Identification and Characterization of 24-Dehydrocholesterol Reductase (DHCR24) in the Two-Spotted Cricket, Gryllus bimaculatus
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. BLAST Search and Phylogenetic Analysis
2.2. Experimental Insects
2.3. Preparation of Azacosterol Diet
2.4. Preparation of Ultrancentrifugal Fractions
2.5. Evaluation of Enzyme Activity
2.6. GC-MS Conditions
2.7. Examination of the Effects of Azacosterol on Enzyme Activity
2.8. Preparation of cDNA
2.9. Examination of Transcriptional Levels of DHCR24 in Tissues
2.10. RNA Interference (RNAi)
2.11. Statistical Analyses
3. Results
3.1. Characterization of GbDHCR24
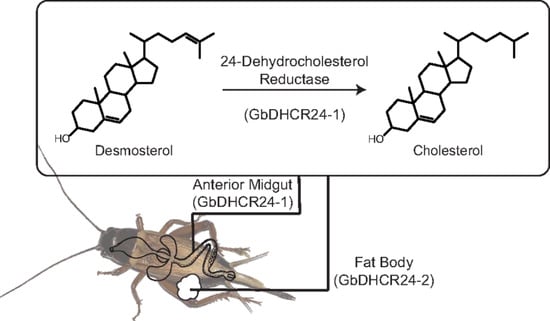
3.2. Tissue Distribution of GbDHCR24 Expression
3.3. Examination of Enzyme Activity of GbDHCR24
3.4. Effects of Azacosterol on Crickets
3.5. Effects of Knockdown of GbDHCR24s
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brown, A.J.; Sharpe, L.J. Chapter 11-Cholesterol Synthesis. In Biochemistry of Lipids, Lipoproteins and Membranes, 6th ed.; Ridgway, N.D., McLeod, R.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 327–358. [Google Scholar] [CrossRef]
- Ačimovič, J.; Rozman, D. Steroidal Triterpenes of Cholesterol Synthesis. Molecules 2013, 18, 4002–4017. [Google Scholar] [CrossRef] [PubMed]
- Zerenturk, E.J.; Sharpe, L.J.; Ikonen, E.; Brown, A.J. Desmosterol and DHCR24: Unexpected New Directions for a Terminal Step in Cholesterol Synthesis. Prog. Lipid Res. 2013, 52, 666–680. [Google Scholar] [CrossRef]
- Mitsche, M.A.; McDonald, J.G.; Hobbs, H.H.; Cohen, J.C. Flux Analysis of Cholesterol Biosynthesis in vivo Reveals Multiple Tissue and Cell-Type Specific Pathways. Elife 2015, 4, e07999. [Google Scholar] [CrossRef] [PubMed]
- Kircher, H.W. Sterols and Insects. In Cholesterol Systems in Insects and Animals; Dupont, J., Ed.; CRC Press: Boca Raton, FL, USA, 1982; pp. 1–50. [Google Scholar]
- Svoboda, J.A. Variability of Metabolism and Function of Sterols in Insects. Crit. Rev. Biochem. Mol. Biol. 1999, 34, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, J.A.; Thompson, M.J.; Robbins, W.E. Identification of Fucosterol as a Metabolite and Probable Intermediate in Conversion of β-sitosterol to Cholesterol in the Tobacco hornworm. Nat. New Biol. 1971, 230, 57–58. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, J.A.; Thompson, M.J.; Robbins, W.E. 24-Methylenecholesterol: Isolation and Ddentification as an Dntermediate in the Conversion of Campesterol to Cholesterol in the Tobacco hornworm. Lipids 1972, 7, 156–158. [Google Scholar] [CrossRef]
- Svoboda, J.A.; Kaplanis, J.N.; Robbins, W.E.; Thompson, M.J. Recent Developments in Insect Steroid Metabolism. Annu. Rev. Entomol. 1975, 20, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, J.A.; Rees, H.H.; Thompson, M.J.; Hoggard, N. Intermediates of Stigmasterol metabolism in Spodoptera Littoralis. Steroids 1989, 53, 329–343. [Google Scholar] [CrossRef]
- Maruyama, S.; Fujimoto, Y.; Morisaki, M.; Ikekawa, N. Mechanism of Campesterol Demethylation in Insect. Tetrahedron Lett. 1982, 23, 1701–1704. [Google Scholar] [CrossRef]
- Nicotra, F.; Ronchetti, F.; Russo, G. The Metabolism of Phytosterols in the Insect Tenebrio molitor: Utilization of 24- Methylenecholesterol and 24,28-Epoxymethylenecholesterol. Lipids 1982, 17, 184–186. [Google Scholar] [CrossRef]
- Ciufo, L.F.; