Classification of Diabetic Retinopathy Disease Levels by Extracting Spectral Features Using Wavelet CNN
Abstract
1. Introduction
- A novel approach is introduced that combines Wavelet CNN to extract spectral features from retinal fundus images to perform DR disease classification across multiple grades.
- The extracted features from the Wavelet CNN are utilised as inputs for an SVM classifier, enabling efficient and effective classification.
2. Related Works
2.1. Motivation
2.2. Wavelet-Based Techniques Applied to Retinal Images
2.3. Doctor Grading Tasks Using EyePACS Dataset
2.4. Research Gaps
- Wavelet features are effective for texture analysis in retinal images. Without them, the ability to quantify and analyse different textures such as microaneurysms, exudates, or drusen is limited.
- Wavelet CNNs can effectively analyse retinal images at multiple scales, capturing both fine details and global features. Without them, the analysis is limited to a fixed resolution that leads to missing important structures at different scales.
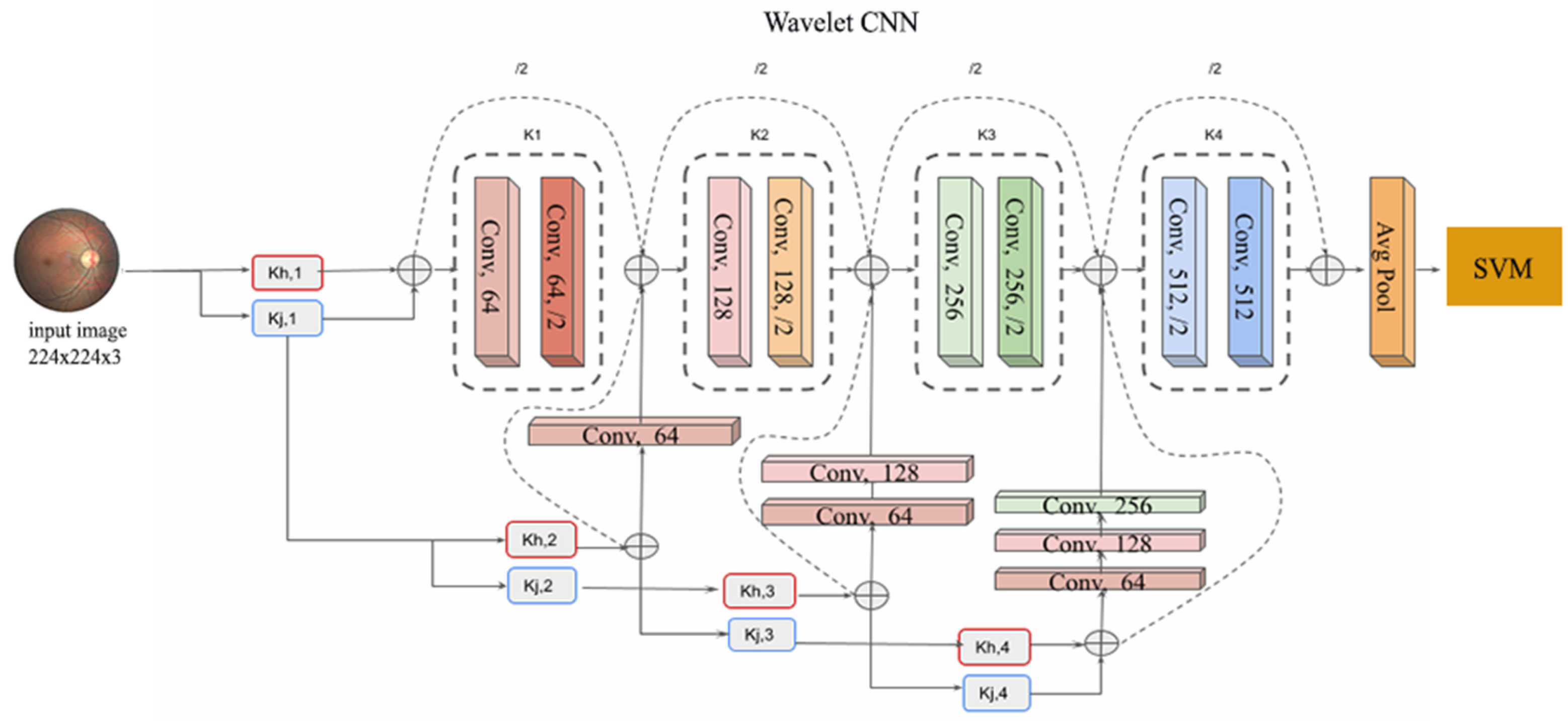
3. Materials and Methods
The Proposed Pipeline
4. Classifiers
4.1. Support Vector Machine
