Novel Biomarkers for Early Detection of Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Challenges in Hepatocellular Carcinoma Screening
2.2. Precision in Hepatocellular Carcinoma Surveillance and Early Detection
2.3. Biomarker Development for Early Detection of Hepatocellular Carcinoma
2.4. Classification of Biomarkers
2.5. Traditional Serum Protein Biomarkers
2.5.1. Alpha-Fetoprotein
2.5.2. Des-γ-Carboxy Prothrombin
2.5.3. Alpha-Fetoprotein-L3
3. Emerging Biomarkers
3.1. Nucleic Acid Biomarkers
3.1.1. Circulating Tumor DNA (ctDNA)
3.1.2. MicroRNAs (miRNAs), Long Noncoding RNA (lncRNAs), and Circular RNA (circRNAs)
3.2. Metabolomic Biomarkers
3.3. Extracellular Vesicles (EV)
3.4. Biomarker Panels
3.5. Urine-Based Biomarkers
3.6. Gut Microbiome
4. The Future of Hepatocellular Carcinoma Biomarker for Early Detection
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Rumgay, H.; Ferlay, J.; de Martel, C.; Georges, D.; Ibrahim, A.S.; Zheng, R.; Wei, W.; Lemmens, V.E.; Soerjomataram, I. Global, regional and national burden of primary liver cancer by subtype. Eur. J. Cancer 2022, 161, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.; Meyer, T.; Sapisochin, G.; Salem, R.; Saborowski, A. Hepatocellular Carcinoma. Lancet 2022, 400, 1345–1362. [Google Scholar] [CrossRef] [PubMed]
- de Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef]
- Yang, J.D.; Heimbach, J.K. New advances in the diagnosis and management of hepatocellular carcinoma. BMJ 2020, 371, m3544. [Google Scholar] [CrossRef]
- Marrero, J.A.; Kulik, L.M.; Sirlin, C.B.; Zhu, A.X.; Finn, R.S.; Abecassis, M.M.; Lewis, R.R.; Julie, K.H. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018, 68, 723–750. [Google Scholar] [CrossRef]
- Pepe, M.S.; Etzioni, R.; Feng, Z.; Potter, J.D.; Thompson, M.L.; Thornquist, M.; Winget, M.; Yasui, Y. Phases of Biomarker Development for Early Detection of Cancer. JNCI J. Natl. Cancer Inst. 2001, 93, 1054–1061. [Google Scholar] [CrossRef]
- Singal, A.G.; Hoshida, Y.; Pinato, D.J.; Marrero, J.; Nault, J.-C.; Paradis, V.; Tayob, N.; Sherman, M.; Lim, Y.S.; Feng, Z.; et al. International Liver Cancer Association (ILCA) White Paper on Biomarker Development for Hepatocellular Carcinoma. Gastroenterology 2021, 160, 2572–2584. [Google Scholar] [CrossRef]
- Singal, A.G.; Kanwal, F.; Llovet, J.M. Global trends in hepatocellular carcinoma epidemiology: Implications for screening, prevention and therapy. Nat. Rev. Clin. Oncol. 2023, 20, 864–884. [Google Scholar] [CrossRef]
- Singal, A.G.; Tiro, J.A.; Murphy, C.C.; Blackwell, J.-M.; Kramer, J.R.; Khan, A.; Liu, Y.; Zhang, S.; Phillips, J.L.; Hernaez, R. Patient-Reported Barriers Are Associated with Receipt of Hepatocellular Carcinoma Surveillance in a Multicenter Cohort of Patients With Cirrhosis. Clin. Gastroenterol. Hepatol. 2021, 19, 987–995.e1. [Google Scholar] [CrossRef]
- Tapper, E.B.; Parikh, N.D. Mortality due to cirrhosis and liver cancer in the United States, 1999–2016: Observational study. BMJ 2018, 362, k2817. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Pillai, A.; Tiro, J. Early Detection, Curative Treatment, and Survival Rates for Hepatocellular Carcinoma Surveillance in Patients with Cirrhosis: A Meta-Analysis. PLoS Med. 2014, 11, e1001624. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Mittal, S.; Yerokun, O.A.; Ahn, C.; Marrero, J.A.; Yopp, A.C.; Parikh, N.D.; Scaglione, S.J. Hepatocellular Carcinoma Screening Associated with Early Tumor Detection and Improved Survival among Patients with Cirrhosis in the US. Am. J. Med. 2017, 130, 1099–1106.e1. [Google Scholar] [CrossRef]
- Wolf, E.; Rich, N.E.; Marrero, J.A.; Parikh, N.D.; Singal, A.G. Use of Hepatocellular Carcinoma Surveillance in Patients with Cirrhosis: A Systematic Review and Meta-Analysis. Hepatology 2021, 73, 713–725. [Google Scholar] [CrossRef]
- Rezaee-Zavareh, M.S.; Liang, J.; Yang, J.D. Ethnic disparities in the epidemiology, treatment, and outcome of patients with hepatocellular carcinoma in the United States. Hepatoma Res. 2023, 9, 18. [Google Scholar] [CrossRef]
- Singal, A.G.; Yopp, A.; Skinner, C.S.; Packer, M.; Lee, W.M.; Tiro, J.A. Utilization of Hepatocellular Carcinoma Surveillance Among American Patients: A Systematic Review. J. Gen. Intern. Med. 2012, 27, 861–867. [Google Scholar] [CrossRef]
- Rich, N.E.; Hester, C.; Odewole, M.; Murphy, C.C.; Parikh, N.D.; Marrero, J.A.; Yopp, A.C.; Singal, A.G. Racial and Ethnic Differences in Presentation and Outcomes of Hepatocellular Carcinoma. Clin. Gastroenterol. Hepatol. 2019, 17, 551–559.e1. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Marrero, J.A.; Yopp, A. Screening Process Failures for Hepatocellular Carcinoma. J. Natl. Compr. Cancer Netw. 2014, 12, 375–382. [Google Scholar] [CrossRef]
- Singal, A.G.; Lok, A.S.; Feng, Z.; Kanwal, F.; Parikh, N.D. Conceptual Model for the Hepatocellular Carcinoma Screening Continuum: Current Status and Research Agenda. Clin. Gastroenterol. Hepatol. 2022, 20, 9–18. [Google Scholar] [CrossRef]
- Parikh, N.D.; Tayob, N.; Al-Jarrah, T.; Kramer, J.; Melcher, J.; Smith, D.; Marquardt, P.; Liu, P.-H.; Tang, R.; Kanwal, F.; et al. Barriers to Surveillance for Hepatocellular Carcinoma in a Multicenter Cohort. JAMA Netw. Open 2022, 5, e2223504. [Google Scholar] [CrossRef]
- Zhao, C.; Jin, M.; Le, R.H.; Le, M.H.; Chen, V.L.; Jin, M.; Wong, G.L.; Wong, V.W.; Lim, Y.; Chuang, W.; et al. Poor adherence to hepatocellular carcinoma surveillance: A systematic review and meta-analysis of a complex issue. Liver Int. 2018, 38, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Farvardin, S.; Patel, J.; Khambaty, M.; Yerokun, O.A.; Mok, H.; Tiro, J.A.; Yopp, A.C.; Parikh, N.D.; Marrero, J.A.; Singal, A.G. Patient-reported barriers are associated with lower hepatocellular carcinoma surveillance rates in patients with cirrhosis. Hepatology 2017, 65, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Tzartzeva, K.; Obi, J.; Rich, N.E.; Parikh, N.D.; Marrero, J.A.; Yopp, A.; Waljee, A.K.; Singal, A.G. Surveillance Imaging and Alpha Fetoprotein for Early Detection of Hepatocellular Carcinoma in Patients with Cirrhosis: A Meta-analysis. Gastroenterology 2018, 154, 1706–1718.e1. [Google Scholar] [CrossRef]
- Singal, A.G.; Patibandla, S.; Obi, J.; Fullington, H.; Parikh, N.D.; Yopp, A.C.; Marrero, J.A. Benefits and Harms of Hepatocellular Carcinoma Surveillance in a Prospective Cohort of Patients with Cirrhosis. Clin. Gastroenterol. Hepatol. 2021, 19, 1925–1932.e1. [Google Scholar] [CrossRef]
- Anstee, Q.M.; Castera, L.; Loomba, R. Impact of non-invasive biomarkers on hepatology practice: Past, present and future. J. Hepatol. 2022, 76, 1362–1378. [Google Scholar] [CrossRef]
- Atiq, O.; Tiro, J.; Yopp, A.C.; Muffler, A.; Marrero, J.A.; Parikh, N.D.; Murphy, C.; McCallister, K.; Singal, A.G. An assessment of benefits and harms of hepatocellular carcinoma surveillance in patients with cirrhosis. Hepatology 2017, 65, 1196–1205. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Qin, H.; Liu, S.; Sheng, J.; Zhang, X. Precision diagnosis of Hepatocellular Carcinoma. Chin. Med. J. 2023, 136, 1155–1165. [Google Scholar] [CrossRef]
- Galun, D.; Mijac, D.; Filipovic, A.; Bogdanovic, A.; Zivanovic, M.; Masulovic, D. Precision Medicine for Hepatocellular Carcinoma: Clinical Perspective. J. Pers. Med. 2022, 12, 149. [Google Scholar] [CrossRef]
- Martin, S.P.; Wang, X.W. The evolving landscape of precision medicine in primary liver cancer. Hepatic Oncol. 2019, 6, HEP12. [Google Scholar] [CrossRef]
- Liu, X.-N.; Cui, D.-N.; Li, Y.-F.; Liu, Y.-H.; Liu, G.; Liu, L. Multiple “Omics” data-based biomarker screening for hepatocellular carcinoma diagnosis. World. J. Gastroenterol. 2019, 25, 4199–4212. [Google Scholar] [CrossRef]
- Marrero, J.A.; Feng, Z.; Wang, Y.; Nguyen, M.H.; Befeler, A.S.; Roberts, L.R.; Reddy, K.R.; Harnois, D.; Llovet, J.M.; Normolle, D.; et al. α-Fetoprotein, Des-γ Carboxyprothrombin, and Lectin-Bound α-Fetoprotein in Early Hepatocellular Carcinoma. Gastroenterology 2009, 137, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, T.; Jin, B.; Li, W.; Wang, Z.; Zhang, H.; Song, Y.; Li, N. Diagnosis accuracy of serum glypican-3 level in patients with hepatocellular carcinoma: A systematic review with meta-analysis. Int. J. Biol. Markers 2018, 33, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Fattovich, G.; Stroffolini, T.; Zagni, I.; Donato, F. Hepatocellular carcinoma in cirrhosis: Incidence and risk factors. Gastroenterology 2004, 127 (Suppl. 1), S35–S50. [Google Scholar] [CrossRef]
- Tu, T.; Budzinska, M.A.; Maczurek, A.E.; Cheng, R.; Di Bartolomeo, A.; Warner, F.J.; McCaughan, G.W.; McLennan, S.V.; Shackel, N.A. Novel Aspects of the Liver Microenvironment in Hepatocellular Carcinoma Pathogenesis and Development. Int. J. Mol. Sci. 2014, 15, 9422–9458. [Google Scholar] [CrossRef]
- Omar, M.A.; Omran, M.M.; Farid, K.; Tabll, A.A.; Shahein, Y.E.; Emran, T.M.; Petrovic, A.; Lucic, N.R.; Smolic, R.; Kovac, T.; et al. Biomarkers for Hepatocellular Carcinoma: From Origin to Clinical Diagnosis. Biomedicines 2023, 11, 1852. [Google Scholar] [CrossRef]
