Identification of Radiomic Signatures in Brain MRI Sequences T1 and T2 That Differentiate Tumor Regions of Midline Gliomas with H3.3K27M Mutation
Abstract
:1. Introduction
2. Patients and Methods
2.1. Subject and Study Design
2.2. Imaging Evaluation of Midline Gliomas and Software for Calculation of Radiomics
2.3. Statistical Analysis
2.4. Software
3. Results
3.1. Demographics and Clinical Features
3.2. Selected Radiomics Measurements in Post-Gadolinium T1 and T2 Sequences
3.3. Identification of Radiomics Useful for Discriminating DMG in T1 and T2 Sequences
3.4. Comparison of Viable Tumor versus Peritumoral Edema
4. Discussion
5. Limitations
6. Future Directions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mackay, A.; Burford, A.; Carvalho, D.; Izquierdo, E.; Fazal-Salom, J.; Taylor, K.R.; Bjerke, L.; Clarke, M.; Vinci, M.; Nandhabalan, M.; et al. Integrated Molecular Meta-Analysis of 1000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell 2017, 32, 520–537.e525. [Google Scholar] [CrossRef] [Green Version]
- Lim-Fat, M.J.; Macdonald, M.; Lapointe, S.; Climans, S.A.; Cacciotti, C.; Chahal, M.; Perreault, S.; Tsang, D.S.; Gao, A.; Yip, S.; et al. Molecular testing for adolescent and young adult central nervous system tumors: A Canadian guideline. Front. Oncol. 2022, 12, 960509. [Google Scholar] [CrossRef]
- Nikam, R.M.; Yue, X.; Kaur, G.; Kandula, V.; Khair, A.; Kecskemethy, H.H.; Averill, L.W.; Langhans, S.A. Advanced Neuroimaging Approaches to Pediatric Brain Tumors. Cancers 2022, 14, 3401. [Google Scholar] [CrossRef] [PubMed]
- Immisch, L.; Papafotiou, G.; Popp, O.; Mertins, P.; Blankenstein, T.; Willimsky, G. H3.3K27M mutation is not a suitable target for immunotherapy in HLA-A2(+) patients with diffuse midline glioma. J. Immunother. Cancer 2022, 10, e005535. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.; Holloway, L.; Metcalfe, P.; Koh, E.S.; Brighi, C. Challenges in Glioblastoma Radiomics and the Path to Clinical Implementation. Cancers 2022, 14, 3897. [Google Scholar] [CrossRef] [PubMed]
- Habib, A.; Jovanovich, N.; Hoppe, M.; Ak, M.; Mamindla, P.; Colen, R.R.; Zinn, P.O. MRI-Based Radiomics and Radiogenomics in the Management of Low-Grade Gliomas: Evaluating the Evidence for a Paradigm Shift. J. Clin. Med. 2021, 10, 1411. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Kucharczyk, M.J.; Daniel, P.; Sabri, S.; Jean-Claude, B.J.; Niazi, T.; Abdulkarim, B. Radiomics in Glioblastoma: Current Status and Challenges Facing Clinical Implementation. Front. Oncol. 2019, 9, 374. [Google Scholar] [CrossRef] [Green Version]