4.2. XGBoost
4.3. Random Forest
4.4. Experimental Setup
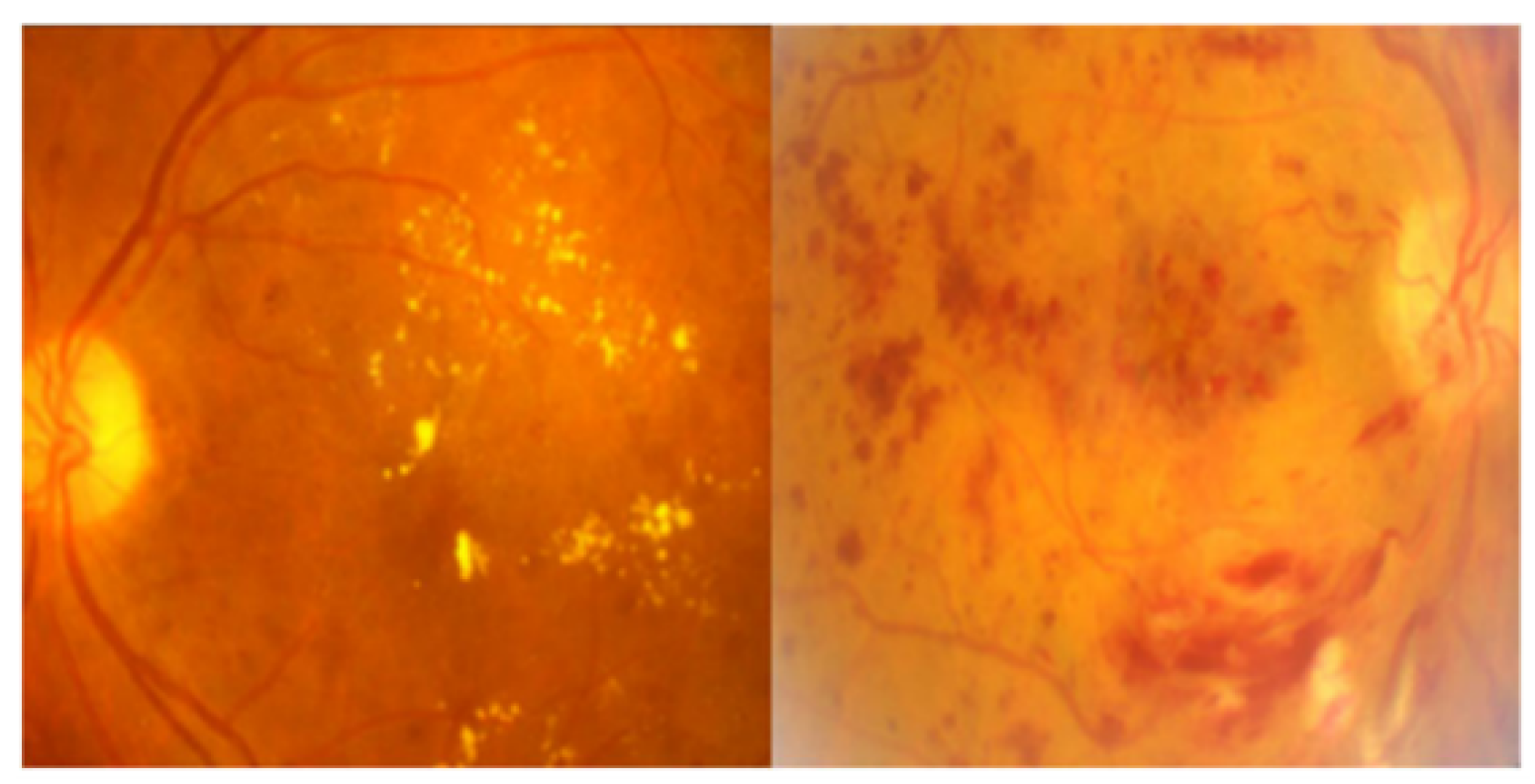
4.5. Dataset Description
4.6. Metrics Used
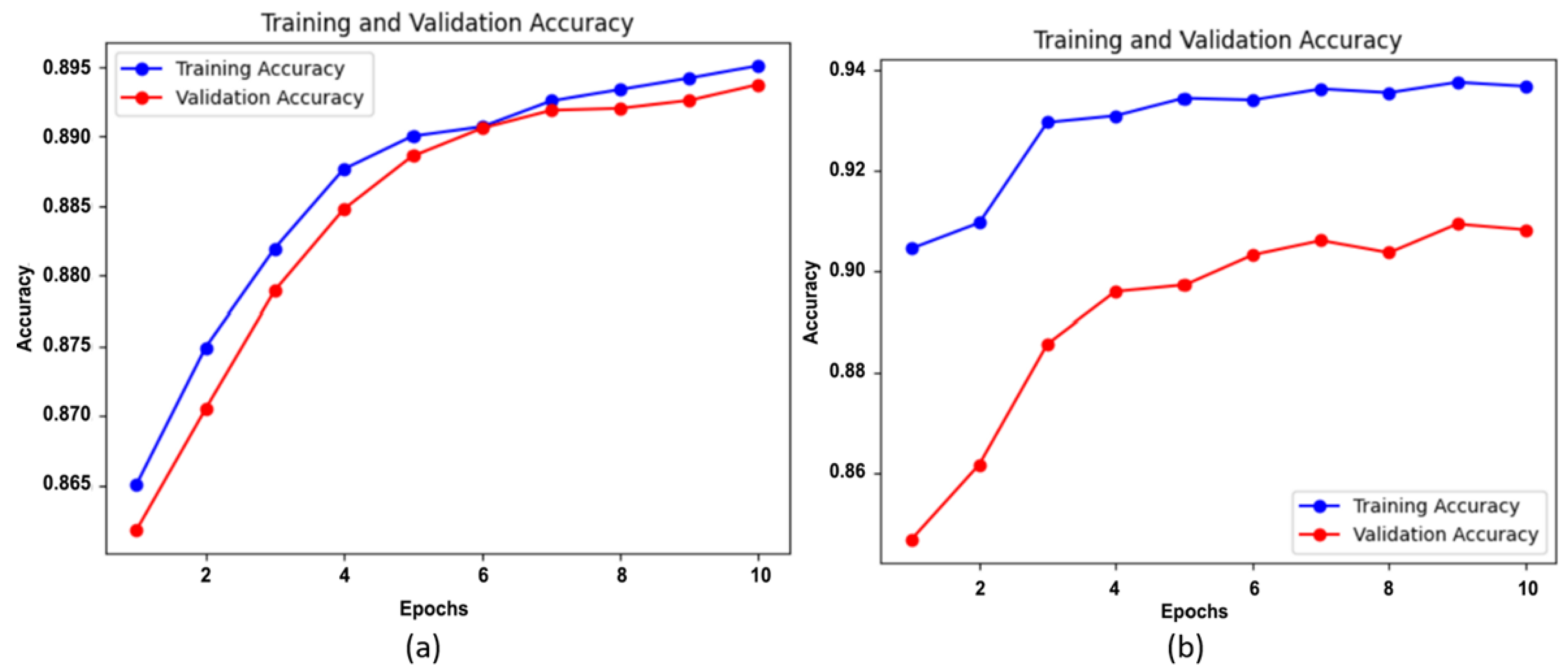
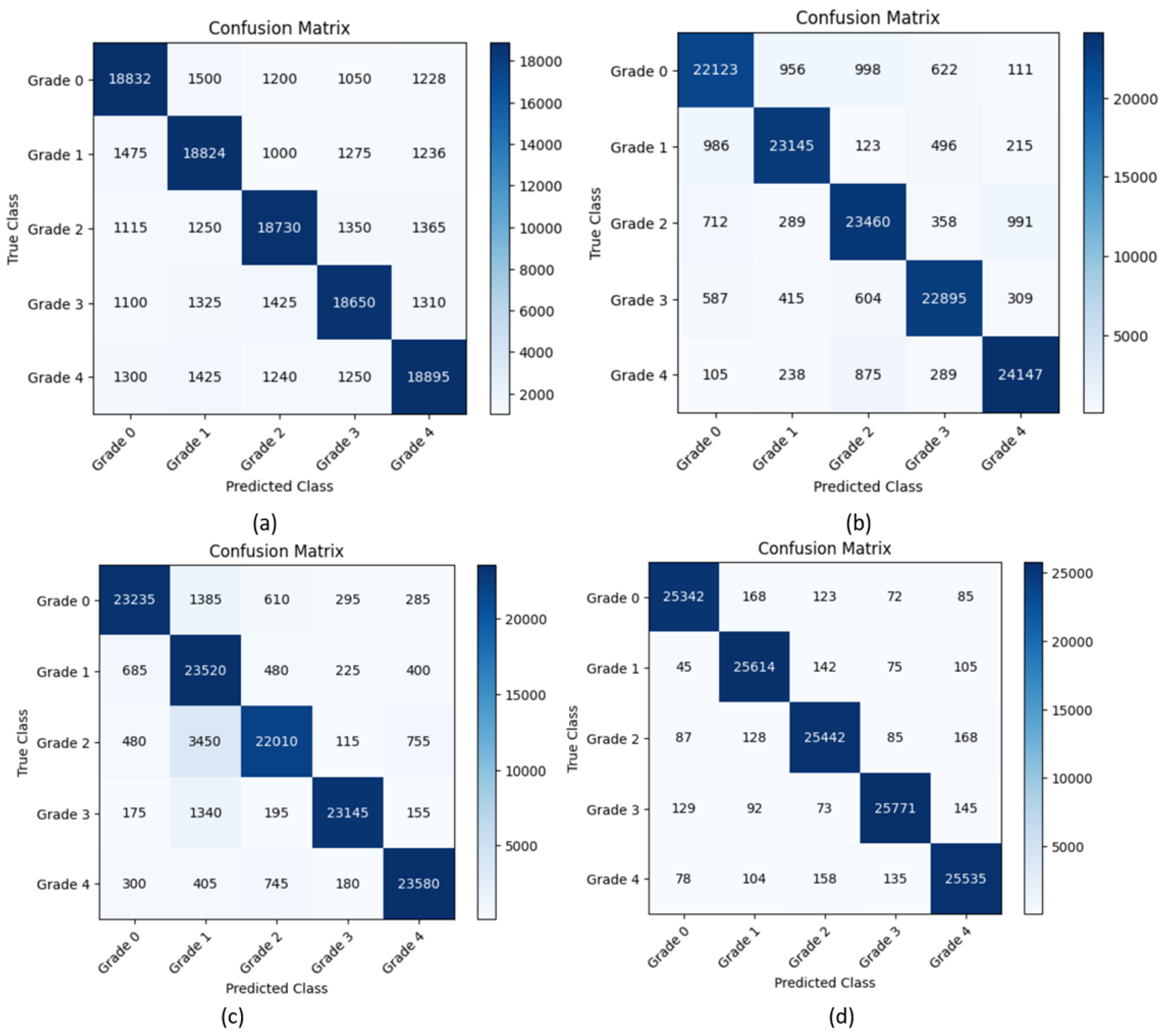
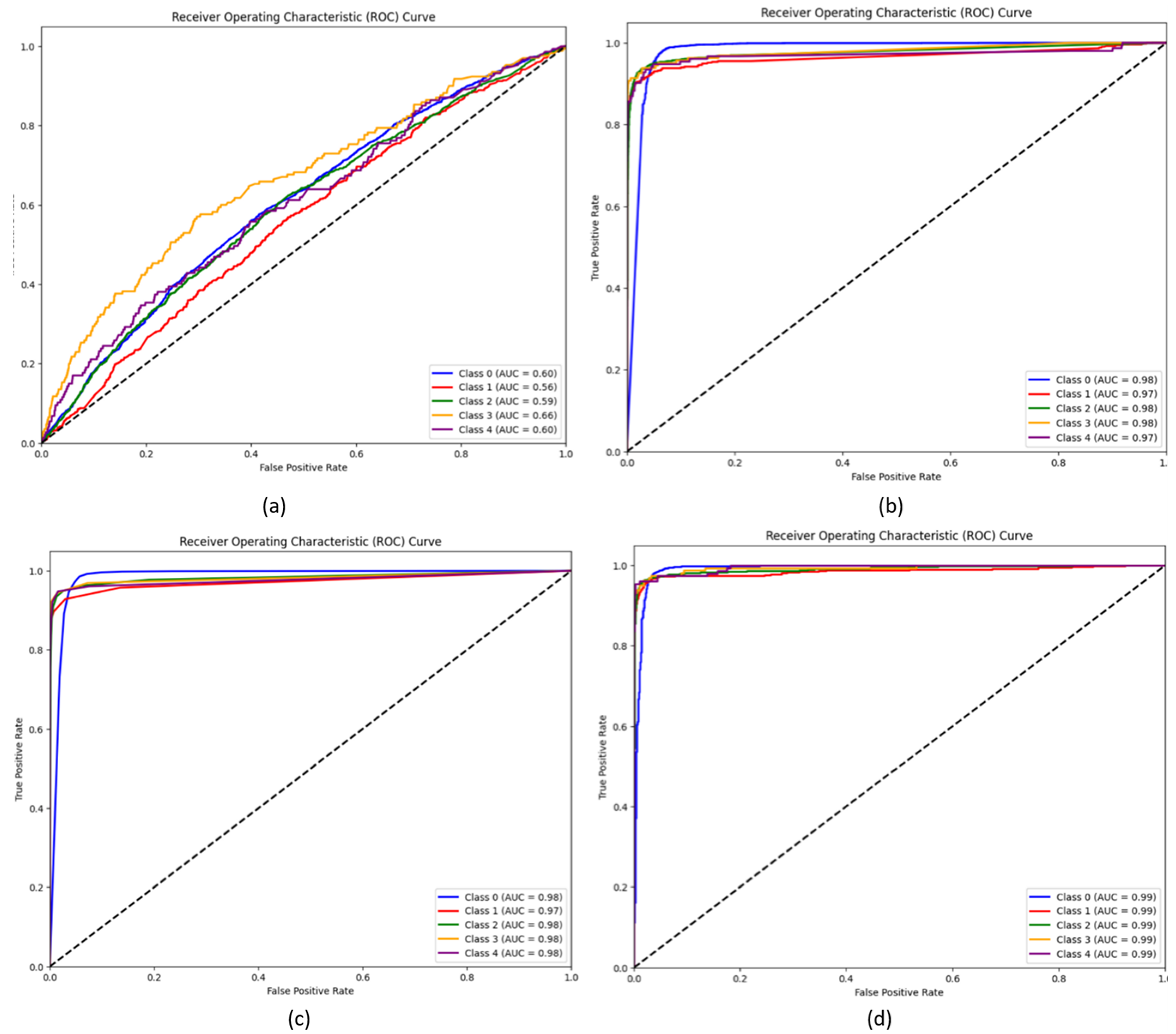
5. Results and Discussion
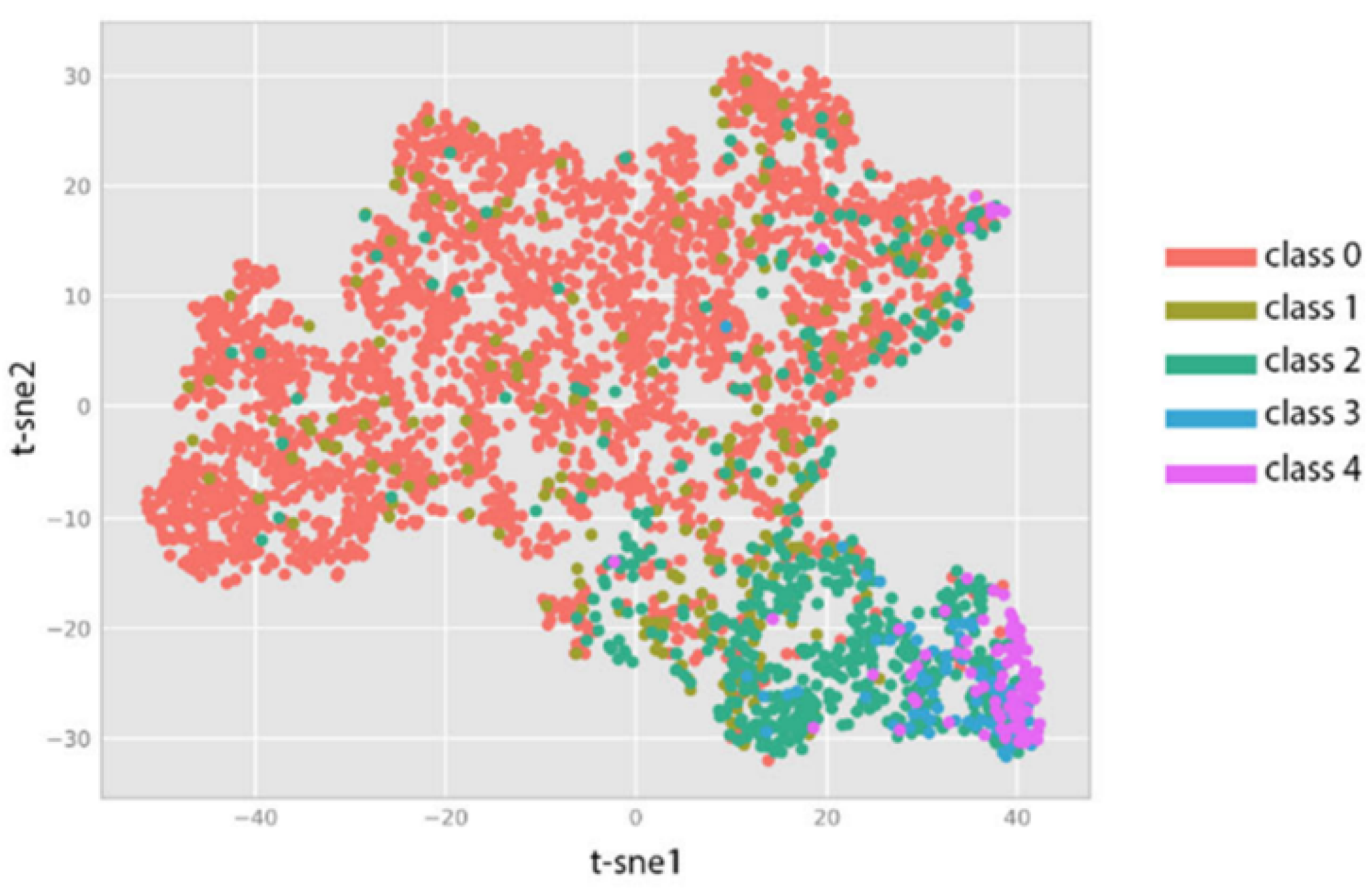
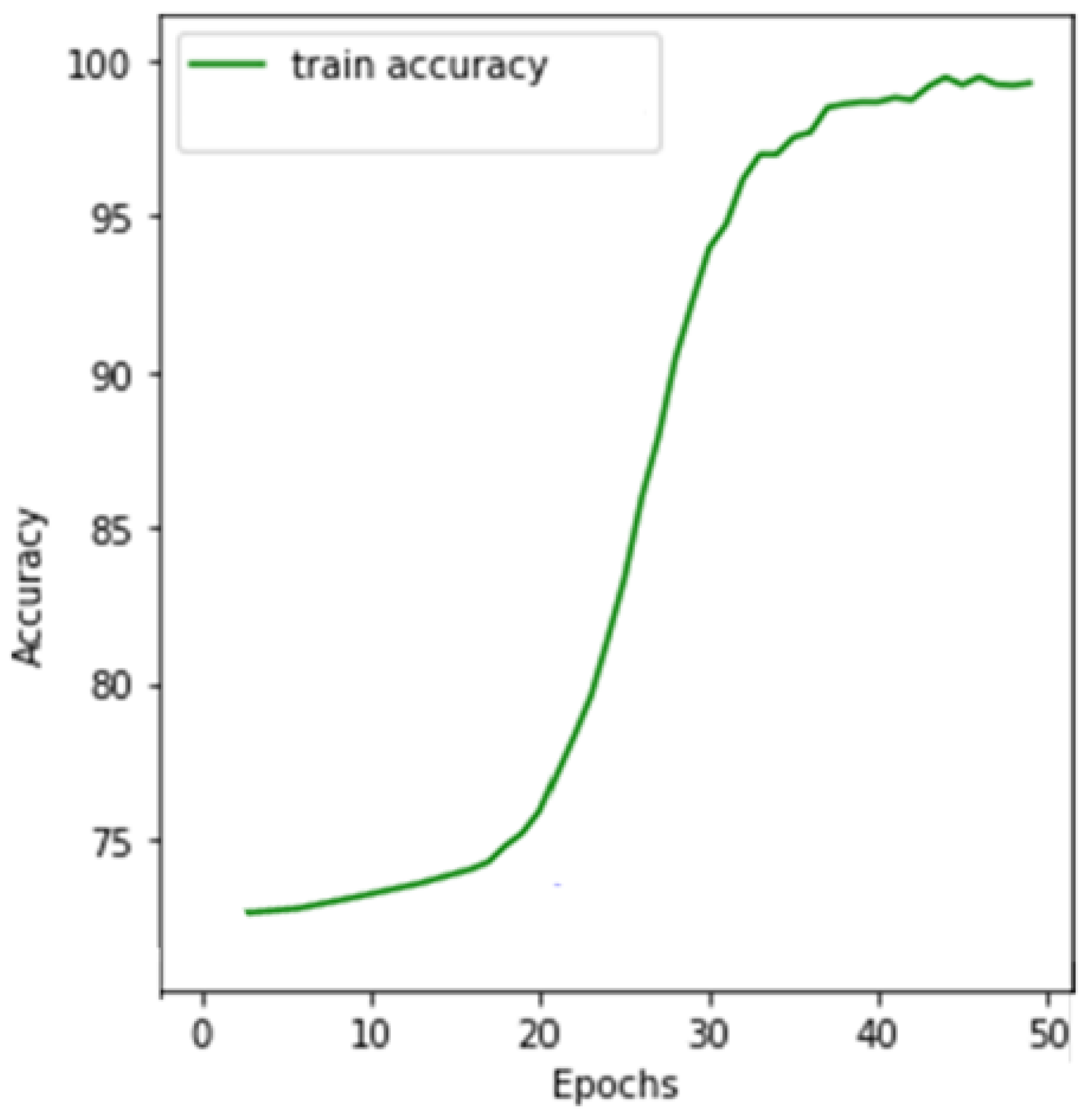
5.1. Experimental Results