- Abelev, G.I.; Perova, S.D.; Khramkova, N.I.; Postnikova, Z.A.; Irlin, I.S. Production of Embryonal α-Globulin by Transplantable Mouse Hepatomas. Transplantation 1963, 1, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Yeo, Y.H.; Lee, Y.-T.; Tseng, H.-R.; Zhu, Y.; You, S.; Agopian, V.G.; Yang, J.D. Alpha-fetoprotein: Past, present, and future. Hepatol. Commun. 2024, 8, e0422. [Google Scholar] [CrossRef]
- Abelev, G.I.; Assecritova, I.V.; Kraevsky, N.A.; Perova, S.D.; Perevodchikova, N.I. Embryonal serum α-globulin in cancer patients: Diagnostic value. Int. J. Cancer 1967, 2, 551–558. [Google Scholar] [CrossRef]
- Choi, J.; Kim, G.; Han, S.; Lee, W.; Chun, S.; Lim, Y. Longitudinal Assessment of Three Serum Biomarkers to Detect Very Early-Stage Hepatocellular Carcinoma. Hepatology 2019, 69, 1983–1994. [Google Scholar] [CrossRef]
- Lee, C.-W.; Tsai, H.-I.; Lee, W.-C.; Huang, S.-W.; Lin, C.-Y.; Hsieh, Y.-C.; Kuo, T.; Chen, C.-W.; Yu, M.-C. Normal Alpha-Fetoprotein Hepatocellular Carcinoma: Are They Really Normal? J. Clin. Med. 2019, 8, 1736. [Google Scholar] [CrossRef]
- Zhang, Y.-S.; Chu, J.-H.; Cui, S.-X.; Song, Z.-Y.; Qu, X.-J. Des-γ-Carboxy Prothrombin (DCP) as a Potential Autologous Growth Factor for the Development of Hepatocellular Carcinoma. Cell Physiol. Biochem. 2014, 34, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Taniguchi, T.; Sannomiya, K.; Takenaka, H.; Tomonari, T.; Okamoto, K.; Kitamura, S.; Okahisa, T.; Tamaki, K.; Mikasa, H.; et al. Novel des-γ-carboxy prothrombin in serum for the diagnosis of hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2013, 28, 1348–1355. [Google Scholar] [CrossRef]
- Lok, A.S.; Sterling, R.K.; Everhart, J.E.; Wright, E.C.; Hoefs, J.C.; Di Bisceglie, A.M.; Morgan, T.R.; Kim, H.Y.; Lee, W.M.; Bonkovsky, H.L.; et al. Des-gamma-carboxy prothrombin and alpha-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology 2010, 138, 493–502. [Google Scholar] [CrossRef]
- Pandyarajan, V.; Govalan, R.; Yang, J.D. Risk Factors and Biomarkers for Chronic Hepatitis B Associated Hepatocellular Carcinoma. Int. J. Mol. Sci. 2021, 22, 479. [Google Scholar] [CrossRef] [PubMed]
- Lamerz, R. AFP isoforms and their clinical significance (overview). Anticancer Res. 1997, 17, 2927–2930. [Google Scholar]
- Taketa, K.; Okada, S.; Win, N.; Hlaing, N.K.T.; Wind, K.M. Evaluation of tumor markers for the detection of hepatocellular carcinoma in Yangon General Hospital, Myanmar. Acta Med. Okayama 2002, 56, 317–320. [Google Scholar] [CrossRef]
- Khien, V.V.; Mao, H.V.; Chinh, T.T.; Ha, P.T.; Bang, M.H.; Lac, B.V.; Tuan, N.A.; Don, L.V.; Taketa, K.; Satomura, S. Clinical Evaluation of Lentil Lectin-Reactive Alpha-Fetoprotein-L3 in Histology-Proven Hepatocellular Carcinoma. Int. J. Biol. Markers 2001, 16, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.-C.; Feng, Y.-L.; Guo, T.; Xie, A.-Y.; Cai, X.-J. Circulating tumor DNA in hepatocellular carcinoma: Trends and challenges. Cell Biosci. 2016, 6, 32. [Google Scholar] [CrossRef]
- Huang, Z.-H.; Hu, Y.; Hua, D.; Wu, Y.-Y.; Song, M.-X.; Cheng, Z.-H. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp. Mol. Pathol. 2011, 91, 702–707. [Google Scholar] [CrossRef]
- Lin, N.; Lin, Y.; Xu, J.; Liu, D.; Li, D.; Meng, H.; Gallant, M.A.; Kubota, N.; Roy, D.; Li, J.S.; et al. A multi-analyte cell-free DNA–based blood test for early detection of hepatocellular carcinoma. Hepatol. Commun. 2022, 6, 1753–1763. [Google Scholar] [CrossRef]
- Chalasani, N.P.; Porter, K.; Bhattacharya, A.; Book, A.J.; Neis, B.M.; Xiong, K.M.; Ramasubramanian, T.S.; Edwards, D.K.; Chen, I.; Johnson, S.; et al. Validation of a Novel Multitarget Blood Test Shows High Sensitivity to Detect Early Stage Hepatocellular Carcinoma. Clin. Gastroenterol. Hepatol. 2022, 20, 173–182.e7. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yu, L.; Gao, X.; Hu, J.; Wang, J.; Dai, Z.; Wang, J.-F.; Zhang, Z.; Lu, S.; Huang, X.; et al. Plasma MicroRNA Panel to Diagnose Hepatitis B Virus–Related Hepatocellular Carcinoma. J. Clin. Oncol. 2011, 29, 4781–4788. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Xu, J.; Wu, C.; Che, X.; Wang, L.; Yu, D.; Zhang, T.; Huang, L.; Li, H.; Tan, W.; Wang, C.; et al. Circulating MicroRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 2011, 50, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Pelizzaro, F.; Cardin, R.; Sartori, A.; Imondi, A.; Penzo, B.; Aliberti, C.; Ponzoni, A.; Vitale, A.; Cillo, U.; Farinati, F. Circulating MicroRNA-21 and MicroRNA-122 as Prognostic Biomarkers in Hepatocellular Carcinoma Patients Treated with Transarterial Chemoembolization. Biomedicines 2021, 9, 890. [Google Scholar] [CrossRef]
- Jiang, Y.; He, J.; Li, Y.; Guo, Y.; Tao, H. The Diagnostic Value of MicroRNAs as a Biomarker for Hepatocellular Carcinoma: A Meta-Analysis. BioMed Res. Int. 2019, 2019, 5179048. [Google Scholar] [CrossRef]
- Han, Z.; Li, K.; Wu, J.; Wang, K.; Qiu, C.; Ye, H.; Cui, C.; Song, C.; Wang, K.; Shi, J.; et al. Diagnostic Value of Rna for Hepatocellular Carcinoma: A Network Meta-Analysis. Biomarkers Med. 2021, 15, 1755–1767. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, W.; Shao, Z. Prognostic and diagnostic significance of circRNAs expression in hepatocellular carcinoma patients: A meta-analysis. Cancer Med. 2019, 8, 1148–1156. [Google Scholar] [CrossRef]
- Hao, Q.; Han, Y.; Xia, W.; Wang, Q.; Qian, H. Systematic Review and Meta-Analysis of the Utility of Circular RNAs as Biomarkers of Hepatocellular Carcinoma. Can. J. Gastroenterol. Hepatol. 2019, 2019, 1–13. [Google Scholar] [CrossRef]
- Yu, G.; Yang, L.; Zhou, J.; Zhang, L.; Xia, L. Abnormally Expressed Circular RNAs are Promising Biomarkers for Diagnosis of Hepatocellular Carcinoma: A Meta-Analysis. Clin. Lab. 2019, 65, 2101–2109. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, X.; Tang, J.; Jiang, R.; Zhang, W.; Ji, J.; Sun, B. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell Physiol. Biochem. 2015, 37, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Sun, Y.; Liu, L.; Zhou, B.; Wang, S.; Gu, D. Circulating LncRNAs Serve as Diagnostic Markers for Hepatocellular Carcinoma. Cell Physiol. Biochem. 2017, 44, 125–132. [Google Scholar] [CrossRef]
- Wang, C.; Ren, T.; Wang, K.; Zhang, S.; Liu, S.; Chen, H.; Yang, P. Identification of long non-coding RNA p34822 as a potential plasma biomarker for the diagnosis of hepatocellular carcinoma. Sci. China Life Sci. 2017, 60, 1047–1050. [Google Scholar] [CrossRef]
- Zeng, C.; Tang, Y.; Jiang, Y.; Zuo, Z.; Tao, H. Long noncoding RNAs as biomarkers for the diagnosis of hepatocellular carcinoma: A meta-analysis. Pathol. Res. Pract. 2021, 224, 153546. [Google Scholar] [CrossRef]
- Lumkul, L.; Jantaree, P.; Jaisamak, K.; Wongkummool, W.; Lapisatepun, W.; Orrapin, S.; Udomruk, S.; Lo Piccolo, L.; Chaiyawat, P. Combinatorial Gene Expression Profiling of Serum HULC, HOTAIR, and UCA1 lncRNAs to Differentiate Hepatocellular Carcinoma from Liver Diseases: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2024, 25, 1258. [Google Scholar] [CrossRef]
- Ganesan, R.; Yoon, S.J.; Suk, K.T. Microbiome and Metabolomics in Liver Cancer: Scientific Technology. Int. J. Mol. Sci. 2022, 24, 537. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, H.; Wang, H. N1-methyladenosine modification in cancer biology: Current status and future perspectives. Comput. Struct. Biotechnol. J. 2022, 20, 6578–6585. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Wang, W.; He, C. m1A methylation modification patterns and metabolic characteristics in hepatocellular carcinoma. BMC Gastroenterol. 2022, 22, 93. [Google Scholar] [CrossRef]
- Omran, M.M.; Farid, K.; Omar, M.A.; Emran, T.M.; El-Taweel, F.M.; Tabll, A.A. A combination of α-fetoprotein, midkine, thioredoxin and a metabolite for predicting hepatocellular carcinoma. Ann. Hepatol. 2020, 19, 179–185. [Google Scholar] [CrossRef]
- Sanchez, J.I.; Fontillas, A.C.; Kwan, S.-Y.; Sanchez, C.I.; Calderone, T.L.; Lee, J.L.; Elsaiey, A.; Cleere, D.W.; Wei, P.; Vierling, J.M.; et al. Metabolomics biomarkers of hepatocellular carcinoma in a prospective cohort of patients with cirrhosis. JHEP Rep. 2024, 6, 101119. [Google Scholar] [CrossRef]
- Chen, R.; Xu, X.; Tao, Y.; Qian, Z.; Yu, Y. Exosomes in hepatocellular carcinoma: A new horizon. Cell Commun. Signal. 2019, 17, 1. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Chen, Y.; Dong, X.; Wang, X. Serum Exosomal Long Noncoding RNAs ENSG00000258332.1 and LINC00635 for the Diagnosis and Prognosis of Hepatocellular Carcinoma. Cancer Epidemiol. Biomark. Prev. 2018, 27, 710–716. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, C.; Zhang, P.; Guo, G.; Jiang, T.; Zhao, X.; Jiang, J.; Huang, X.; Tong, H.; Tian, Y. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med. 2018, 7, 1670–1679. [Google Scholar] [CrossRef]