- Kandemirli, S.G.; Kocak, B.; Naganawa, S.; Ozturk, K.; Yip, S.S.F.; Chopra, S.; Rivetti, L.; Aldine, A.S.; Jones, K.; Cayci, Z.; et al. Machine Learning-Based Multiparametric Magnetic Resonance Imaging Radiomics for Prediction of H3K27M Mutation in Midline Gliomas. World Neurosurg. 2021, 151, e78–e85. [Google Scholar] [CrossRef]
- Li, Q.; Dong, F.; Jiang, B.; Zhang, M. Exploring MRI Characteristics of Brain Diffuse Midline Gliomas with the H3 K27M Mutation Using Radiomics. Front. Oncol. 2021, 11, 646267. [Google Scholar] [CrossRef]
- Su, X.; Chen, N.; Sun, H.; Liu, Y.; Yang, X.; Wang, W.; Zhang, S.; Tan, Q.; Su, J.; Gong, Q.; et al. Automated machine learning based on radiomics features predicts H3 K27M mutation in midline gliomas of the brain. Neuro Oncol. 2020, 22, 393–401. [Google Scholar] [CrossRef]
- Di Ruscio, V.; Del Baldo, G.; Fabozzi, F.; Vinci, M.; Cacchione, A.; de Billy, E.; Megaro, G.; Carai, A.; Mastronuzzi, A. Pediatric Diffuse Midline Gliomas: An Unfinished Puzzle. Diagnostics 2022, 12, 2064. [Google Scholar] [CrossRef] [PubMed]
- Chilaca-Rosas, M.-F.; Garcia-Lezama, M.; Moreno-Jimenez, S.; Roldan-Valadez, E. Diagnostic Performance of Selected MRI-Derived Radiomics Able to Discriminate Progression-Free and Overall Survival in Patients with Midline Glioma and the H3F3AK27M Mutation. Diagnostics 2023, 13, 849. [Google Scholar] [CrossRef]
- Comité Consultivo Nacional de Normalización de Innovación, Desarrollo, Tecnologías e Información en Salud. NORMA Oficial Mexicana NOM-012-SSA3-2012. Que Establece los Criterios para la Ejecución de Proyectos de Investigación para la Salud en Seres Humanos. Diario Oficial de la Federación. 2013. Available online: http://diariooficial.gob.mx/nota_detalle.php?codigo=5284148&fecha=04/01/2013#gsc.tab=0 (accessed on 21 June 2023).
- Association, W.M. World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects. JAMA 2013, 310, 2191–2194. [Google Scholar]
- Wen, P.Y.; Huse, J.T. 2016 World Health Organization Classification of Central Nervous System Tumors. Continuum (Minneap Minn). CONTINUUM Lifelong Learn. Neurol. 2017, 23, 1531–1547. [Google Scholar] [CrossRef] [PubMed]
