A Metagenomics Pipeline to Characterize Self-Collected Vaginal Microbiome Samples
Abstract
1. Introduction
2. Materials and Methods
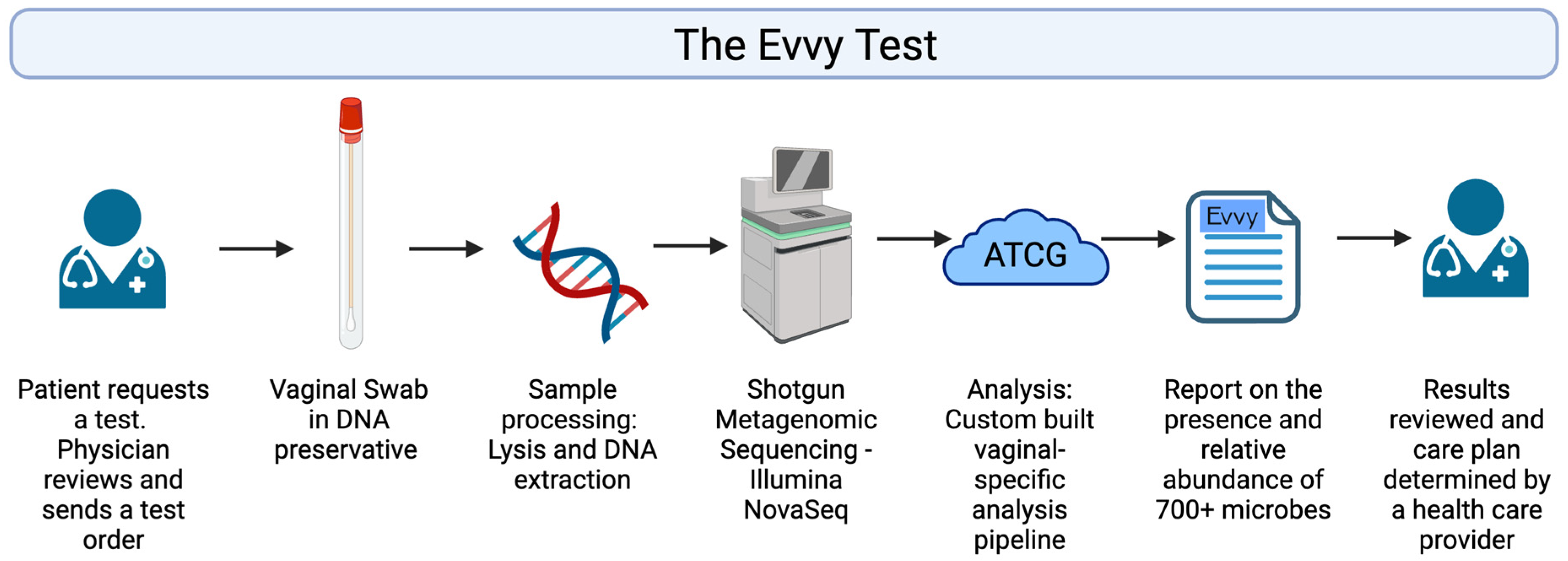
2.1. Evvy Test Workflow
2.2. Isolate Sequencing
2.3. Limit of Detection and Precision Studies
2.4. Mock Community Analysis
2.5. Study Participants, Ethics, and Sample Collection and Transportation
2.6. Shotgun Metagenomics Analysis of Vaginal Samples
3. Results
3.1. Sensitivity and Specificity
3.2. Limit of Detection and Precision
3.3. Mock Community Analysis
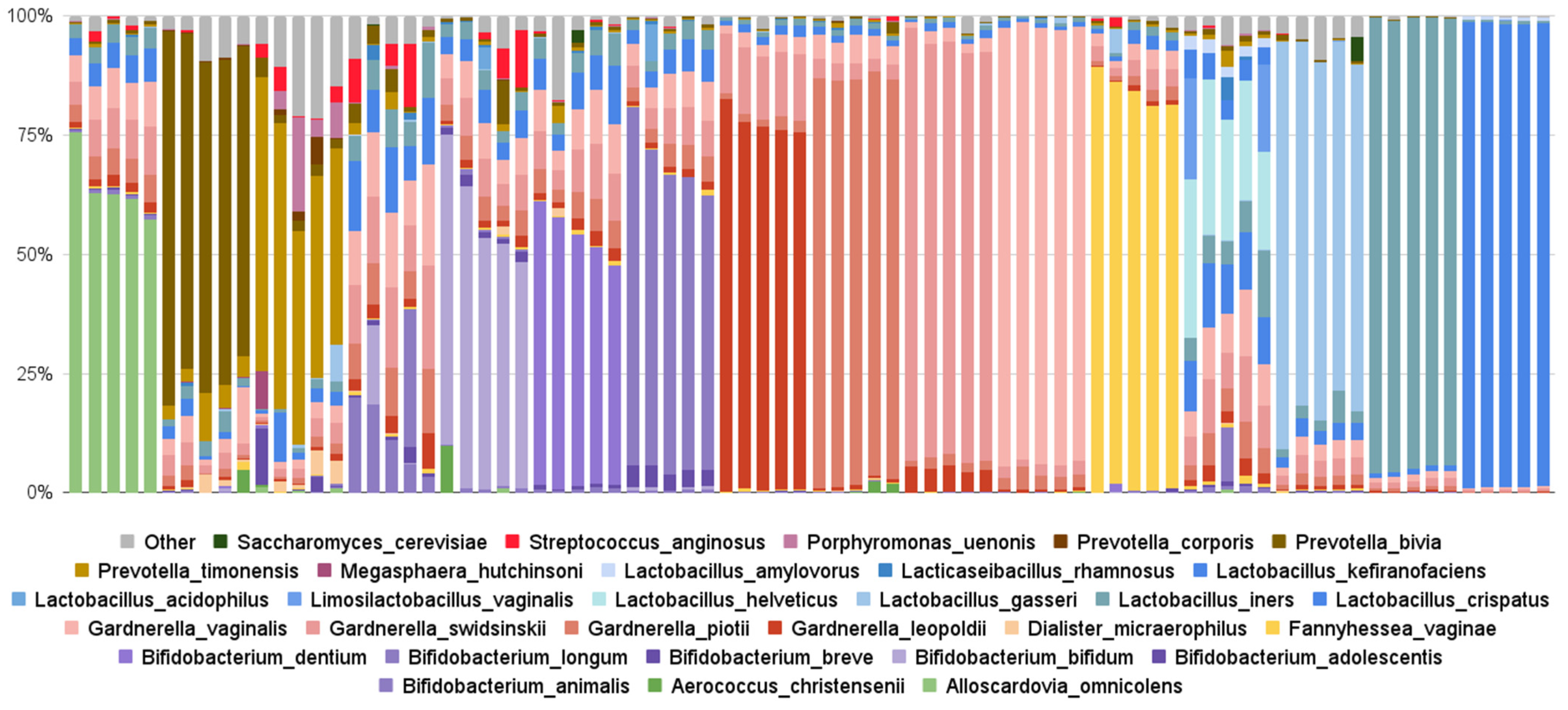
3.3.1. Findings from Evvy’s Vaginal Metagenomics Test
3.3.2. Co-Occurrence of Pathogens Detected
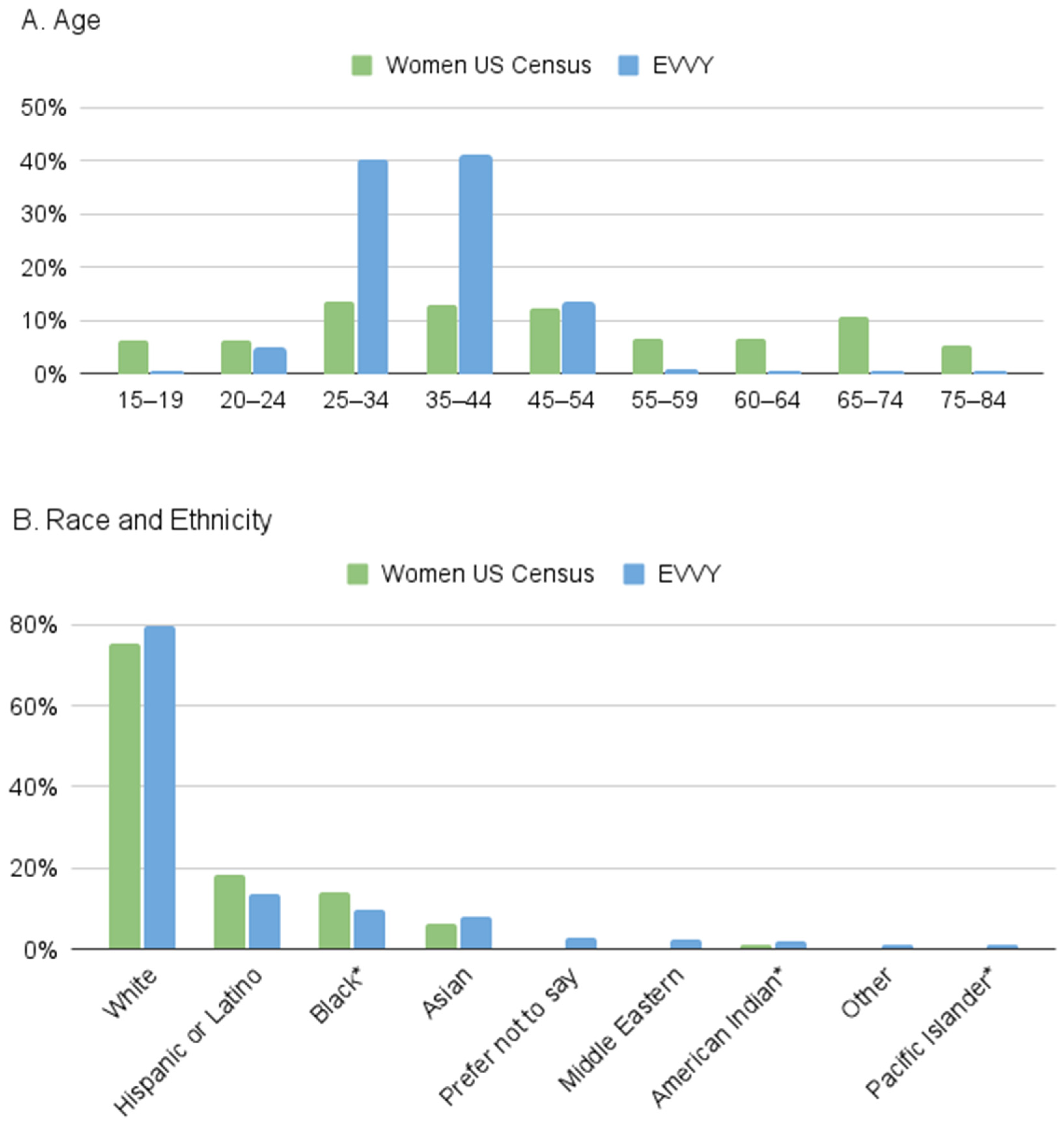
3.3.3. Comparison to US Census Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muzny, C.A.; Balkus, J.; Mitchell, C.; Sobel, J.D.; Workowski, K.; Marrazzo, J.; Schwebke, J.R. Diagnosis and Management of Bacterial Vaginosis: Summary of Evidence Reviewed for the 2021 Centers for Disease Control and Prevention Sexually Transmitted Infections Treatment Guidelines. Clin. Infect. Dis. 2022, 74 (Suppl. S2), S144–S151. [Google Scholar] [CrossRef] [PubMed]
- Peebles, K.; Velloza, J.; Balkus, J.E.; McClelland, R.S.; Barnabas, R.V. High Global Burden and Costs of Bacterial Vaginosis: A Systematic Review and Meta-Analysis. Sex. Transm. Dis. 2019, 46, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B.; Muraglia, R.; Dietz, J.P.; Sobel, J.D.; Wagner, J. Prevalence of recurrent vulvovaginal candidiasis in 5 European countries and the United States: Results from an internet panel survey. J. Low Genit. Tract. Dis. 2013, 17, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Flagg, E.W.; Meites, E.; Phillips, C.; Papp, J.; Torrone, E.A. Prevalence of Trichomonas vaginalis Among Civilian, Noninstitutionalized Male and Female Population Aged 14 to 59 Years: United States, 2013 to 2016. Sex. Transm. Dis. 2019, 46, e93–e96. [Google Scholar] [CrossRef]
