Early Detection of Lymph Node Metastasis Using Primary Head and Neck Cancer Computed Tomography and Fluorescence Lifetime Imaging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
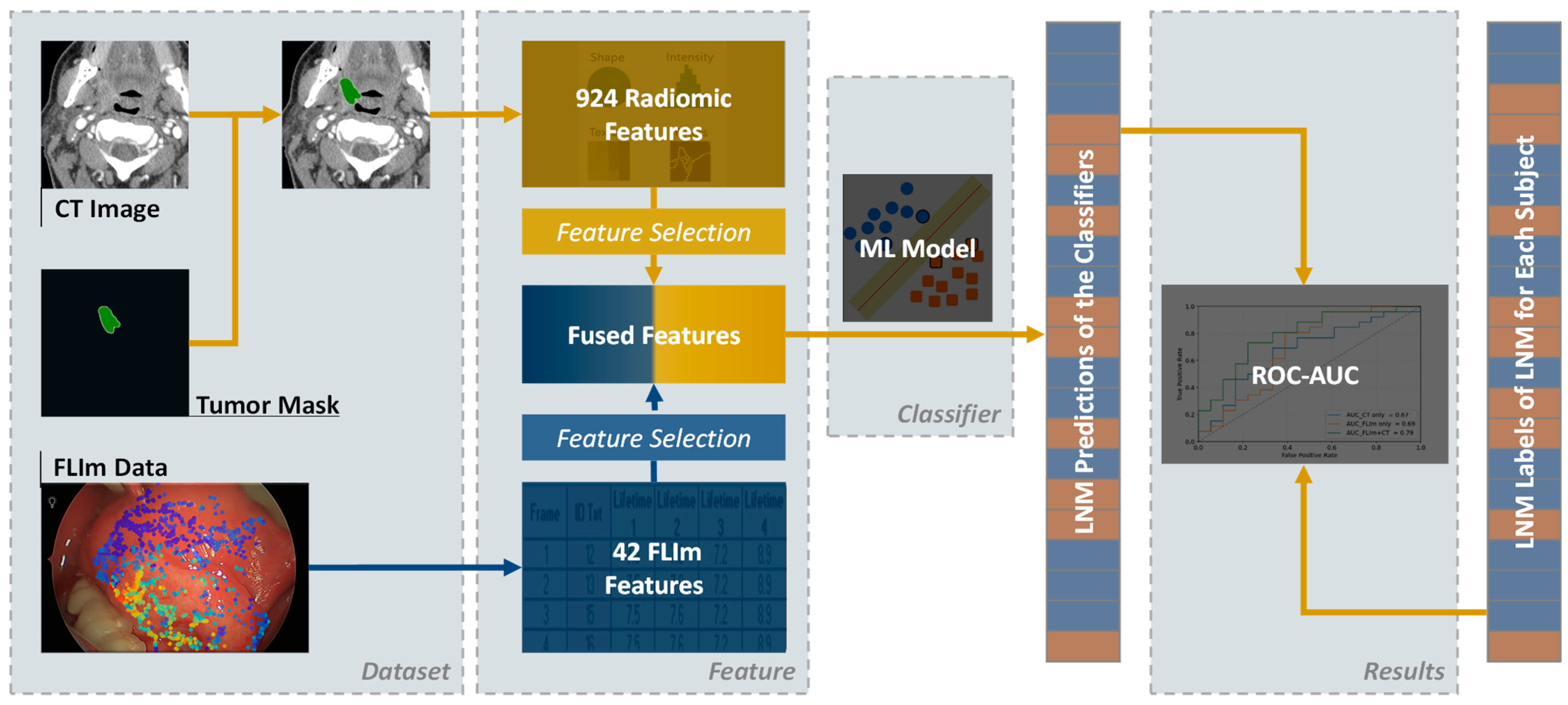
2.2. Workflow
2.3. Data Acquisition and Feature Extraction
2.3.1. CT Data
2.3.2. FLIm Data
2.4. Model Training and Validation
2.4.1. Conditional Random Grouping
2.4.2. Nested Cross-Validation
2.4.3. Weighted Samples
2.4.4. Feature Reduction and Selection
2.4.5. Classifier Model Construction and Evaluation
3. Results
3.1. Predicting LNM Using CT and FLIm
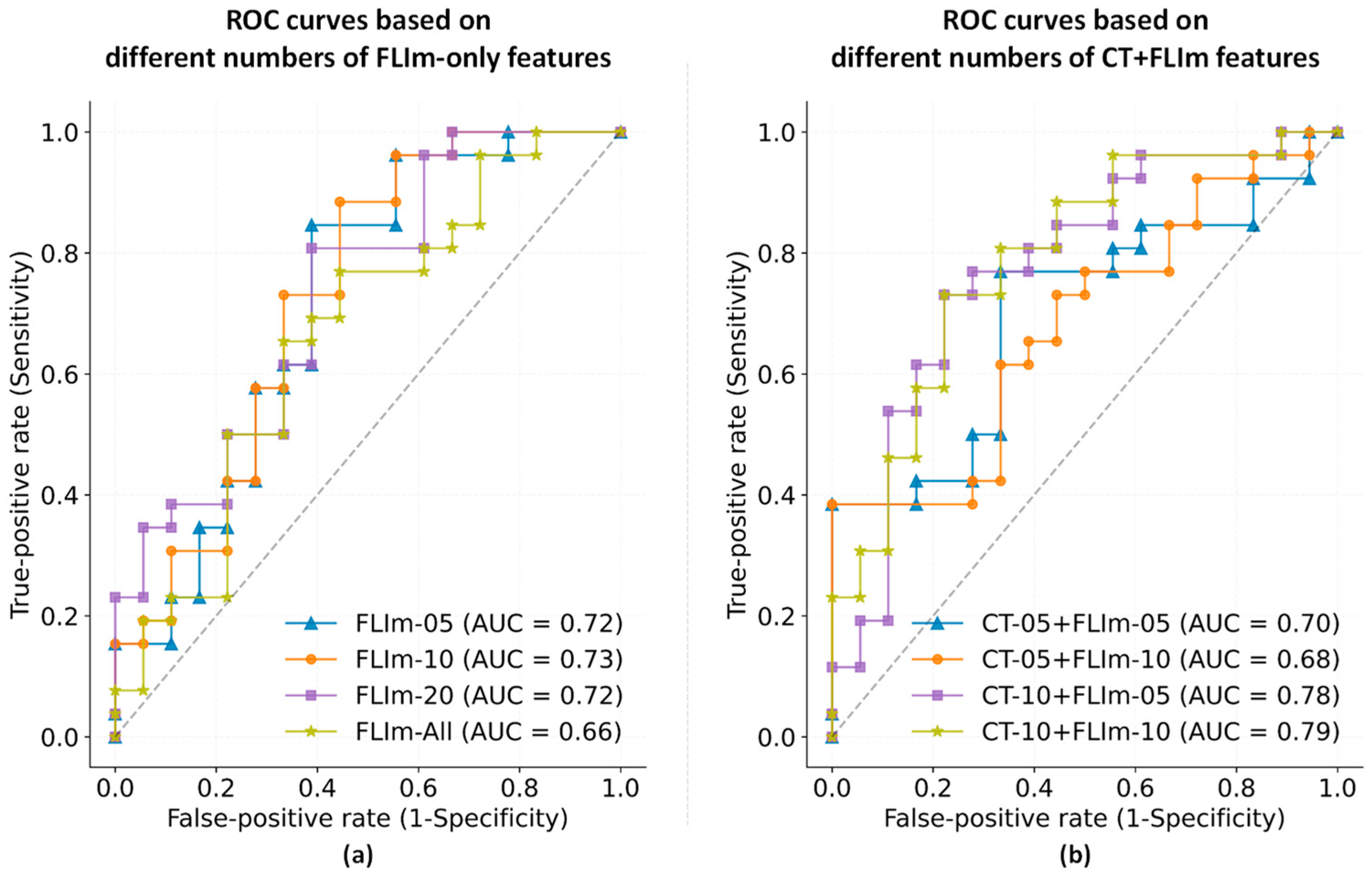
3.2. Feature Selection
3.3. Performance in Relation to Tumor Size and Tumor Anatomical Site
4. Discussion
4.1. Overall
4.2. Limitation
4.2.1. Limited Performance for CT-Only Classifier
4.2.2. Unbalanced Performance over Tumor Sizes and Anatomical Sites
4.2.3. Feature Fusion Method
4.3. Future Perspectives
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jawa, Y.; Yadav, P.; Gupta, S.; Mathan, S.V.; Pandey, J.; Saxena, A.K.; Kateriya, S.; Tiku, A.B.; Mondal, N.; Bhattacharya, J.; et al. Current insights and advancements in head and neck Cancer: Emerging biomarkers and therapeutics with cues from single cell and 3D model omics profiling. Front. Oncol. 2021, 11, 676948. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Xiao, W.; Chen, X.; Zhao, Z.; Lin, Y.; Chen, G. Epidemiological trends of head and neck cancer: A population-based study. BioMed Res. Int. 2021, 2021, 1738932. [Google Scholar]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Nishio, N.; Fujimoto, Y.; Hiramatsu, M.; Maruo, T.; Tsuzuki, H.; Mukoyama, N.; Yokoi, S.; Wada, A.; Furukawa, M.K.; Furukawa, M.; et al. Diagnosis of cervical lymph node metastases in head and neck cancer with ultrasonic measurement of lymph node volume. Auris Nasus Larynx 2019, 46, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Sproll, K.C.; Hermes, I.; Felder, G.; Stoecklein, N.H.; Seidl, M.; Kaiser, P.; Kaisers, W. Comparative analysis of diagnostic ultrasound and histopathology for detecting cervical lymph node metastases in head and neck cancer. J. Cancer Res. Clin. Oncol. 2023, 149, 17319–17333. [Google Scholar] [CrossRef]