- Feiden, S.; Feiden, W. WHO classification of tumours of the CNS: Revised edition of 2007 with critical comments on the typing und grading of common-type diffuse gliomas. Pathologe 2008, 29, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Nioche, C.; Orlhac, F.; Boughdad, S.; Reuze, S.; Goya-Outi, J.; Robert, C.; Pellot-Barakat, C.; Soussan, M.; Frouin, F.; Buvat, I. LIFEx: A Freeware for Radiomic Feature Calculation in Multimodality Imaging to Accelerate Advances in the Characterisation of Tumor Heterogeneity. Cancer Res. 2018, 78, 4786–4789. [Google Scholar] [CrossRef] [Green Version]
- Venn, J. On the diagrammatic and mechanical representation of propositions and reasonings. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1880, 10, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Mejia, M.; Roldan-Valadez, E. Comparisons of Apparent Diffusion Coefficient Values in Penumbra, Infarct, and Normal Brain Regions in Acute Ischemic Stroke: Confirmatory Data Using Bootstrap Confidence Intervals, Analysis of Variance, and Analysis of Means. J. Stroke Cerebrovasc. Dis. 2016, 25, 515–522. [Google Scholar] [CrossRef]
- Mahmood, T.; Riaz, M.; Iqbal, A.; Mulenga, K. An improved statistical approach to compare means. AIMS Math. 2023, 8, 4596–4629. [Google Scholar] [CrossRef]
- Roldan-Valadez, E.; Anaya-Sanchez, S.; Rivera-Sotelo, N.; Moreno-Jimenez, S. Diffusion tensor imaging-derived biomarkers performance in glioblastoma tumor regions: Exploratory data analysis using zombie plots and diagnostic tests. Gac. Med. Mex. 2022, 158, 55–62. [Google Scholar] [CrossRef]
- Durand-Munoz, C.; Flores-Alvarez, E.; Moreno-Jimenez, S.; Roldan-Valadez, E. Pre-operative apparent diffusion coefficient values and tumour region volumes as prognostic biomarkers in glioblastoma: Correlation and progression-free survival analyses. Insights Imaging 2019, 10, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores-Alvarez, E.; Anselmo Rios Piedra, E.; Cruz-Priego, G.A.; Durand-Munoz, C.; Moreno-Jimenez, S.; Roldan-Valadez, E. Correlations between DTI-derived metrics and MRS metabolites in tumour regions of glioblastoma: A pilot study. Radiol. Oncol. 2020, 54, 394–408. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, Z.; Qu, L.; Zhang, P.; Duan, Y.; Cheng, D.; Xu, X.; Sun, T.; Ding, J.; Xie, C.; Liu, X.; et al. Prediction of H3K27M-mutant brainstem glioma by amide proton transfer-weighted imaging and its derived radiomics. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 4426–4436. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; She, D.; Xing, Z.; Lin, X.; Wang, F.; Song, Y.; Cao, D. Multiparametric MRI-Based Radiomics Model for Predicting H3 K27M Mutant Status in Diffuse Midline Glioma: A Comparative Study Across Different Sequences and Machine Learning Techniques. Front. Oncol. 2022, 12, 796583. [Google Scholar] [CrossRef]