- Sun, L.; Su, Y.; Liu, X.; Xu, M.; Chen, X.; Zhu, Y.; Guo, Z.; Bai, T.; Dong, L.; Wei, C.; et al. Serum and exosome long non coding RNAs as potential biomarkers for hepatocellular carcinoma. J. Cancer 2018, 9, 2631–2639. [Google Scholar] [CrossRef]
- Xu, H.; Dong, X.; Chen, Y.; Wang, X. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin. Chem. Lab. Med. 2018, 56, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Xu, H.-F.; Liu, M.-Y.; Xu, Y.-J.; He, J.-C.; Zhou, Y.; Cang, S.-D. Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J. Gastroenterol. 2019, 25, 1890–1898. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Baek, G.O.; Ahn, H.R.; Sung, S.; Seo, C.W.; Cho, H.J.; Nam, S.W.; Cheong, J.Y.; Eun, J.W. Serum small extracellular vesicle-derived LINC00853 as a novel diagnostic marker for early hepatocellular carcinoma. Mol. Oncol. 2020, 14, 2646–2659. [Google Scholar] [CrossRef]
- Lee, Y.; Fujiwara, N.; Yang, J.D.; Hoshida, Y. Risk stratification and early detection biomarkers for precision HCC screening. Hepatology 2023, 78, 319–362. [Google Scholar] [CrossRef]
- Cho, H.J.; Eun, J.W.; Baek, G.O.; Seo, C.W.; Ahn, H.R.; Kim, S.S.; Cho, S.W.; Cheong, J.Y. Serum exosomal microRNA, miR-10b-5p, as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. J. Clin. Med. 2020, 9, 281. [Google Scholar] [CrossRef]
- von Felden, J.; Garcia-Lezana, T.; Dogra, N.; Gonzalez-Kozlova, E.; Ahsen, M.E.; Craig, A.; Gifford, S.; Wunsch, B.; Smith, J.T.; Kim, S.; et al. Unannotated small RNA clusters associated with circulating extracellular vesicles detect early stage liver cancer. Gut 2022, 71, 2069–2080. [Google Scholar] [CrossRef]
- Sun, N.; Zhang, C.; Lee, Y.; Tran, B.V.; Wang, J.; Kim, H.; Lee, J.; Zhang, R.Y.; Wang, J.J.; Hu, J.; et al. HCC EV ECG score: An extracellular vesicle-based protein assay for detection of early-stage hepatocellular carcinoma. Hepatology 2023, 77, 774–788. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Lee, Y.-T.; Zhang, R.Y.; Kao, R.; Teng, P.-C.; Yang, Y.; Yang, P.; Wang, J.J.; Smalley, M.; Chen, P.-J.; et al. Purification of HCC-specific extracellular vesicles on nanosubstrates for early HCC detection by digital scoring. Nat. Commun. 2020, 11, 4489. [Google Scholar] [CrossRef] [PubMed]
- Tiyuri, A.; Baghermanesh, S.S.; Davatgaran-Taghipour, Y.; Eslami, S.S.; Shaygan, N.; Parsaie, H.; Barati, M.; Jafari, D. Diagnostic accuracy of serum derived exosomes for hepatocellular carcinoma: A systematic review and meta-analysis. Expert. Rev. Mol. Diagn. 2023, 23, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Shahini, E.; Pasculli, G.; Solimando, A.G.; Tiribelli, C.; Cozzolongo, R.; Giannelli, G. Updating the Clinical Application of Blood Biomarkers and Their Algorithms in the Diagnosis and Surveillance of Hepatocellular Carcinoma: A Critical Review. Int. J. Mol. Sci. 2023, 24, 4286. [Google Scholar] [CrossRef]
- Cagnin, S.; Donghia, R.; Martini, A.; Pesole, P.L.; Coletta, S.; Shahini, E.; Boninsegna, G.; Biasiolo, A.; Pontisso, P.; Giannelli, G. Galad Score as a Prognostic Marker for Patients with Hepatocellular Carcinoma. Int. J. Mol. Sci. 2023, 24, 16485. [Google Scholar] [CrossRef]
- Yang, J.D.; Addissie, B.D.; Lavu, S.; Cvinar, J.L.; Giama, N.H.; Moser, C.D.; Miyabe, K.; Allotey, L.K.; Algeciras-Schimnich, A.; Theobald, J.P.; et al. GALAD Score for Hepatocellular Carcinoma Detection in Comparison with Liver Ultrasound and Proposal of GALADUS Score. Cancer Epidemiol. Biomarkers Prev. 2019, 28, 531–538. [Google Scholar] [CrossRef]
- Beudeker, B.J.; Fu, S.; Balderramo, D.; Mattos, A.Z.; Carrera, E.; Diaz, J.; Prieto, J.; Banales, J.; Vogel, A.; Arrese, M.; et al. Validation and optimization of AFP-based biomarker panels for early HCC detection in Latin America and Europe. Hepatol. Commun. 2023, 7, e0264. [Google Scholar] [CrossRef]
- El-Serag, H.B.; Jin, Q.; Nabihah, T.; Salem, E.; Luster, M.; Alsarraj, A.; Khaderi, S.; Singal, A.G.; Marrero, J.A.; Asrani, S.K.; et al. HES V2.0 outperforms GALAD for detection of HCC: A phase 3 biomarker study in the United States. Hepatology 2024, 10, 1097. [Google Scholar] [CrossRef]
- Dinges, S.S.; Hohm, A.; Vandergrift, L.A.; Nowak, J.; Habbel, P.; Kaltashov, I.A.; Cheng, L.L. Cancer metabolomic markers in urine: Evidence, techniques and recommendations. Nat. Rev. Urol. 2019, 16, 339–362. [Google Scholar] [CrossRef]