- Amabebe, E.; Anumba, D.O.C. The Vaginal Microenvironment: The Physiologic Role of Lactobacilli. Front. Med. 2018, 5, 181. [Google Scholar] [CrossRef]
- Sobel, J.D.; Subramanian, C.; Foxman, B.; Fairfax, M.; Gygax, S.E. Mixed vaginitis-more than coinfection and with therapeutic implications. Curr. Infect. Dis. Rep. 2013, 15, 104–108. [Google Scholar] [CrossRef]
- Nugent, R.P.; Krohn, M.A.; Hillier, S.L. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J. Clin. Microbiol. 1991, 29, 297–301. [Google Scholar] [CrossRef]
- Amsel, R.; Totten, P.A.; Spiegel, C.A.; Chen, K.C.; Eschenbach, D.; Holmes, K.K. Nonspecific vaginitis. Diagnostic criteria and microbial and epidemiologic associations. Am. J. Med. 1983, 74, 14–22. [Google Scholar] [CrossRef]
- Schwiertz, A.; Taras, D.; Rusch, K.; Rusch, V. Throwing the dice for the diagnosis of vaginal complaints? Ann. Clin. Microbiol. Antimicrob. 2006, 5, 4. [Google Scholar] [CrossRef]
- Muzny, C.A.; Cerca, N.; Elnaggar, J.H.; Taylor, C.M.; Sobel, J.D.; Van Der Pol, B. State of the Art for Diagnosis of Bacterial Vaginosis. J. Clin. Microbiol. 2023, 61, e0083722. [Google Scholar] [CrossRef]
- Lebeer, S.; Ahannach, S.; Gehrmann, T.; Wittouck, S.; Eilers, T.; Oerlemans, E.; Condori, S.; Dillen, J.; Spacova, I.; Vander Donck, L.; et al. A citizen-science-enabled catalogue of the vaginal microbiome and associated factors. Nat. Microbiol. 2023, 8, 2183–2195. [Google Scholar] [CrossRef] [PubMed]
- Ravel, J.; Moreno, I.; Simon, C. Bacterial vaginosis and its association with infertility, endometritis, and pelvic inflammatory disease. Am. J. Obstet. Gynecol. 2021, 224, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Trevethan, R. Sensitivity, Specificity, and Predictive Values: Foundations, Pliabilities, and Pitfalls in Research and Practice. Front. Public Health 2017, 5, 307. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wang, G.; Lau, H.C.; Yu, J. Metagenomic Sequencing for Microbial DNA in Human Samples: Emerging Technological Advances. Int. J. Mol. Sci. 2022, 23, 181. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef]
- Paavonen, J.A.; Brunham, R.C. Vaginitis in Nonpregnant Patients: ACOG Practice Bulletin Number 215. Obstet. Gynecol. 2020, 135, 1229–1230. [Google Scholar] [CrossRef]
- Healthcare, A. 2022 Survey of Physician Appointment Wait Times and Medicare and Medicaid Acceptance Rates. Available online: https://www.wsha.org/wp-content/uploads/mha2022waittimesurveyfinal.pdf (accessed on 28 March 2024).
- Peterson, C.L.; Alexander, D.; Chen, J.C.; Adam, H.; Walker, M.; Ali, J.; Forbes, J.; Taboada, E.; Barker, D.O.R.; Graham, M.; et al. Clinical Metagenomics Is Increasingly Accurate and Affordable to Detect Enteric Bacterial Pathogens in Stool. Microorganisms 2022, 10, 441. [Google Scholar] [CrossRef]
- Mu, S.; Hu, L.; Zhang, Y.; Liu, Y.; Cui, X.; Zou, X.; Wang, Y.; Lu, B.; Zhou, S.; Liang, X.; et al. Prospective Evaluation of a Rapid Clinical Metagenomics Test for Bacterial Pneumonia. Front. Cell. Infect. Microbiol. 2021, 11, 684965. [Google Scholar] [CrossRef]
- Baptista, P.V.; Stockdale, C.; Sobel, J.D.; Aidé, S. International Society for the Study of Vulvovaginal Disease Recommendations for the Diagnosis Andtreatment of Vaginitis; Admedic: Lisbon, Portugal, 2023. [Google Scholar] [CrossRef]
- Thomas-White, K.; Wever, F.; Navarro, P. Incidence and Symptom Profiling of Vaginitis Containing Aerobic and Anaerobic Pathogens. Am. J. Obstet. Gynecol. 2024, 230, S641–S642. [Google Scholar]