- Brancato, V.; Cerrone, M.; Lavitrano, M.; Salvatore, M.; Cavaliere, C. A Systematic Review of the Current Status and Quality of Radiomics for Glioma Differential Diagnosis. Cancers 2022, 14, 2731. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Ren, J.; Niu, G.; Liu, A.; Wu, Q.; Xie, S.; Ma, X.; Li, B.; Wang, P.; Shen, J.; et al. Multiparametric MR radiomics in brain glioma: Models comparation to predict biomarker status. BMC Med. Imaging 2022, 22, 137. [Google Scholar] [CrossRef]
- Sun, C.; Fan, L.; Wang, W.; Wang, W.; Liu, L.; Duan, W.; Pei, D.; Zhan, Y.; Zhao, H.; Sun, T.; et al. Radiomics and Qualitative Features from Multiparametric MRI Predict Molecular Subtypes in Patients with Lower-Grade Glioma. Front. Oncol. 2021, 11, 756828. [Google Scholar] [CrossRef]
- Muller, M.; Winz, O.; Gutsche, R.; Leijenaar, R.T.H.; Kocher, M.; Lerche, C.; Filss, C.P.; Stoffels, G.; Steidl, E.; Hattingen, E.; et al. Static FET PET radiomics for the differentiation of treatment-related changes from glioma progression. J. Neuro-Oncol. 2022, 159, 519–529. [Google Scholar] [CrossRef]
- Zaragori, T.; Verger, A. Reply: (18)F-FDOPA PET for the Non-invasive Prediction of Glioma Molecular Parameters: A Radiomics Study. J. Nucl. Med. 2022, 63, 801–803. [Google Scholar] [CrossRef]
- Langen, K.J.; Mottaghy, F.M. (18)F-FDOPA PET for the Non-invasive Prediction of Glioma Molecular Parameters: A Radiomics Study. J. Nucl. Med. 2022, 63, 801. [Google Scholar] [CrossRef] [PubMed]
- Ahrari, S.; Zaragori, T.; Rozenblum, L.; Oster, J.; Imbert, L.; Kas, A.; Verger, A. Relevance of Dynamic (18)F-DOPA PET Radiomics for Differentiation of High-Grade Glioma Progression from Treatment-Related Changes. Biomedicines 2021, 9, 1924. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Yao, S.; Wang, Z.M.; Zhu, D.M.; Bie, Y.N.; Zhang, S.Z.; Chen, W.L. Multiparametric MRI Features Predict the SYP Gene Expression in Low-Grade Glioma Patients: A Machine Learning-Based Radiomics Analysis. Front. Oncol. 2021, 11, 663451. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Wang, F.F.; Luo, S.; Chen, G.; Tang, G. Diagnostic performance of radiomics using machine learning algorithms to predict MGMT promoter methylation status in glioma patients: A meta-analysis. Diagn. Interv. Radiol. 2021, 27, 716–724. [Google Scholar] [CrossRef]