- Gao, Y. Urine—An untapped goldmine for biomarker discovery? Sci. China Life Sci. 2013, 56, 1145–1146. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Zeng, Y.; Luo, Y.; Guo, S.; Bao, L.; Zhang, Q. Urine miR-93-5p is a promising biomarker for early detection of HBV-related hepatocellular carcinoma. Eur. J. Surg. Oncol. 2022, 48, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.K.; Hamilton, J.P.; Lin, S.Y.; Chang, T.-T.; Hann, H.-W.; Hu, C.-T.; Lou, Y.; Lin, Y.-J.; Gade, T.P.; Park, G.; et al. Urine DNA biomarkers for hepatocellular carcinoma screening. Br. J. Cancer 2022, 126, 1432–1438. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.Y.; Xia, W.; Kim, A.K.; Chen, D.; Schleyer, S.; Choi, L.; Wang, Z.; Hamilton, J.P.; Luu, H.; Hann, H.-W.; et al. Novel urine cell-free DNA methylation markers for hepatocellular carcinoma. Sci. Rep. 2023, 13, 21585. [Google Scholar] [CrossRef]
- Dawuti, W.; Zheng, X.; Liu, H.; Zhao, H.; Dou, J.; Sun, L.; Chu, J.; Lin, R.; Lü, G. Urine surface-enhanced Raman spectroscopy combined with SVM algorithm for rapid diagnosis of liver cirrhosis and hepatocellular carcinoma. Photodiagnosis Photodyn. Ther. 2022, 38, 102811. [Google Scholar] [CrossRef] [PubMed]
- Dou, J.; Dawuti, W.; Zheng, X.; Zhang, R.; Zhou, J.; Lin, R.; Lü, G. Urine fluorescence spectroscopy combined with machine learning for screening of hepatocellular carcinoma and liver cirrhosis. Photodiagnosis Photodyn. Ther. 2022, 40, 103102. [Google Scholar] [CrossRef]
- Yavuz, B.G.; Datar, S.; Chamseddine, S.; Mohamed, Y.I.; LaPelusa, M.; Lee, S.S.; Hu, Z.I.; Koay, E.J.; Cao, H.S.T.; Jalal, P.K.; et al. The Gut Microbiome as a Biomarker and Therapeutic Target in Hepatocellular Carcinoma. Cancers 2023, 15, 4875. [Google Scholar] [CrossRef]
- Li, X.; Yi, Y.; Wu, T.; Chen, N.; Gu, X.; Xiang, L.; Jiang, Z.; Li, J.; Jin, H. Integrated microbiome and metabolome analysis reveals the interaction between intestinal flora and serum metabolites as potential biomarkers in hepatocellular carcinoma patients. Front. Cell Infect. Microbiol. 2023, 13, 1170748. [Google Scholar] [CrossRef]
- Huang, H.; Ren, Z.; Gao, X.; Hu, X.; Zhou, Y.; Jiang, J.; Lu, H.; Yin, S.; Ji, J.; Zhou, L.; et al. Integrated analysis of microbiome and host transcriptome reveals correlations between gut microbiota and clinical outcomes in HBV-related hepatocellular carcinoma. Genome Med. 2020, 12, 102. [Google Scholar] [CrossRef]
- Mohieldeen, K.; Hamoda, S.A.F.; Ahmed, S.M.; Najeeb, A.; Ellakany, W.I. Gut microbiome in cirrhotic hepatitis C virus patients with and without hepatocellular carcinoma. Egypt. Liver J. 2021, 11, 79. [Google Scholar] [CrossRef]
- Behary, J.; Amorim, N.; Jiang, X.-T.; Raposo, A.; Gong, L.; McGovern, E.; Ibrahim, R.; Chu, F.; Stephens, C.; Jebeili, H.; et al. Gut microbiota impact on the peripheral immune response in non-alcoholic fatty liver disease related hepatocellular carcinoma. Nat. Commun. 2021, 12, 187. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, Y.; Amir, A.; Nosenko, R.; Uzan-Yulzari, A.; Veitsman, E.; Cohen-Ezra, O.; Davidov, Y.; Weiss, P.; Bradichevski, T.; Segev, S.; et al. Alterations in the Gut Microbiome in the Progression of Cirrhosis to Hepatocellular Carcinoma. mSystems 2020, 5, e00153-20. [Google Scholar] [CrossRef] [PubMed]
- Ponziani, F.R.; Bhoori, S.; Castelli, C.; Putignani, L.; Rivoltini, L.; Del Chierico, F.; Sanguinetti, M.; Morelli, D.; Sterbini, F.P.; Petito, V.; et al. Hepatocellular Carcinoma Is Associated With Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology 2019, 69, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.M.; Mohs, A.; Gui, W.; Galvez, E.J.C.; Candels, L.S.; Hoenicke, L.; Muthukumarasamy, U.; Holland, C.H.; Elfers, C.; Kilic, K.; et al. Imbalanced gut microbiota fuels hepatocellular carcinoma development by shaping the hepatic inflammatory microenvironment. Nat. Commun. 2022, 13, 3964. [Google Scholar] [CrossRef]
- Ren, Z.; Li, A.; Jiang, J.; Zhou, L.; Yu, Z.; Lu, H.; Xie, H.; Chen, X.; Shao, L.; Zhang, R.; et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 2019, 68, 1014–1023. [Google Scholar] [CrossRef]
- Zheng, R.; Wang, G.; Pang, Z.; Ran, N.; Gu, Y.; Guan, X.; Yuan, Y.; Zuo, X.; Pan, H.; Zheng, J.; et al. Liver cirrhosis contributes to the disorder of gut microbiota in patients with hepatocellular carcinoma. Cancer Med. 2020, 9, 4232–4250. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, J.; Liu, Y.; Zeng, Y.; Jiang, Z.; Yan, H.; Lin, J.; Zhou, W.; Ou, Q.; Ao, L. Identification reproducible microbiota biomarkers for the diagnosis of cirrhosis and hepatocellular carcinoma. AMB Express 2023, 13, 35. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Li, J.; He, B.; Chen, B.; Liu, F.; Chen, Z.; Zheng, J.; Shi, Z.; Zhang, T.; Deng, L.; et al. Gut microbiome alteration as a diagnostic tool and associated with inflammatory response marker in primary liver cancer. Hepatol. Int. 2022, 16, 99–111. [Google Scholar] [CrossRef]