- D’Cruz, A.K.; Vaish, R.; Kapre, N.; Dandekar, M.; Gupta, S.; Hawaldar, R.; Agarwal, J.P.; Pantvaidya, G.; Chaukar, D.; Deshmukh, A.; et al. Elective versus therapeutic neck dissection in node-negative oral cancer. N. Engl. J. Med. 2015, 373, 521–529. [Google Scholar] [CrossRef]
- Pisani, P.; Airoldi, M.; Allais, A.; Valletti, P.A.; Battista, M.; Benazzo, M.; Briatore, R.; Cacciola, S.; Cocuzza, S.; Colombo, A.; et al. Metastatic disease in head \& neck oncology. Acta Otorhinolaryngol. Ital. 2020, 40, S1. [Google Scholar]
- Han, A.Y.; John, M.A.S. Predictors of nodal metastasis in cutaneous head and neck cancers. Curr. Oncol. Rep. 2022, 24, 1145–1152. [Google Scholar] [CrossRef]
- Strüder, D.; Momper, T.; Irmscher, N.; Krause, M.; Liese, J.; Schraven, S.; Zimpfer, A.; Zonnur, S.; Burmeister, A.-S.; Schneider, B.; et al. Establishment and characterization of patient-derived head and neck cancer models from surgical specimens and endoscopic biopsies. J. Exp. Clin. Cancer Res. 2021, 40, 246. [Google Scholar] [CrossRef]
- Hoang, J.K.; Vanka, J.; Ludwig, B.J.; Glastonbury, C.M. Evaluation of cervical lymph nodes in head and neck cancer with CT and MRI: Tips, traps, and a systematic approach. AJR Am. J. Roentgenol. 2013, 200, W17–W25. [Google Scholar] [CrossRef]
- Kim, J.H.; Choi, K.Y.; Lee, S.-H.; Lee, D.J.; Park, B.J.; Yoon, D.Y.; Rho, Y.-S. The value of CT, MRI, and PET-CT in detecting retropharyngeal lymph node metastasis of head and neck squamous cell carcinoma. BMC Med. Imaging 2020, 20, 88. [Google Scholar] [CrossRef] [PubMed]
- Morisada, M.V.; Bewley, A.F.; Broadhead, K.; Assadsangabi, R.; Paydar, A.; Birkeland, A.C.; Abouyared, M.; Qi, L.; Ivanovic, V. CT predictors of sub-centimeter occult lymph node metastases in oral cavity squamous cell carcinoma: A case-control study. Neuroradiol. J. 2023, 37, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Jović, A.; Fila, J.; Gršić, K.; Ivkić, M.; Ozretić, D. Diffusion-weighted MRI: Impact of the size of the ROI in detecting metastases in subcentimeter lymph nodes in head and neck squamous cell carcinoma. Neuroradiology 2020, 62, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Ionna, F.; Pavone, E.; Aversa, C.; Maffia, F.; Spinelli, R.; Carraturo, E.; Salzano, G.; Maglitto, F.; Sarcinella, M.; Fusco, R.; et al. Sentinel Lymph Node Biopsy (SLNB) for Early-Stage Head and Neck Squamous-Cell Carcinoma of the Tongue: Twenty Years of Experience at I.N.T. “G.Pascale”. Cancers 2024, 16, 1153. [Google Scholar] [CrossRef] [PubMed]
- Kerawala, C.J.; Heliotos, M. Prevention of complications in neck dissection. Head Neck Oncol. 2009, 1, 35. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Giannitto, C.; Mercante, G.; Ammirabile, A.; Cerri, L.; De Giorgi, T.; Lofino, L.; Vatteroni, G.; Casiraghi, E.; Marra, S.; Esposito, A.A.; et al. Radiomics-based machine learning for the diagnosis of lymph node metastases in patients with head and neck cancer: Systematic review. Head Neck 2023, 45, 482–491. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, T.; Yang, Z.; Zhou, Y.; Kang, Z.; Wang, Y.; Huang, Z. Radiomics based on magnetic resonance imaging for preoperative prediction of lymph node metastasis in head and neck cancer: Machine learning study. Head Neck 2022, 44, 2786–2795. [Google Scholar] [CrossRef]
- Zhai, T.-T.; Langendijk, J.A.; van Dijk, L.V.; van Der Schaaf, A.; Sommers, L.; Vemer-van Den Hoek, J.G.M.; Bijl, H.P.; Halmos, G.B.; Witjes, M.J.H.; Oosting, S.F.; et al. Pre-treatment radiomic features predict individual lymph node failure for head and neck cancer patients. Radiother. Oncol. 2020, 146, 58–65. [Google Scholar] [CrossRef]
- Chen, L.; Zhou, Z.; Sher, D.; Zhang, Q.; Shah, J.; Pham, N.L.; Jiang, S.; Wang, J. Combining many-objective radiomics and 3D convolutional neural network through evidential reasoning to predict lymph node metastasis in head and neck cancer. Phys. Med. Biol. 2019, 64, 75011. [Google Scholar] [CrossRef]
- Andrearczyk, V.; Oreiller, V.; Hatt, M.; Depeursinge, A. Head and Neck Tumor Segmentation and Outcome Prediction; Springer Nature: Cham, Switzerland, 2023; Volume 13626. [Google Scholar]
- Myronenko, A.; Siddiquee, M.M.R.; Yang, D.; He, Y.; Xu, D. Automated Head and Neck Tumor Segmentation from 3D PET/CT HECKTOR 2022, Challenge Report; Springer: Berlin/Heidelberg, Germany, 2022; pp. 31–37. [Google Scholar]