5.2. Comparison with Pretrained CNN Models
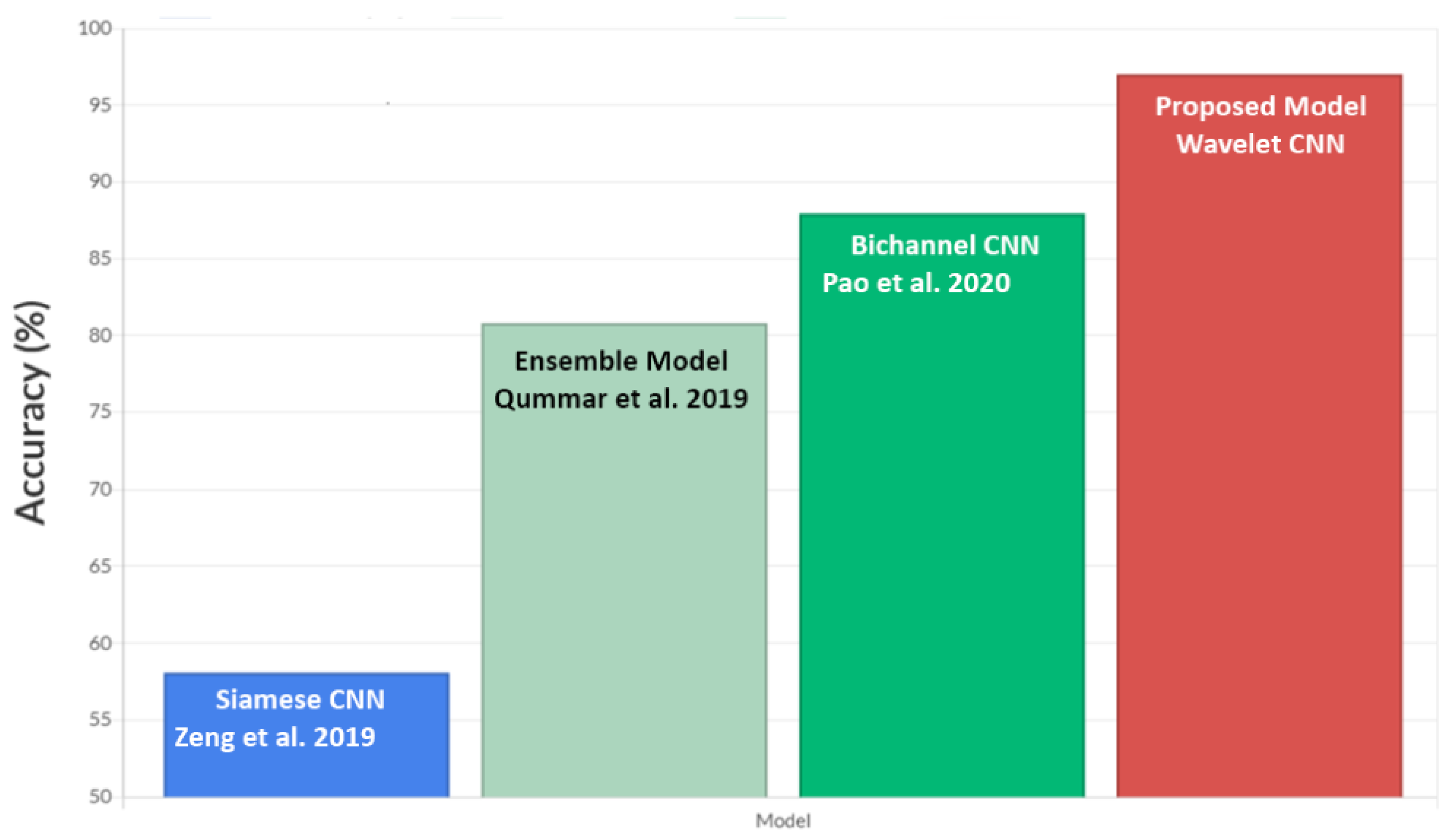
5.3. Comparison with Other Models Using EyePACS Dataset
5.4. Discussion
- SVM can effectively learn complex decision boundaries to transform the data into a higher-dimensional space. This allows SVM to capture intricate relationships and patterns that might be missed by random forest and XGBoost.