- Deng, D.B.; Liao, Y.T.; Zhou, J.F.; Cheng, L.N.; He, P.; Wu, S.N.; Wang, W.S.; Zhou, Q. Non-Invasive Prediction of Survival Time of Midline Glioma Patients Using Machine Learning on Multiparametric MRI Radiomics Features. Front. Neurol. 2022, 13, 866274. [Google Scholar] [CrossRef]
- Hu, G.; Hu, X.; Yang, K.; Yu, Y.; Jiang, Z.; Liu, Y.; Liu, D.; Hu, X.; Xiao, H.; Zou, Y.; et al. Radiomics-Based Machine Learning to Predict Recurrence in Glioma Patients Using Magnetic Resonance Imaging. J. Comput. Assist. Tomogr. 2022, 47, 129–135. [Google Scholar] [CrossRef]
- Liu, C.; Li, Y.; Xia, X.; Wang, J.; Hu, C. Application of radiomics feature captured from MRI for prediction of recurrence for glioma patients. J. Cancer 2022, 13, 965–974. [Google Scholar] [CrossRef]
- Shaheen, A.; Bukhari, S.T.; Nadeem, M.; Burigat, S.; Bagci, U.; Mohy-Ud-Din, H. Overall Survival Prediction of Glioma Patients with Multiregional Radiomics. Front. Neurosci. 2022, 16, 911065. [Google Scholar] [CrossRef]
- Zhou, H.; Xu, R.; Mei, H.; Zhang, L.; Yu, Q.; Liu, R.; Fan, B. Application of Enhanced T1WI of MRI Radiomics in Glioma Grading. Int. J. Clin. Pract. 2022, 2022, 3252574. [Google Scholar] [CrossRef]
- Yan, J.; Zhang, B.; Zhang, S.; Cheng, J.; Liu, X.; Wang, W.; Dong, Y.; Zhang, L.; Mo, X.; Chen, Q.; et al. Quantitative MRI-based radiomics for noninvasively predicting molecular subtypes and survival in glioma patients. NPJ Precis. Oncol. 2021, 5, 72. [Google Scholar] [CrossRef]
- Malik, N.; Geraghty, B.; Dasgupta, A.; Maralani, P.J.; Sandhu, M.; Detsky, J.; Tseng, C.L.; Soliman, H.; Myrehaug, S.; Husain, Z.; et al. MRI radiomics to differentiate between low grade glioma and glioblastoma peritumoral region. J. Neuro-Oncol. 2021, 155, 181–191. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Liang, Y.; Wei, R.; Zhang, W.; Yao, W.; Luo, S.; Pang, X.; Wang, Y.; Jiang, X.; et al. Radiomics can differentiate high-grade glioma from brain metastasis: A systematic review and meta-analysis. Eur. Radiol. 2022, 32, 8039–8051. [Google Scholar] [CrossRef] [PubMed]
- Flores-Alvarez, E.; Durand-Munoz, C.; Cortes-Hernandez, F.; Munoz-Hernandez, O.; Moreno-Jimenez, S.; Roldan-Valadez, E. Clinical Significance of Fractional Anisotropy Measured in Peritumoral Edema as a Biomarker of Overall Survival in Glioblastoma: Evidence Using Correspondence Analysis. Neurol. India 2019, 67, 1074–1081. [Google Scholar] [CrossRef] [PubMed]