- Skafte-Holm, A.; Humaidan, P.; Bernabeu, A.; Lledo, B.; Jensen, J.S.; Haahr, T. The Association between Vaginal Dysbiosis and Reproductive Outcomes in Sub-Fertile Women Undergoing IVF-Treatment: A Systematic PRISMA Review and Meta-Analysis. Pathogens 2021, 10, 295. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Yu, Y.; Zhou, J.; Liu, J.; Gao, J. Relationship of Lactobacillus Vaginal Microbiota Changes and the Risk of Preterm Birth: A Systematic Review and Meta-Analysis. J. Womens Health 2024, 33, 228–238. [Google Scholar] [CrossRef] [PubMed]
- Kosti, I.; Lyalina, S.; Pollard, K.S.; Butte, A.J.; Sirota, M. Meta-Analysis of Vaginal Microbiome Data Provides New Insights Into Preterm Birth. Front. Microbiol. 2020, 11, 476. [Google Scholar] [CrossRef] [PubMed]
- Torcia, M.G. Interplay among Vaginal Microbiome, Immune Response and Sexually Transmitted Viral Infections. Int. J. Mol. Sci. 2019, 20, 266. [Google Scholar] [CrossRef]
- Laniewski, P.; Herbst-Kralovetz, M.M. Connecting microbiome and menopause for healthy ageing. Nat. Microbiol. 2022, 7, 354–358. [Google Scholar] [CrossRef]
- Sharifian, K.; Shoja, Z.; Jalilvand, S. The interplay between human papillomavirus and vaginal microbiota in cervical cancer development. Virol. J. 2023, 20, 73. [Google Scholar] [CrossRef]
- Yang, X.; Da, M.; Zhang, W.; Qi, Q.; Zhang, C.; Han, S. Role of Lactobacillus in cervical cancer. Cancer Manag. Res. 2018, 10, 1219–1229. [Google Scholar] [CrossRef]

| Positive | Negative | Total | Predictive Value | ||
|---|---|---|---|---|---|
| NovaSeq | Positive | 95 | 11 | 106 | 89.6% (95/106) PPV |
| Negative | 7 | 99 | 106 | 93.4% (99/106) NPV | |
| Total | 102 | 110 | |||
| Sensitivity 93.1% (95/102) | Specificity 90% (99/110) |
| Top 10 Species by Frequency of Detection | Percentage of Tests Detected | Top 10 Species by Relative Abundance | Average Abundance | Percent of Tests Detected |
|---|---|---|---|---|
| Gardnerella vaginalis | 99% | Lactobacillus crispatus | 33% | 93% |
| Gardnerella swidinskii | 99% | Lactobacillus helveticus | 23% | 0.1% |
| Lactobacillus iners | 95% | Bifidobacterium dentium | 19% | 0.9% |
| Gardnerella piotii | 95% | Lactobacillus iners | 18% | 95% |
| Lactobacillus crispatus | 93% | Lactobacillus gasseri | 15% | 14% |
| Gardnerella leopoldii | 78% | Bifidobacterium longum | 13% | 2% |
| Prevotella bivia | 46% | Bifidobacterium bifidum | 13% | 0.7% |
| Fannyhessae vaginae | 33% | Gardnerella swidinskii | 12% | 99% |
| Bifidobacterium animalis | 31% | Alloscardovia omnicolens | 12% | 2% |
| Prevotella timonensis | 27% | Gardnerella vaginalis | 11% | 99% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas-White, K.; Hilt, E.E.; Olmschenk, G.; Gong, M.; Phillips, C.D.; Jarvis, C.; Sanford, N.; White, J.; Navarro, P. A Metagenomics Pipeline to Characterize Self-Collected Vaginal Microbiome Samples. Diagnostics 2024, 14, 2039. https://doi.org/10.3390/diagnostics14182039
Thomas-White K, Hilt EE, Olmschenk G, Gong M, Phillips CD, Jarvis C, Sanford N, White J, Navarro P. A Metagenomics Pipeline to Characterize Self-Collected Vaginal Microbiome Samples. Diagnostics. 2024; 14(18):2039. https://doi.org/10.3390/diagnostics14182039
Chicago/Turabian StyleThomas-White, Krystal, Evann E. Hilt, Genevieve Olmschenk, Maryann Gong, Caleb D. Phillips, Courtney Jarvis, Nicholas Sanford, Jennifer White, and Pita Navarro. 2024. "A Metagenomics Pipeline to Characterize Self-Collected Vaginal Microbiome Samples" Diagnostics 14, no. 18: 2039. https://doi.org/10.3390/diagnostics14182039
APA StyleThomas-White, K., Hilt, E. E., Olmschenk, G., Gong, M., Phillips, C. D., Jarvis, C., Sanford, N., White, J., & Navarro, P. (2024). A Metagenomics Pipeline to Characterize Self-Collected Vaginal Microbiome Samples. Diagnostics, 14(18), 2039. https://doi.org/10.3390/diagnostics14182039