- SVM is less sensitive to outliers compared to ensemble methods like random forest and XGBoost. SVM focuses on maximising the margin between different classes, which makes it more robust to noisy and outlier data points. On the other hand, decision trees in random forest and XGBoost can be influenced by outliers and may create biased splits.
- The hyperparameters of SVM, such as C, which defines the penalty for misclassification, and Gamma, which defines the influence of a single training example, reduce the risk of overfitting or underfitting.
6. Conclusions
7. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eisma, J.H.; Dulle, J.E.; Fort, P.E. Current knowledge on diabetic retinopathy from human donor tissues. World J. Diabetes 2015, 6, 312. [Google Scholar] [CrossRef] [PubMed]
- Cho, N.H.; Shaw, J.; Karuranga, S.; Huang, Y.; da Rocha Fernandes, J.; Ohlrogge, A.; Malanda, B. IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract. 2018, 138, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Yahaya, S.W.; Lotfi, A.; Mahmud, M. Towards a data-driven adaptive anomaly detection system for human activity. Pattern Recognit. Lett. 2021, 145, 200–207. [Google Scholar] [CrossRef]
- Fabietti, M.I.; Mahmud, M.; Lotfi, A.; Leparulo, A.; Fontana, R.; Vassanelli, S.; Fassolato, C. Detection of Healthy and Unhealthy Brain States from Local Field Potentials Using Machine Learning. In Proceedings of the International Conference on Brain Informatics, Padua, Italy, 15–17 July 2022; pp. 27–39. [Google Scholar]
- Akter, T.; Ali, M.H.; Satu, M.S.; Khan, M.I.; Mahmud, M. Towards autism subtype detection through identification of discriminatory factors using machine learning. In Proceedings of the International Conference on Brain Informatics, Virtual, 17–19 September 2021; pp. 401–410. [Google Scholar]
- Ahmed, S.; Hossain, M.F.; Nur, S.B.; Shamim Kaiser, M.; Mahmud, M. Toward Machine Learning-Based Psychological Assessment of Autism Spectrum Disorders in School and Community. In Proceedings of the Trends in Electronics and Health Informatics: TEHI 2021; Springer Nature Singapore: Singapore, 2022; pp. 139–149. [Google Scholar]
- Mahmud, M.; Kaiser, M.S.; Rahman, M.A.; Wadhera, T.; Brown, D.J.; Shopland, N.; Burton, A.; Hughes-Roberts, T.; Al Mamun, S.; Ieracitano, C.; et al. Towards explainable and privacy-preserving artificial intelligence for personalisation in autism spectrum disorder. In Proceedings of the International Conference on Human-Computer Interaction; Springer: Cham, Switzerland, 2022; pp. 356–370. [Google Scholar]
- Biswas, M.; Rahman, A.; Kaiser, M.S.; Al Mamun, S.; Ebne Mizan, K.S.; Islam, M.S.; Mahmud, M. Indoor navigation support system for patients with neurodegenerative diseases. In Proceedings of the International Conference on Brain Informatics, Virtual, 17–19 September 2021; pp. 411–422. [Google Scholar]
- Mahmud, M.; Kaiser, M.S. Machine learning in fighting pandemics: A COVID-19 case study. In COVID-19: Prediction, Decision-Making, and Its Impacts; Springer: Singapore, 2021; pp. 77–81. [Google Scholar]
- Farhin, F.; Sultana, I.; Islam, N.; Kaiser, M.S.; Rahman, M.S.; Mahmud, M. Attack detection in internet of things using software defined network and fuzzy neural network. In Proceedings of the 2020 Joint 9th International Conference on Informatics, Electronics & Vision (ICIEV) and 2020 4th International Conference on Imaging, Vision & Pattern Recognition (icIVPR), Kitakyushu, Japan, 26–29 August 2020; pp. 1–6. [Google Scholar]
- Ahmed, S.; Hossain, M.F.; Kaiser, M.S.; Noor, M.B.T.; Mahmud, M.; Chakraborty, C. Artificial intelligence and machine learning for ensuring security in smart cities. In Data-Driven Mining, Learning and Analytics for Secured Smart Cities; Springer: Cham, Switzerland, 2021; pp. 23–47. [Google Scholar]
- Islam, N.; Farhin, F.; Sultana, I.; Kaiser, M.S.; Rahman, M.S.; Mahmud, M.; Hosen, A.S.M.S.; Cho, G.H. Towards machine learning based intrusion detection in IoT networks. Comput. Mater. Contin 2021, 69, 1801–1821. [Google Scholar] [CrossRef]
- Esha, N.H.; Tasmim, M.R.; Huq, S.; Mahmud, M.; Kaiser, M.S. Trust IoHT: A trust management model for internet of healthcare things. In Proceedings of the International Conference on Data Science and Applications: ICDSA 2019; Springer: Singapore, 2021; pp. 47–57. [Google Scholar]
- Zohora, M.F.; Tania, M.H.; Kaiser, M.S.; Mahmud, M. Forecasting the risk of type ii diabetes using reinforcement learning. In Proceedings of the 2020 Joint 9th International Conference on Informatics, Electronics & Vision (ICIEV) and 2020 4th International Conference on Imaging, Vision & Pattern Recognition (icIVPR), Kitakyushu, Japan, 26–29 August 2020; pp. 1–6. [Google Scholar]
- Deepa, B.; Murugappan, M.; Sumithra, M.; Mahmud, M.; Al-Rakhami, M.S. Pattern Descriptors Orientation and MAP Firefly Algorithm Based Brain Pathology Classification Using Hybridized Machine Learning Algorithm. IEEE Access 2021, 10, 3848–3863. [Google Scholar] [CrossRef]
