PCA; thus, its output is fed into clustering algorithms. DBSCAN achieved an accuracy of 64%, a sensitivity of 78.13%, and a specificity of 38.89%. Moumita et al. [
28] designed three schemes based on decision-forest and SysFor algorithms through ForestPA features for PD diagnosis. The approach requires minimal decision trees to achieve good accuracy. Increasing the density of decision trees for dynamic training and testing new samples proves to be the best method for PD detection. The decision tree with ForestPA features reached an accuracy of 94.12%. Sarkar et al. [
29] collected and analysed acoustic data from 40 people: 20 with PD and 20 that were normal. The researchers used support-vector machines (SVM) and K-nearest-neighbour (KNN) classifiers to analyse and diagnose samples. To diagnose a speech disorder in PD patients, Little et al. [
30] proposed an algorithm to measure dysphonia and analyse speech methods. The main objective was to distinguish between PD and normal patients through two features, namely recurrence and fractal scaling, which distinguish distorted sounds from normal sounds and applied the pitch period entropy (PPE) method to diagnose a dataset consisting of 23 patients with PD and 8 healthy subjects through the extraction of dysphonia features. The method achieved an accuracy of 91.4%. Canturk et al. [
31] presented four methods for selecting features and classified the selected features into six categories. The system achieved an accuracy of 57.5% with LOSO CV and an accuracy of 68.94% with fold CV. Li et al. [
32] extracted hybrid features, and then diagnosed these features through SVM; the algorithm achieved 82.5% accuracy. Benba et al. [
33] extracted features by Mel-frequency cepstral coefficients and rated them by SVM, and the algorithm achieved 82.5% accuracy with LOSO CV. They also applied human factor cepstral coefficients to extract features from vowels, achieving an accuracy of 87.5% with LOSO CV. Almeida et al. [
34] extracted the features of phonemic pronunciation using several methods, and PD was detected on the basis of several classifiers. Das et al. [
35] applied partial least squares to reduce dimensions and applied a self-organising map (SOM) for clustering. Finally, PD was detected by the unified PD rating scale. Yuvaraj et al. [
36] studied emotional information such as happiness, anger, fear, sadness, and disgust to diagnose PD and distinguish it from normal states through the use of EEG signals. Spectral decomposition was also applied with KNN and SVM classifiers, and the researchers observed that emotions were reduced in PD patients [
37,
38]. Yuvaraj et al. [
39] applied higher-order spectra to extract features from electroencephalography signals to diagnose PD from normal cases. All classifiers achieved promising results. Sivaranjini et al. [
40] used the AlexNet model to diagnose MR images to distinguish PD from normal cases; the model reached an accuracy of 88.9%. Ali et al. [
41] extracted features and selected the most important ones, ranking them by the chi-square statistical method for PD diagnosis. Senturk et al. [
42] applied methods to choose features and remove unessential ones, and the selected features were diagnosed through ML methods for early detection of PD. Gupta et al. [
43] employed an optimised version of the crow search algorithm for early detection of PD, in which the system achieved superior accuracy.
Through previous studies, it is noted that there are limitations in the techniques used and the failure to achieve satisfactory results for the automated and early diagnosis of PD.
5. Discussion
PD is a health problem that threatens the elderly and is caused by neurodegeneration due to the death of neurons that secrete dopamine. The manual diagnosis of PD is still lacking, doctors’ opinions differ, and the number of doctors in developing countries is small. Thus, automated diagnosis by artificial intelligence solves these challenges. In this study, many systems have been developed that go through many stages of image processing. The PD dataset went through data optimization and feature correlation rents. The RFE algorithm was applied to give the percentage contribution of each feature to the target feature. The features of the dataset were subjected to the t-SNE and PCA algorithms to select the most important features and fed to five classifiers for classification. When analyzing acoustic signals for the diagnosis of Parkinson’s disease, t-SNE and PCA are both dimensionality reduction techniques that can be employed. However, they serve different purposes and have distinct advantages and limitations. Complementary information: t-SNE and PCA capture different aspects of the data. While t-SNE is effective at visualizing clusters and preserving the local structure, it may not capture the overall variance or global patterns in the data. PCA, on the other hand, focuses on explaining the maximum variance in the data, which can provide valuable insights into the most significant features. By combining both techniques, you can benefit from their complementary information. Interpretability: PCA produces orthogonal components that are interpretable as linear combinations of the original features. These components represent the directions of maximum variance in the data. In contrast, t-SNE does not provide straightforward interpretations or explicit relationships with the original features. Therefore, PCA can help in understanding the underlying factors contributing to the acoustic signals related to Parkinson’s disease. This section discusses the evaluation of algorithms developed on the PD dataset for early diagnosis of PD at each category level. The systems achieved better results after applying the t-SNE and PCA algorithms. This means that the existence of some features is negatively associated with the target feature and affects the efficiency of the system.
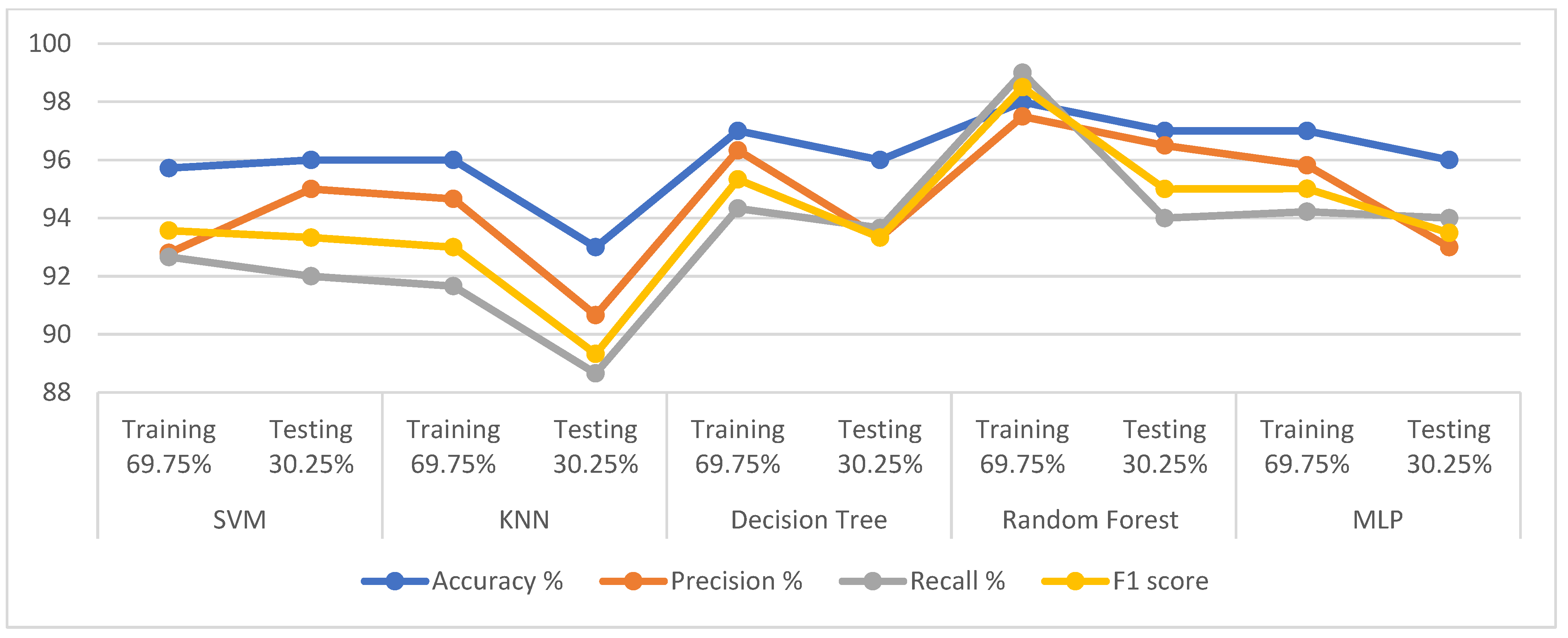
First, when the classifiers are fed the dataset produced by the t-SNE algorithm,
Table 10 and
Figure 5 describe the results of diagnosing the classes of the dataset, which are healthy (class 0) and PD (class 1). In the healthy class, it is noted that SVM, KNN, DT, RF, and MLP achieved the following results: for precision by 96%, 96%, 97%, 97%, and 98%, respectively; also for recall by 99%, 96%, 99%, 99%, and 97%, respectively; while the for F1-score 98%, 96%, 98%, 98%, and 97%, respectively. For the PD class, it is noted that SVM, KNN, DT, RF, and MLP achieved the following results: for precision 94%, 88%, 96%, 96%, and 90.50%, respectively; and for recall at 88.50%, 85%, 92%, 89%, and 92.50%, respectively; while the F1-score by 91%, 86%, 91%, 93%, and 91.50%, respectively.
Our findings suggest that acoustic signals can be used to detect Parkinson’s disease automatically and early. This could lead to earlier diagnosis and treatment, which could improve the quality of life for people with Parkinson’s disease.
The systems optimize the early diagnosis of PD by evaluating selected features and hyperparameter tuning of ML algorithms for diagnosing PD based on voice disorders. The study found that the proposed techniques were superior to existing studies, indicating the superiority of the proposed techniques. This contributes to the evaluation and advancement of existing knowledge in the field of Parkinson’s disease diagnosis.