- Cisneros-Sanchez, A.K.; Flores-Alvarez, E.; Melendez-Mier, G.; Roldan-Valadez, E. Basic principles of mathematical growth modeling applied to high-grade gliomas: A brief clinical review for clinicians. Neurol. India 2018, 66, 1575–1583. [Google Scholar] [CrossRef] [PubMed]
- Cortez-Conradis, D.; Favila, R.; Isaac-Olive, K.; Martinez-Lopez, M.; Rios, C.; Roldan-Valadez, E. Diagnostic performance of regional DTI-derived tensor metrics in glioblastoma multiforme: Simultaneous evaluation of p, q, L, Cl, Cp, Cs, R.A., R.D., A.D., mean diffusivity and fractional anisotropy. Eur. Radiol. 2013, 23, 1112–1121. [Google Scholar] [CrossRef]
- Yi, Z.; Long, L.; Zeng, Y.; Liu, Z. Current Advances and Challenges in Radiomics of Brain Tumors. Front. Oncol. 2021, 11, 732196. [Google Scholar] [CrossRef]
- Jiang, P.; Sinha, S.; Aldape, K.; Hannenhalli, S.; Sahinalp, C.; Ruppin, E. Big data in basic and translational cancer research. Nat. Rev. Cancer 2022, 22, 625–639. [Google Scholar] [CrossRef]
- Raita, Y.; Camargo, C.A., Jr.; Liang, L.; Hasegawa, K. Big Data, Data Science, and Causal Inference: A Primer for Clinicians. Front. Med. 2021, 8, 678047. [Google Scholar] [CrossRef]
- Tomaszewski, M.R.; Gillies, R.J. The Biological Meaning of Radiomic Features. Radiology 2021, 298, 505–516. [Google Scholar] [CrossRef]
- van Houdt, P.J.; Yang, Y.; van der Heide, U.A. Quantitative Magnetic Resonance Imaging for Biological Image-Guided Adaptive Radiotherapy. Front. Oncol. 2020, 10, 615643. [Google Scholar] [CrossRef]
- Tang, P.L.Y.; Mendez Romero, A.; Jaspers, J.P.M.; Warnert, E.A.H. The potential of advanced M.R. techniques for precision radiotherapy of glioblastoma. MAGMA 2022, 35, 127–143. [Google Scholar] [CrossRef] [PubMed]
- Roldan-Valadez, E.; Rios, C.; Motola-Kuba, D.; Matus-Santos, J.; Villa, A.R.; Moreno-Jimenez, S. Choline-to-N-acetyl aspartate and lipids-lactate-to-creatine ratios together with age assemble a significant Cox’s proportional-hazards regression model for prediction of survival in high-grade gliomas. Br. J. Radiol. 2016, 89, 20150502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, X.L.; Luo, H.J.; Ren, J.L.; Yin, P.; Liu, Y.; Niu, J.; Hong, N. Multisequence magnetic resonance imaging-based radiomics models for the prediction of microsatellite instability in endometrial cancer. Radiol. Med. 2023, 128, 242–251. [Google Scholar] [CrossRef] [PubMed]




| Clinical Characteristics | n = 12 (100%) |
|---|---|
| Age | Median ten years (range 19–29) years |
| Sex (M:F) | 2:1 |
| Paediatric patients | 9 (75) |