- Mammoottil, M.J.; Kulangara, L.J.; Cherian, A.S.; Mohandas, P.; Hasikin, K.; Mahmud, M. Detection of Breast Cancer from Five-View Thermal Images Using Convolutional Neural Networks. J. Healthc. Eng. 2022, 2022, 4295221. [Google Scholar] [CrossRef] [PubMed]
- Kumar, I.; Kumar, A.; Kumar, V.A.; Kannan, R.; Vimal, V.; Singh, K.U.; Mahmud, M. Dense Tissue Pattern Characterization Using Deep Neural Network. Cogn. Comput. 2022, 14, 1728–1751. [Google Scholar] [CrossRef]
- Farhin, F.; Kaiser, M.S.; Mahmud, M. Secured smart healthcare system: Blockchain and bayesian inference based approach. In Proceedings of the International Conference on Trends in Computational and Cognitive Engineering: Proceedings of TCCE 2020; Springer: Singapore, 2021; pp. 455–465. [Google Scholar]
- Kaiser, M.S.; Zenia, N.; Tabassum, F.; Mamun, S.A.; Rahman, M.A.; Islam, M.S.; Mahmud, M. 6G access network for intelligent internet of healthcare things: Opportunity, challenges, and research directions. In Proceedings of the International Conference on Trends in Computational and Cognitive Engineering: Proceedings of TCCE 2020; Springer: Singapore, 2021; pp. 317–328. [Google Scholar]
- Rabby, G.; Azad, S.; Mahmud, M.; Zamli, K.Z.; Rahman, M.M. A flexible keyphrase extraction technique for academic literature. Procedia Comput. Sci. 2018, 135, 553–563. [Google Scholar] [CrossRef]
- Ghosh, T.; Banna, M.H.A.; Angona, T.M.; Nahian, M.J.A.; Uddin, M.N.; Kaiser, M.S.; Mahmud, M. An Attention-Based Mood Controlling Framework for Social Media Users. In Proceedings of the International Conference on Brain Informatics, Virtual, 17–19 September 2021; pp. 245–256. [Google Scholar]
- Rahman, M.A.; Brown, D.J.; Shopland, N.; Burton, A.; Mahmud, M. Explainable multimodal machine learning for engagement analysis by continuous performance test. In Proceedings of the International Conference on Human-Computer Interaction, Virtual, 26 June–1 July 2022; pp. 386–399. [Google Scholar]
- Ahuja, N.J.; Thapliyal, M.; Bisht, A.; Stephan, T.; Kannan, R.; Al-Rakhami, M.S.; Mahmud, M. An Investigative Study on the Effects of Pedagogical Agents on Intrinsic, Extraneous and Germane Cognitive Load: Experimental Findings with Dyscalculia and Non-Dyscalculia Learners. IEEE Access 2021, 10, 3904–3922. [Google Scholar] [CrossRef]
- Sundar, S.; Sumathy, S. An effective deep learning model for grading abnormalities in retinal fundus images using variational auto-encoders. Int. J. Imaging Syst. Technol. 2023, 33, 92–107. [Google Scholar] [CrossRef]
- Sundar, S.; Sumathy, S. Classification of Diabetic Retinopathy disease levels by extracting topological features using Graph Neural Networks. IEEE Access 2023, 11, 51435–51444. [Google Scholar] [CrossRef]
- Sundar, S.; Sumathy, S. RetU-Net: An Enhanced U-Net Architecture for Retinal Lesion Segmentation. Int. J. Artif. Intell. Tools 2023, 32, 2350013. [Google Scholar] [CrossRef]
- Dai, L.; Sheng, B.; Chen, T.; Wu, Q.; Liu, R.; Cai, C.; Wu, L.; Yang, D.; Hamzah, H.; Liu, Y.; et al. A deep learning system for predicting time to progression of diabetic retinopathy. Nat. Med. 2024, 30, 584–594. [Google Scholar] [CrossRef]
- Choi, J.Y.; Ryu, I.H.; Kim, J.K.; Lee, I.S.; Yoo, T.K. Development of a generative deep learning model to improve epiretinal membrane detection in fundus photography. BMC Med. Inform. Decis. Mak. 2024, 24, 25. [Google Scholar] [CrossRef]
- Gangwar, A.K.; Ravi, V. Diabetic retinopathy detection using transfer learning and deep learning. In Proceedings of the Evolution in Computational Intelligence: Frontiers in Intelligent Computing: Theory and Applications (FICTA 2020); Springer: Singapore, 2021; Volume 1, pp. 679–689. [Google Scholar]
- Gargeya, R.; Leng, T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology 2017, 124, 962–969. [Google Scholar] [CrossRef]
- Li, X.; Hu, X.; Yu, L.; Zhu, L.; Fu, C.W.; Heng, P.A. CANet: Cross-disease attention network for joint diabetic retinopathy and diabetic macular edema grading. IEEE Trans. Med. Imaging 2019, 39, 1483–1493. [Google Scholar] [CrossRef]
- Choi, J.Y.; Yoo, T.K.; Seo, J.G.; Kwak, J.; Um, T.T.; Rim, T.H. Multi-categorical deep learning neural network to classify retinal images: A pilot study employing small database. PLoS ONE 2017, 12, e0187336. [Google Scholar] [CrossRef]
- Prakash, N.; Murugappan, M.; Hemalakshmi, G.; Jayalakshmi, M.; Mahmud, M. Deep transfer learning for COVID-19 detection and infection localization with superpixel based segmentation. Sustain. Cities Soc. 2021, 75, 103252. [Google Scholar] [CrossRef]
- Guo, T.; Zhang, T.; Lim, E.; Lopez-Benitez, M.; Ma, F.; Yu, L. A review of wavelet analysis and its applications: Challenges and opportunities. IEEE Access 2022, 10, 58869–58903. [Google Scholar] [CrossRef]
- Agboola, H.A.; Zaccheus, J.E. Wavelet image scattering based glaucoma detection. BMC Biomed. Eng. 2023, 5, 1. [Google Scholar] [CrossRef]