- Feng, J.; Wu, Y.; Dai, P.; Wang, D.; Liu, L.; Chai, B. Gut microbial signatures of patients with primary hepatocellular carcinoma and their healthy first-degree relatives. J. Appl. Microbiol. 2023, 134, lxad221. [Google Scholar] [CrossRef]
- Jiang, N.; Song, X.; Peng, Y.-M.; Wang, W.-N.; Song, Z. Association of disease condition with changes in intestinal flora, and plasma endotoxin and vascular endothelial growth factor levels in patients with liver cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 3605–3613. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, X.; Cai, L.; Cai, X. Dysbiosis of the gut microbiome in elderly patients with hepatocellular carcinoma. Sci. Rep. 2023, 13, 7797. [Google Scholar] [CrossRef] [PubMed]
- Jinato, T.; Anuntakarun, S.; Satthawiwat, N.; Chuaypen, N.; Tangkijvanich, P. Distinct alterations of gut microbiota between viral- and non-viral-related hepatocellular carcinoma. Appl. Microbiol. Biotechnol. 2024, 108, 34. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhou, H.; Xiang, Y.; Cui, F. The diagnostic potential of gut microbiome for early hepatitis B virus-related hepatocellular carcinoma. Eur. J. Gastroenterol. Hepatol. 2021, 33, e167–e175. [Google Scholar] [CrossRef] [PubMed]
- Pomyen, Y.; Chaisaingmongkol, J.; Rabibhadana, S.; Pupacdi, B.; Sripan, D.; Chornkrathok, C.; Budhu, A.; Budhisawasdi, V.; Lertprasertsuke, N.; Chotirosniramit, A.; et al. Gut dysbiosis in Thai intrahepatic cholangiocarcinoma and hepatocellular carcinoma. Sci. Rep. 2023, 13, 11406. [Google Scholar] [CrossRef] [PubMed]
- McMahon, B.; Cohen, C.; Brown Jr, R.S.; El-Serag, H.; Ioannou, G.N.; Lok, A.S.; Roberts, L.R.; Singal, A.G.; Block, T. Opportunities to address gaps in early detection and improve outcomes of liver cancer. JNCI Cancer Spectr. 2023, 2, pkad034. [Google Scholar] [CrossRef]
- Yang, Z.; Qi, Y.; Wang, Y.; Chen, X.; Wang, Y.; Zhang, X. Identifying Network Biomarkers in Early Diagnosis of Hepatocellular Carcinoma via miRNA–Gene Interaction Network Analysis. Curr. Issues Mol. Biol. 2023, 45, 7374–7387. [Google Scholar] [CrossRef]
- Johnson, P.J.; Bhatti, E.; Toyoda, H.; He, S. Serologic Detection of Hepatocellular Carcinoma: Application of Machine Learning and Implications for Diagnostic Models. JCO Clin. Cancer Inform. 2024, 8, e2300199. [Google Scholar] [CrossRef]
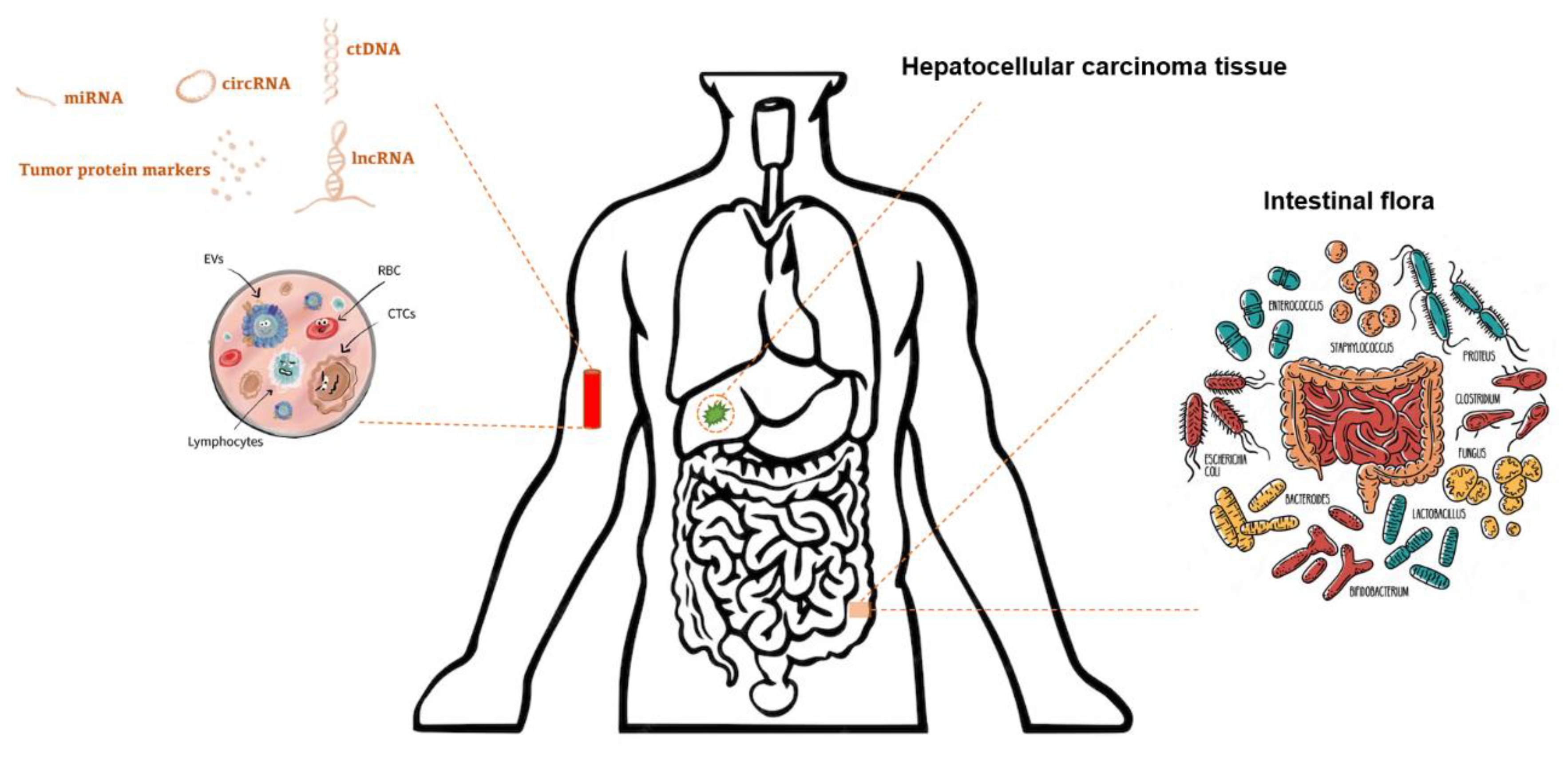
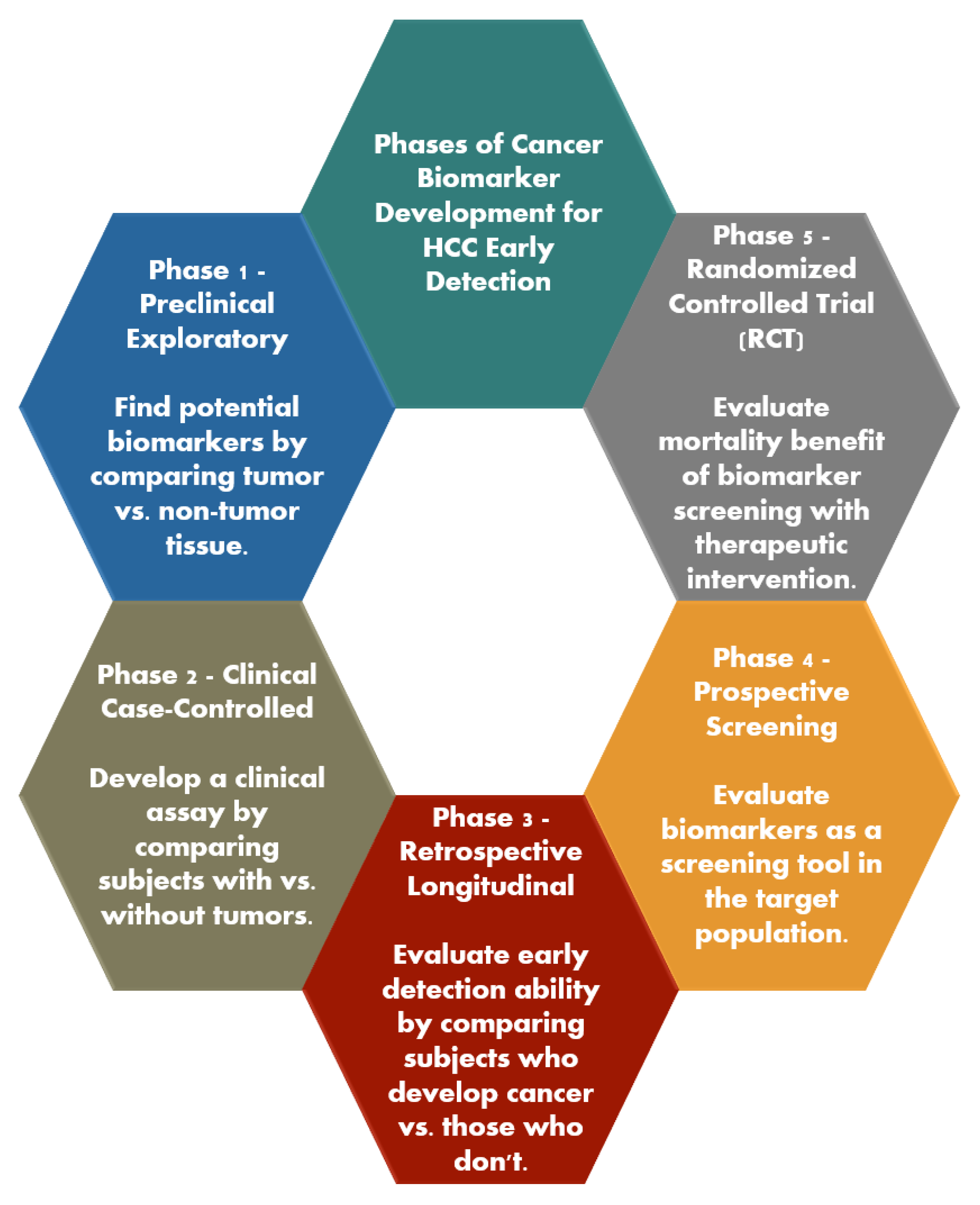
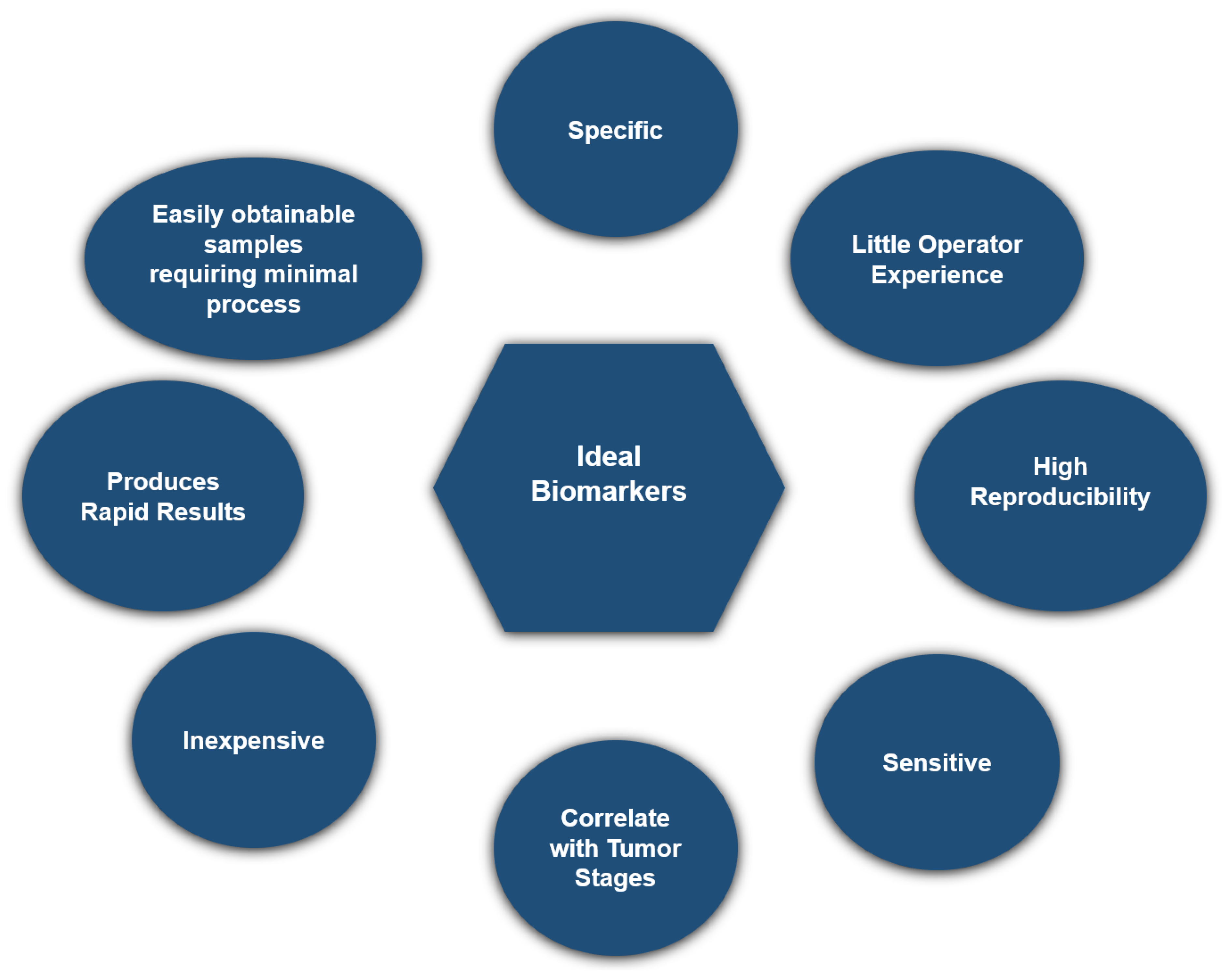
| Category | Biomarker Examples | Advantages | Limitations |
|---|---|---|---|
| Proteins | AFP, AFP-L3, DCP, Glypican-3, osteopontin, GALAD score, | Widely available assays, non-invasive or minimally invasive collection, relatively simple detection methods | Low sensitivity and specificity for early-stage HCC—Elevated levels in non-HCC conditions |
| Emerging Biomarkers | Nucleic Acid Biomarkers: miRNAs and lncRNAs, ctDNA | Potentially high sensitivity and specificity, potential for personalized medicine | Extracting and examining certain nucleic acids from blood may be technically demanding and need specialized equipment. |
| Exosomes, EV-associated biomarkers, Integrated Omics | Non-invasive, tumor specificity, stability, targeted therapeutics | These methodologies are currently in the process of being developed, and more investigation is required to verify their efficacy in clinical environments. | |
| Metabolites: amino acids, bile acids | Non-invasive approach, early detection potential, potential for multi-marker panels | Limited understanding of specific metabolite roles, standardization challenges | |
| Urine/Stool samples miRNAs | Non-invasive, early detection potential, Indicates the conditions of the intestines and the possibility of detecting cancer at an early stage. | Low sensitivity and specificity |
| Disease/Condition/Model (Reference) | Bacterial Genera (Increase/Decrease) | Key Findings |
|---|---|---|
| HCC Progression (Mouse Model) [104] | Dysbiotic microbiota, Akkermansia muciniphila ↓ | TLR4 activation → Increase in mMDSCs → Suppression of T-cells → Weakens the body’s immune response → Allows HCC to progress: reversible with antibiotics or Akkermansia muciniphila. |