- Marsden, M.; Weyers, B.W.; Bec, J.; Sun, T.; Gandour-Edwards, R.F.; Birkeland, A.C.; Abouyared, M.; Bewley, A.F.; Farwell, D.G.; Marcu, L. Intraoperative margin assessment in oral and oropharyngeal cancer using label-free fluorescence lifetime imaging and machine learning. IEEE Trans. Biomed. Eng. 2020, 68, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.A.; Weyers, B.W.; Bec, J.; Fereidouni, F.; Qi, J.; Gui, D.; Bewley, A.F.; Abouyared, M.; Farwell, D.G.; Birkeland, A.C.; et al. Anatomy-Specific Classification Model Using Label-free FLIm to Aid Intraoperative Surgical Guidance of Head and Neck Cancer. IEEE Trans. Biomed. Eng. 2023, 70, 2863–2873. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed]
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging—“how-to” guide and critical reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Gorpas, D.; Phipps, J.; Bec, J.; Ma, D.; Dochow, S.; Yankelevich, D.; Sorger, J.; Popp, J.; Bewley, A.; Gandour-Edwards, R.; et al. Autofluorescence lifetime augmented reality as a means for real-time robotic surgery guidance in human patients. Sci. Rep. 2019, 9, 1187. [Google Scholar] [CrossRef]
- Marsden, M.; Fukazawa, T.; Deng, Y.-C.; Weyers, B.W.; Bec, J.; Farwell, D.G.; Marcu, L. FLImBrush: Dynamic visualization of intraoperative free-hand fiber-based fluorescence lifetime imaging. Biomed. Opt. Express 2020, 11, 5166–5180. [Google Scholar] [CrossRef]
- Liu, J.; Sun, Y.; Qi, J.; Marcu, L. A novel method for fast and robust estimation of fluorescence decay dynamics using constrained least-squares deconvolution with Laguerre expansion. Phys. Med. Biol. 2012, 57, 843. [Google Scholar] [CrossRef]
- Pearson, K.L.I.I.I. On lines and planes of closest fit to systems of points in space. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1901, 2, 559–572. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Olawaiye, A.B.; Baker, T.P.; Washington, M.K.; Mutch, D.G. The new (Version 9) American Joint Committee on Cancer tumor, node, metastasis staging for cervical cancer. CA. Cancer J. Clin. 2021, 71, 287–298. [Google Scholar] [CrossRef]
- Rosenthal, E.L.; Warram, J.M.; De Boer, E.; Chung, T.K.; Korb, M.L.; Brandwein-Gensler, M.; Strong, T.V.; Schmalbach, C.E.; Morlandt, A.B.; Agarwal, G.; et al. Safety and tumor specificity of cetuximab-IRDye800 for surgical navigation in head and neck cancer. Clin. Cancer Res. 2015, 21, 3658–3666. [Google Scholar] [CrossRef]
- Gao, R.W.; Teraphongphom, N.T.; van den Berg, N.S.; Martin, B.A.; Oberhelman, N.J.; Divi, V.; Kaplan, M.J.; Hong, S.S.; Lu, G.; Ertsey, R.; et al. Determination of tumor margins with surgical specimen mapping using near-infrared fluorescence. Cancer Res. 2018, 78, 5144–5154. [Google Scholar] [CrossRef] [PubMed]
- Predina, J.D.; Newton, A.D.; Keating, J.; Dunbar, A.; Connolly, C.; Baldassari, M.; Mizelle, J.; Xia, L.; Deshpande, C.; Kucharczuk, J.; et al. A phase I clinical trial of targeted intraoperative molecular imaging for pulmonary adenocarcinomas. Ann. Thorac. Surg. 2018, 105, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Kann, B.H.; Aneja, S.; Loganadane, G.V.; Kelly, J.R.; Smith, S.M.; Decker, R.H.; Yu, J.B.; Park, H.S.; Yarbrough, W.G.; Malhotra, A.; et al. Pretreatment identification of head and neck cancer nodal metastasis and extranodal extension using deep learning neural networks. Sci. Rep. 2018, 8, 14036. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, J.H.; Rath, T.J.; Byrd, J.K.; Albergotti, W.G.; Wang, H.; Duvvuri, U.; Kim, S.; Johnson, J.T.; Branstetter IV, B.F.; Ferris, R.L. Accuracy of computed tomography to predict extracapsular spread in p16-positive squamous cell carcinoma. Laryngoscope 2015, 125, 1613–1618. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Chen, L.; Sher, D.; Zhang, Q.; Shah, J.; Pham, N.-L.; Jiang, S.; Wang, J. Predicting lymph node metastasis in head and neck cancer by combining many-objective radiomics and 3-dimensioal convolutional neural network through evidential reasoning. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 1–4. [Google Scholar]
- Eza, D.; Taheri, T. Low-grade sebaceous adenocarcinoma of salivary gland. A case report. Histopathology 2022, 81, 111. [Google Scholar]
- Serra-Majem, L. Head and neck cancer: Smoking, drinking, eating and… sexual practices. Eur. J. Epidemiol. 2016, 31, 333–335. [Google Scholar] [CrossRef]
- Jethwa, A.R.; Khariwala, S.S. Tobacco-related carcinogenesis in head and neck cancer. Cancer Metastasis Rev. 2017, 36, 411–423. [Google Scholar] [CrossRef]


| Demographic Data | Overall, in This Study |
|---|---|
| No. of patients | 46 |
| Sex | |
| Male | 37 (80.4%) |
| Female | 9 (19.6%) |
| Anatomy/primary tumor site | |
| Oral cavity | 20 (43.5%) |
| Superior tongue | 14(30.4%) |
| Gingiva | 3 (6.5%) |
| Floor of mouth | 2 (4.3%) |
| Palate | 1 (2.2%) |
| Oropharyngeal cancer | 26 (56.5%) |
| Palatine tonsil | 14 (30.4%) |
| Base of tongue | 7 (15.2%) |
| Pharynx | 2 (4.3%) |
| Retrimolar trigone | 2 (4.3%) |
| Vallecula | 1 (2.2%) |
| Lymph node metastasis | |
| Positive | 27 (58.7%) |
| Negative | 19 (41.3%) |
| Age at surgery (y) * | 63.3 ± 11.9 |
| 20~29 | 1 (2.2%) |
| 30~39 | 1 (2.2%) |
| 40~49 | 4 (8.7%) |
| 50~59 | 10 (21.7%) |
| 60~69 | 16 (34.8%) |
| 70~79 | 11 (23.9%) |
| 80~89 | 3 (6.5%) |
| CT + FLIm | CT-Only | FLIm-Only | |
|---|---|---|---|
| Test bACC | 0.65 | 0.58 | 0.61 |
| Test AUC | 0.79 | 0.67 | 0.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, N.; Hassan, M.A.; Ehrlich, K.; Weyers, B.W.; Biddle, G.; Ivanovic, V.; Raslan, O.A.A.; Gui, D.; Abouyared, M.; Bewley, A.F.; et al. Early Detection of Lymph Node Metastasis Using Primary Head and Neck Cancer Computed Tomography and Fluorescence Lifetime Imaging. Diagnostics 2024, 14, 2097. https://doi.org/10.3390/diagnostics14182097
Yuan N, Hassan MA, Ehrlich K, Weyers BW, Biddle G, Ivanovic V, Raslan OAA, Gui D, Abouyared M, Bewley AF, et al. Early Detection of Lymph Node Metastasis Using Primary Head and Neck Cancer Computed Tomography and Fluorescence Lifetime Imaging. Diagnostics. 2024; 14(18):2097. https://doi.org/10.3390/diagnostics14182097
Chicago/Turabian StyleYuan, Nimu, Mohamed A. Hassan, Katjana Ehrlich, Brent W. Weyers, Garrick Biddle, Vladimir Ivanovic, Osama A. A. Raslan, Dorina Gui, Marianne Abouyared, Arnaud F. Bewley, and et al. 2024. "Early Detection of Lymph Node Metastasis Using Primary Head and Neck Cancer Computed Tomography and Fluorescence Lifetime Imaging" Diagnostics 14, no. 18: 2097. https://doi.org/10.3390/diagnostics14182097