| Young Adults Patients | 3 (25) |
| Anatomical location | |
| 3 (25) |
| 3 (25) |
| 6 (50) |
| Volume | Median 18.63 mL (range 4.82–140.15) |
| Volume voxel | Median 7368.5 voxels (range 1545–28,582) |
| KNF | |
| ≥80 | 3 (25) |
| ≤70 | 9 (75) |
| Surgical intervention | |
| 10 (83.4) |
| 1 (8.3) |
| 1 (8.3) |
| Radiation therapy dose | Median: 55 Gy (range 54–55 Gy) |
| Radiation therapy scheme | |
| 10 (83.3) |
| 2 (16.7) |
| Chemotherapy | |
| 9 (75) |
| 3 (25) |
| n | Percentile | IQR | p-Value | |||
|---|---|---|---|---|---|---|
| Median | 75 | 25 | ||||
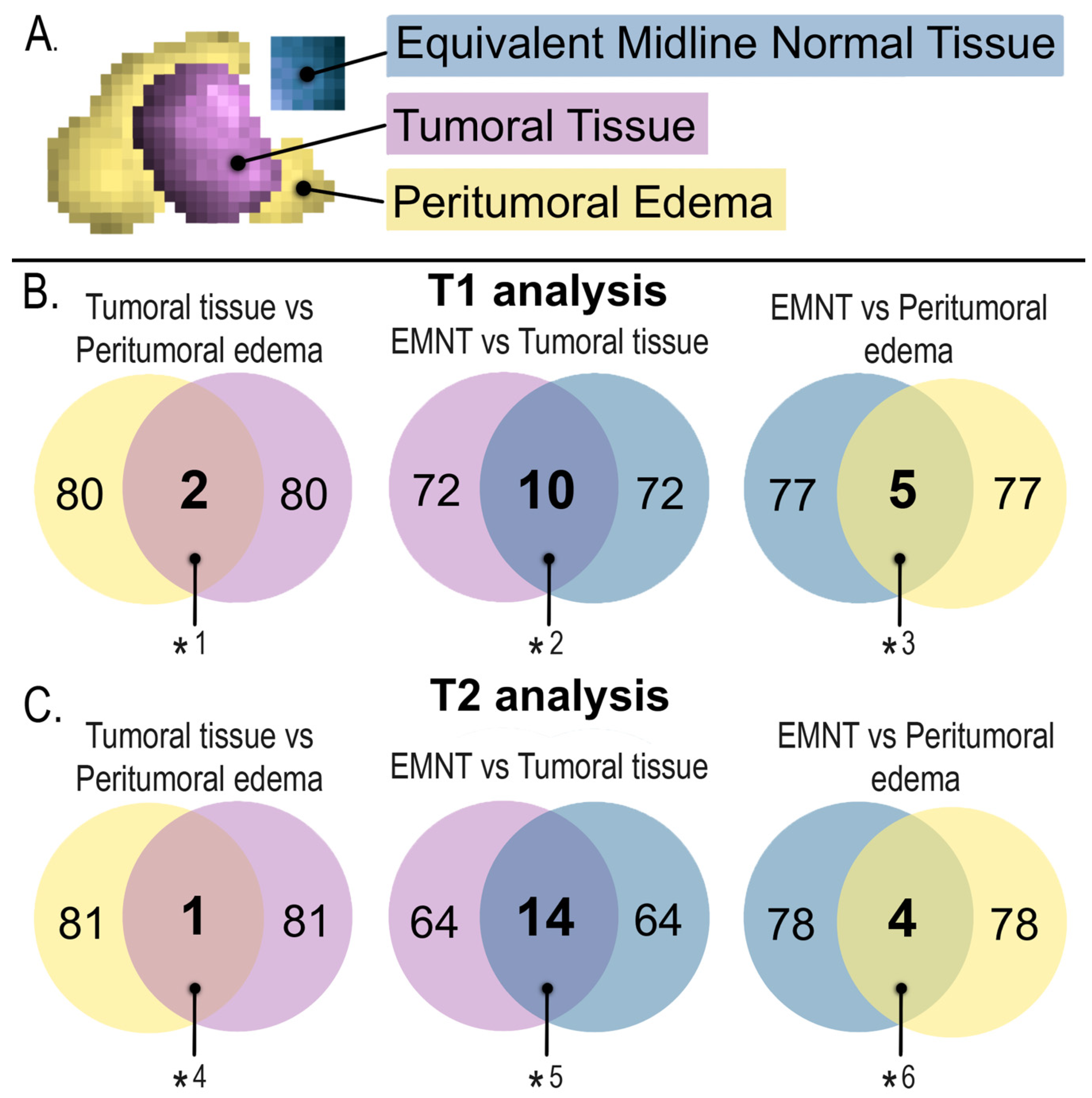
| Radiomics that showed a significant difference between EMNT vs. tumoral tissue | ||||||
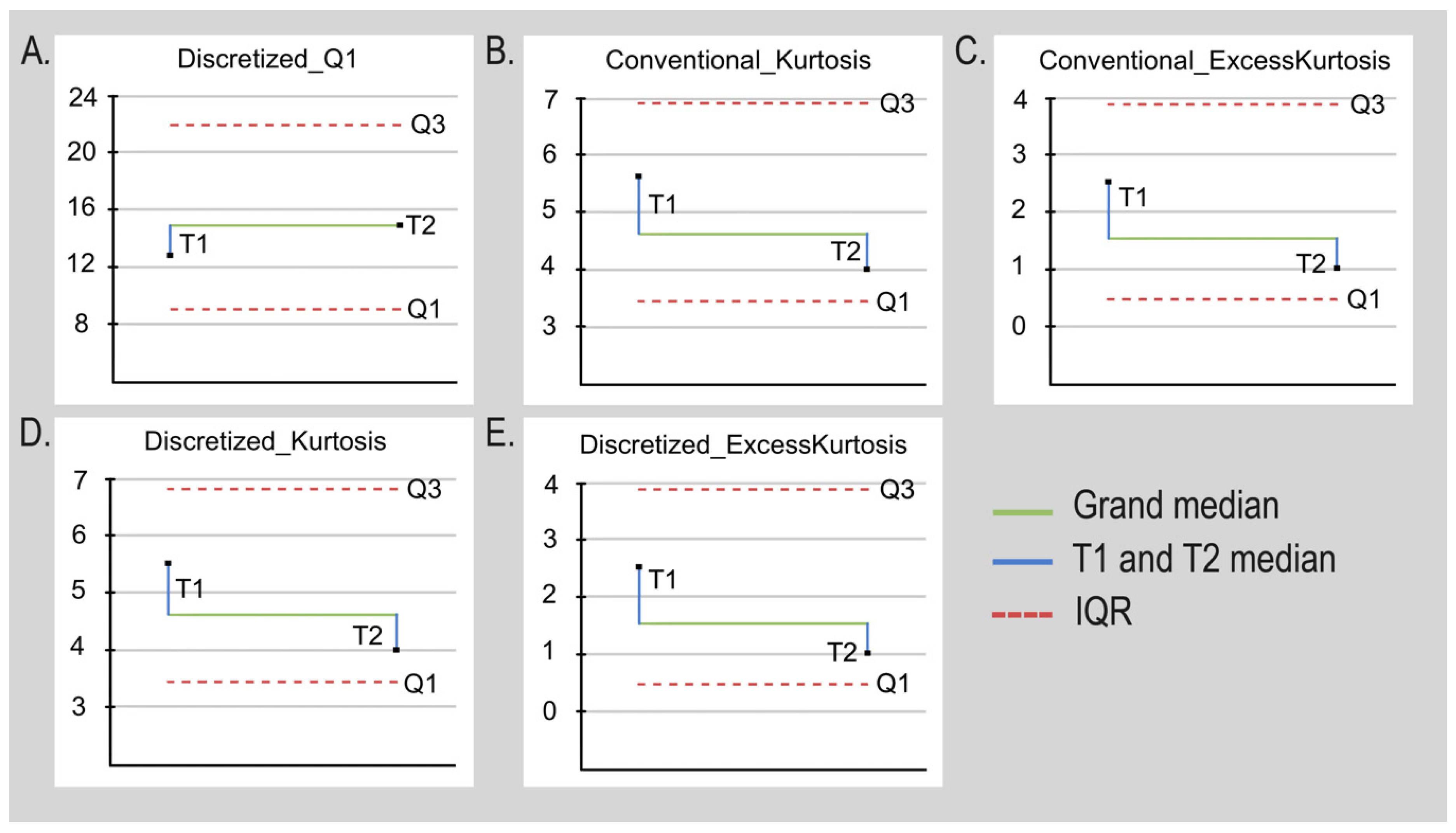
| 24 | 64.756 | 123.139 | 40.629 | 82.51 | 0.003 |
| 24 | 0.115 | 0.592 | −1.122 | 1.714 | 0.027 |
| 24 | 5.197 | 9.514 | 3.401 | 6.113 | 0.043 |
| 24 | 2.197 | 6.514 | 0.401 | 6.113 | 0.043 |
| 24 | 50.362 | 101.225 | 27.328 | 73.897 | 0.005 |
| 24 | 13.000 | 18.750 | 9.250 | 9.5 | 0.047 |
| 24 | 0.120 | 0.592 | −1.128 | 1.72 | 0.027 |
| 24 | 5.166 | 9.152 | 3.396 | 5.756 | 0.043 |
| 24 | 2.166 | 6.152 | 0.396 | 5.756 | 0.043 |
| 24 | 20.000 | 36.711 | 13.528 | 23.183 | 0.020 |
| Radiomics that showed a significant difference between EMNT vs. peritumoral edema | ||||||
| 24 | 5.197 | 9.514 | 3.401 | 6.113 | 0.017 |
| 24 | 2.197 | 6.514 | 0.401 | 6.113 | 0.017 |
| 24 | 5.166 | 9.152 | 3.396 | 5.756 | 0.017 |
| 24 | 2.166 | 6.152 | 0.396 | 5.756 | 0.017 |
| 24 | 0.099 | 0.148 | 0.062 | 0.086 | 0.050 |
| Radiomics that showed a significant difference between tumor tissue versus peritumoral edema | ||||||
| 24 | 0.115 | 0.592 | −1.122 | 1.714 | 0.028 |
| 24 | 0.120 | 0.592 | −1.128 | 1.72 | 0.028 |
| n | Percentile | IQR | p-Value | |||
|---|---|---|---|---|---|---|
| Median | 75 | 25 | ||||
| Radiomics that showed a significant difference between EMNT vs. tumoral tissue | ||||||
| 28 | 19.929 | 24.208 | 12.756 | 11.452 | 0.037 |
| 28 | 16.000 | 21.500 | 11.000 | 10.5 | 0.037 |
| 28 | 20.000 | 23.500 | 12.250 | 11.25 | 0.044 |
| 28 | 23.500 | 27.500 | 15.000 | 12.5 | 0.044 |
| 24 | 0.004 | 0.030 | 0.002 | 0.028 | 0.005 |
| 24 | 435.251 | 672.443 | 135.606 | 536.837 | 0.004 |
| 24 | 0.004 | 0.027 | 0.002 | 0.025 | 0.009 |
| 24 | 405.484 | 604.182 | 127.266 | 476.916 | 0.005 |
| 24 | 0.007 | 0.050 | 0.004 | 0.046 | 0.007 |
| 24 | 592.797 | 1021.915 | 181.131 | 840.784 | 0.004 |
| 25 | 0.005 | 0.027 | 0.004 | 0.023 | 0.026 |