| MASLD-related Cirrhosis with HCC vs. Without HCC [101,103] | Bacteroides ↑, Enterococcus ↑, Ruminococcaceae ↑, Bifidobacterium ↓ | Increased inflammation markers (fecal calprotectin, IL-8, IL-13, etc.). Microbiota triggered immunosuppressive response: ↑ regulatory T cells, ↓ CD8+ T cell activity. |
| Early HCC vs. Cirrhosis [105,106] | Actinobacteria ↑, Gemmiger ↑, Parabacteroides ↑, Lipopolysaccharide-producing bacteria ↑, Butyrate-producing bacteria ↓. | Increased microbial diversity, potential for non-invasive diagnostics. A model with 30 microbial markers had AUROC of 80.6%, distinguishing early HCC from non-HCC. |
| Cirrhotic Cases with HCC vs. Without HCC and Healthy Controls [102] | Clostridium ↑, CF231 ↑, Alphaproteobacteria ↓. | Cirrhotic cases (with/without HCC) had lower bacterial richness than healthy individuals. Key classifiers of HCC–cirrhosis from healthy controls: Veillonella dispar, Faecalibacterium prausnitzii, Ruminococcus gnavus. |
| HCC vs. their healthy first-degree relatives [109] | Lachnospiraceae ↑, Veillonella ↑, Ruminococcaceae UCG-014 ↑, Peptostreptococcaceae ↓, Romboutsia ↓, Citrobacter ↓. | Gut microbial composition in HCC patients is significantly altered. Romboutsia, Veillonella, and Peptostreptococcacae are potential biomarkers for HCC detection |
| Early vs. Middle vs. Advanced Liver cancer [110] | Early: Clostridiales ↑, Firmicutes ↑, Streptococcus ↑. Middle: Ruminococcaceae ↑, Pasteurellaceae ↑, Tanticharoenia ↑, Vagococcus ↑. Advanced: Bifidobacteriales ↑, Actinobacteria ↑, Barnesiella ↑, Porphyromonadaceae ↑, Pseudomonadales ↑. | Changes in microbiota with liver cancer progression: Barnesiella increased, Ruminococcaceae decreased. |
| HCC in elderly patients (60–80 years-old) [111] | ↓: A Blautia, Fusicatenibacter, Anaerostipes, CAG-56, Eggerthella, Lachnospiraceae_FCS020_group, Olsenella. ↑: Escherichia-Shigella, Fusobacterium, Megasphaera, Veillonella, Tyzzerella_4, Prevotella_2, Cronobacter | Age affects gut microbiota composition in HCC cases, and specific microbiota can be used as indicators for screening and diagnosing changes in elderly HCC patients. |
| Cirrhotic HCV Cases with HCC vs. Without HCC and Control [100] | Bacteroides ↑, Lactobacilli ↑, Prevotella ↓, Prevotella/Bacteroides ↓. | HCV-related cirrhosis and HCC show microbial dysbiosis, with HCC patients having higher proinflammatory bacteria compared to cirrhosis. |
| Viral HCC vs. Non-Viral HCC [112] | Viral HCC: Faecalibacterium ↑, Agathobacter ↑, Coprococcus ↑. Non-Viral HCC: Bacteroides ↑, Streptococcus ↑, Ruminococcus gnavus ↑, Parabacteroides ↑, Erysipelatoclostridium ↑. Short-chain fatty acid-producing bacteria ↓. | Gut dysbiosis linked to hepatocarcinogenesis and varies by HCC etiology. Microbiota signatures distinguish Viral-HCC and non-Viral HCC, offering potential for diagnosis and therapy. |
| HBV-related HCC vs. HBV-related Cirrhosis [113] | Veillonella ↓, Streptococcus ↓, Fusobacterium ↓, Blautia ↑, Agathobacter ↑ | Certain bacterial genera may drive progression from cirrhosis to HCC in HBV cases, with gut microbiome showing potential for early HCC diagnosis. |
| HCC vs. iCCA [114] | iCCA: Ruminococcus gnavus ↓, Veillonella ↑. HCC: Blautia ↑. | Greater gut microbiome heterogeneity in iCCA vs. HCC and healthy controls. High Veillonella in iCCA linked to amino acid biosynthesis and glycolysis, while Blautia in HCC linked to phospholipid and thiamine metabolism. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Attia, A.M.; Rezaee-Zavareh, M.S.; Hwang, S.Y.; Kim, N.; Adetyan, H.; Yalda, T.; Chen, P.-J.; Koltsova, E.K.; Yang, J.D. Novel Biomarkers for Early Detection of Hepatocellular Carcinoma. Diagnostics 2024, 14, 2278. https://doi.org/10.3390/diagnostics14202278
Attia AM, Rezaee-Zavareh MS, Hwang SY, Kim N, Adetyan H, Yalda T, Chen P-J, Koltsova EK, Yang JD. Novel Biomarkers for Early Detection of Hepatocellular Carcinoma. Diagnostics. 2024; 14(20):2278. https://doi.org/10.3390/diagnostics14202278
Chicago/Turabian StyleAttia, Abdelrahman M., Mohammad Saeid Rezaee-Zavareh, Soo Young Hwang, Naomy Kim, Hasmik Adetyan, Tamar Yalda, Pin-Jung Chen, Ekaterina K. Koltsova, and Ju Dong Yang. 2024. "Novel Biomarkers for Early Detection of Hepatocellular Carcinoma" Diagnostics 14, no. 20: 2278. https://doi.org/10.3390/diagnostics14202278
APA StyleAttia, A. M., Rezaee-Zavareh, M. S., Hwang, S. Y., Kim, N., Adetyan, H., Yalda, T., Chen, P.-J., Koltsova, E. K., & Yang, J. D. (2024). Novel Biomarkers for Early Detection of Hepatocellular Carcinoma. Diagnostics, 14(20), 2278. https://doi.org/10.3390/diagnostics14202278