- Abdel-Hamid, L. Retinal image quality assessment using transfer learning: Spatial images vs. wavelet detail subbands. Ain Shams Eng. J. 2021, 12, 2799–2807. [Google Scholar] [CrossRef]
- Parashar, D.; Agrawal, D.K. Automated classification of glaucoma stages using flexible analytic wavelet transform from retinal fundus images. IEEE Sens. J. 2020, 20, 12885–12894. [Google Scholar] [CrossRef]
- Chandrasekaran, R.; Loganathan, B. Retinopathy grading with deep learning and wavelet hyper-analytic activations. Vis. Comput. 2023, 39, 2741–2756. [Google Scholar] [CrossRef]
- Attallah, O. GabROP: Gabor wavelets-based CAD for retinopathy of prematurity diagnosis via convolutional neural networks. Diagnostics 2023, 13, 171. [Google Scholar] [CrossRef]
- Parashar, D.; Agrawal, D.K. Automatic classification of glaucoma stages using two-dimensional tensor empirical wavelet transform. IEEE Signal Process. Lett. 2020, 28, 66–70. [Google Scholar] [CrossRef]
- Rasti, R.; Mehridehnavi, A.; Rabbani, H.; Hajizadeh, F. Automatic diagnosis of abnormal macula in retinal optical coherence tomography images using wavelet-based convolutional neural network features and random forests classifier. J. Biomed. Opt. 2018, 23, 035005. [Google Scholar] [CrossRef]
- Oliveira, A.; Pereira, S.; Silva, C.A. Retinal vessel segmentation based on fully convolutional neural networks. Expert Syst. Appl. 2018, 112, 229–242. [Google Scholar] [CrossRef]
- Li, D.; Zhang, L.; Sun, C.; Yin, T.; Liu, C.; Yang, J. Robust retinal image enhancement via dual-tree complex wavelet transform and morphology-based method. IEEE Access 2019, 7, 47303–47316. [Google Scholar] [CrossRef]
- Zeng, X.; Chen, H.; Luo, Y.; Ye, W. Automated diabetic retinopathy detection based on binocular siamese-like convolutional neural network. IEEE Access 2019, 7, 30744–30753. [Google Scholar] [CrossRef]
- Qummar, S.; Khan, F.G.; Shah, S.; Khan, A.; Shamshirband, S.; Rehman, Z.U.; Khan, I.A.; Jadoon, W. A deep learning ensemble approach for diabetic retinopathy detection. IEEE Access 2019, 7, 150530–150539. [Google Scholar] [CrossRef]
- Pao, S.I.; Lin, H.Z.; Chien, K.H.; Tai, M.C.; Chen, J.T.; Lin, G.M. Detection of diabetic retinopathy using bichannel convolutional neural network. J. Ophthalmol. 2020, 2020, 9139713. [Google Scholar] [CrossRef]
- Cervantes, J.; Garcia-Lamont, F.; Rodríguez-Mazahua, L.; Lopez, A. A comprehensive survey on support vector machine classification: Applications, challenges and trends. Neurocomputing 2020, 408, 189–215. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Litjens, G.; Debats, O.; Barentsz, J.; Karssemeijer, N.; Huisman, H. Computer-aided detection of prostate cancer in MRI. IEEE Trans. Med. Imaging 2014, 33, 1083–1092. [Google Scholar] [CrossRef]
- Trigui, R.; Mitéran, J.; Walker, P.M.; Sellami, L.; Hamida, A.B. Automatic classification and localization of prostate cancer using multi-parametric MRI/MRS. Biomed. Signal Process. Control 2017, 31, 189–198. [Google Scholar] [CrossRef]
- Wan, S.; Liang, Y.; Zhang, Y. Deep convolutional neural networks for diabetic retinopathy detection by image classification. Comput. Electr. Eng. 2018, 72, 274–282. [Google Scholar] [CrossRef]
| Grade | Abnormality | No. of Images before Augmentation | No. of Images after Augmentation |
|---|---|---|---|
| Grade 0 | No apparent retinopathy | 25,810 | 25,810 |
| Grade 1 | Mild non-proliferative diabetic retinopathy (NPDR) | 2443 | 25,810 |
| Grade 2 | Moderate NPDR | 5292 | 25,810 |
| Grade 3 | Severe NPDR | 873 | 25,810 |
| Grade 4 | Proliferative diabetic retinopathy (PDR) | 708 | 25,810 |
| Precision | Recall | F1-score | Accuracy | AUC Score | |
|---|---|---|---|---|---|
| Wavelet CNN | 0.857 | 0.849 | 0.853 | 0.73 | 0.602 |
| Wavelet CNN + XGBoost | 0.9412 | 0.8973 | 0.9186 | 0.8924 | 0.976 |
| Wavelet CNN + Random Forest | 0.940 | 0.937 | 0.939 | 0.9095 | 0.978 |
| WaveletCNN + SVM | 0.9831 | 0.9822 | 0.9831 | 0.9895 | 0.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sundar, S.; Subramanian, S.; Mahmud, M. Classification of Diabetic Retinopathy Disease Levels by Extracting Spectral Features Using Wavelet CNN. Diagnostics 2024, 14, 1093. https://doi.org/10.3390/diagnostics14111093
Sundar S, Subramanian S, Mahmud M. Classification of Diabetic Retinopathy Disease Levels by Extracting Spectral Features Using Wavelet CNN. Diagnostics. 2024; 14(11):1093. https://doi.org/10.3390/diagnostics14111093
Chicago/Turabian StyleSundar, Sumod, Sumathy Subramanian, and Mufti Mahmud. 2024. "Classification of Diabetic Retinopathy Disease Levels by Extracting Spectral Features Using Wavelet CNN" Diagnostics 14, no. 11: 1093. https://doi.org/10.3390/diagnostics14111093
APA StyleSundar, S., Subramanian, S., & Mahmud, M. (2024). Classification of Diabetic Retinopathy Disease Levels by Extracting Spectral Features Using Wavelet CNN. Diagnostics, 14(11), 1093. https://doi.org/10.3390/diagnostics14111093