Second, when the classifiers are fed the dataset after dimensionality reduction by the PCA algorithm,
Table 11 and
Figure 6 describe the diagnostic results for each class. In the healthy class, it is noted that SVM, KNN, DT, RF, and MLP achieved the following results: for precision by 80%, 88.50%, 94%, 100%, and 98%, respectively; also for recall by 97%, 84%, 96%, 99%, and 100%, respectively; while the for F1-score 88%, 85.50%, 95%, 99%, and 99%, respectively. For the PD class, it is noted that SVM, KNN, DT, RF, and MLP achieved the following results: for precision 87%, 90%, 94%, 90%, and 97.50%, respectively; and for recall at 88%, 84%, 87.50%, 97%, and 95%, respectively; while the F1-score by 87.50%, 90%, 90.50%, 93%, and 95.50%, respectively.
Table 12 and
Figure 7 present a comparison of the results of the proposed classification models with existing models discussed in the literature. Our proposed model provides better results over existing studies, whereas the previous studies achieved an accuracy score between 95.43% and 78.23%, while the proposed systems reached an accuracy of 98%, 96%, and 98% for the RF, DT, and MLP classifiers, respectively. The measure of recall (sensitivity) in the previous systems was between 95.4% and 71%, while those of the proposed systems were 98%, 93.66%, and 96% for RF, DT, and MLP classifiers, respectively.
Table 12.
Comparison of performance of the proposed system with previous studies.
Table 12.
Comparison of performance of the proposed system with previous studies.
| Previous Studies | Accuracy% | Recall% | Precision% | F1-Score |
|---|
| Khan et al. [63] | 90 | 93 | - | - |
| Benba et al. [64] | 82.5 | 80 | - | - |
| Behroozi et al. [65] | 87.5 | 90 | - | - |
| Li et al. [66] | 82.5 | 85 | - | - |
| Parisi et al. [67] | 78.23 | 72.22 | - | - |
| Cantürk and Karabiber [68] | 68.94 | 74.03 | - | - |
| Mostafa et al. [69] | 95.43 | 95.4 | - | - |
| Wroge et al. [70] | 85 | 71 | | |
| Proposed model using RF | 99 | 98 | 95 | 96 |
| Proposed model using DT | 95 | 90.33 | 94 | 92 |
| Proposed model using MLP | 98 | 96 | 97.66 | 96.66 |
Figure 7.
Display of a comparison of the performance of some of our classifiers with some previous studies [
31,
32,
63,
64,
65,
67,
69,
70].
Figure 7.
Display of a comparison of the performance of some of our classifiers with some previous studies [
31,
32,
63,
64,
65,
67,
69,
70].
Reliability refers to the consistency and stability of measurements or techniques, while validity refers to the accuracy and appropriateness of the measurements or techniques in assessing the intended construct or phenomenon. Fortunately, the information provided does explicitly mention the reliability and validity of the measures used in the study. The study reported the accuracy of the proposed techniques through the accuracy of the results assessed thanks to the reliability and validity of the measures used. The study stated that the proposed techniques were able to identify PD with an accuracy of up to 98%. Overall, the study provides some promising evidence that acoustic signals can be used to detect PD automatically and early.
The implications of the findings are significant for the field of Parkinson’s disease diagnosis and treatment. By leveraging machine-learning algorithms and analyzing acoustic signals, they have demonstrated the potential for automated and early detection of PD. This approach offers several advantages over traditional diagnosis methods, which often involve time-consuming physical and psychological tests and specialist examinations of the patient’s nervous system. By extracting a set of features from recordings of a person’s voice, we were able to train machine-learning models to distinguish between Parkinson’s cases and healthy individuals. This noninvasive approach has the potential to revolutionize the early detection of PD, enabling timely interventions and improved patient outcomes. The study also introduces two techniques, t-SNE and PCA, for the dimensionality reduction of the dataset. By reducing the number of features while retaining the most informative ones, these techniques help improve the efficiency and performance of the classification algorithms. The experimental results presented in the paper demonstrate the effectiveness of the proposed techniques. These findings have practical implications for healthcare professionals involved in PD diagnosis. The proposed techniques can be applied to develop automated systems that assist in the early screening and diagnosis of PD based on voice analysis. Such systems could potentially be integrated into routine clinical practice, enabling cost-effective and widespread screening for PD, particularly in populations where access to specialized neurological examinations may be limited. Furthermore, the study contributes to the existing knowledge by demonstrating the efficacy of specific machine-learning algorithms (RF and MLP) and dimensionality reduction techniques (t-SNE and PCA) in the context of PD diagnosis. This knowledge can inform future studies and inspire further study into the development of advanced diagnostic tools and methodologies for Parkinson’s disease.
Here are some potential limitations and biases to consider: the study’s methodology relies on analyzing voice recordings to diagnose Parkinson’s disease. It is essential to discuss the specifics of the data-collection process, including the recording equipment used, the recording environment, and any potential limitations or sources of error introduced during data acquisition. The RFE algorithm selected relevant features from the voice recordings. The criteria used for feature selection and the potential impact of excluding certain features. Biases may arise if certain features are overrepresented or if crucial features are unintentionally omitted.