| 25 | 426.704 | 582.915 | 258.133 | 324.782 | 0.026 |
| 25 | 0.003 | 0.015 | 0.002 | 0.013 | 0.016 |
| 25 | 242.679 | 348.291 | 175.596 | 172.695 | 0.033 |
| Radiomics that showed a significant difference between EMNT vs. peritumoral edema | ||||||
| 28 | 3.716 | 4.966 | 3.454 | 1.512 | 0.028 |
| 28 | 0.716 | 1.966 | 0.454 | 1.512 | 0.028 |
| 28 | 3.706 | 4.956 | 3.452 | 1.504 | 0.034 |
| 28 | 0.706 | 1.956 | 0.452 | 1.504 | 0.034 |
| Radiomics that showed a significant difference between tumor tissue versus peritumoral edema | ||||||
| 24 | 0.007 | 0.050 | 0.004 | 0.046 | 0.032 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chilaca-Rosas, M.-F.; Contreras-Aguilar, M.-T.; Garcia-Lezama, M.; Salazar-Calderon, D.-R.; Vargas-Del-Angel, R.-G.; Moreno-Jimenez, S.; Piña-Sanchez, P.; Trejo-Rosales, R.-R.; Delgado-Martinez, F.-A.; Roldan-Valadez, E. Identification of Radiomic Signatures in Brain MRI Sequences T1 and T2 That Differentiate Tumor Regions of Midline Gliomas with H3.3K27M Mutation. Diagnostics 2023, 13, 2669. https://doi.org/10.3390/diagnostics13162669
Chilaca-Rosas M-F, Contreras-Aguilar M-T, Garcia-Lezama M, Salazar-Calderon D-R, Vargas-Del-Angel R-G, Moreno-Jimenez S, Piña-Sanchez P, Trejo-Rosales R-R, Delgado-Martinez F-A, Roldan-Valadez E. Identification of Radiomic Signatures in Brain MRI Sequences T1 and T2 That Differentiate Tumor Regions of Midline Gliomas with H3.3K27M Mutation. Diagnostics. 2023; 13(16):2669. https://doi.org/10.3390/diagnostics13162669
Chicago/Turabian StyleChilaca-Rosas, Maria-Fatima, Manuel-Tadeo Contreras-Aguilar, Melissa Garcia-Lezama, David-Rafael Salazar-Calderon, Raul-Gabriel Vargas-Del-Angel, Sergio Moreno-Jimenez, Patricia Piña-Sanchez, Raul-Rogelio Trejo-Rosales, Felipe-Alfredo Delgado-Martinez, and Ernesto Roldan-Valadez. 2023. "Identification of Radiomic Signatures in Brain MRI Sequences T1 and T2 That Differentiate Tumor Regions of Midline Gliomas with H3.3K27M Mutation" Diagnostics 13, no. 16: 2669. https://doi.org/10.3390/diagnostics13162669
APA StyleChilaca-Rosas, M.-F., Contreras-Aguilar, M.-T., Garcia-Lezama, M., Salazar-Calderon, D.-R., Vargas-Del-Angel, R.-G., Moreno-Jimenez, S., Piña-Sanchez, P., Trejo-Rosales, R.-R., Delgado-Martinez, F.-A., & Roldan-Valadez, E. (2023). Identification of Radiomic Signatures in Brain MRI Sequences T1 and T2 That Differentiate Tumor Regions of Midline Gliomas with H3.3K27M Mutation. Diagnostics, 13(16), 2669. https://doi.org/10.3390/diagnostics13162669






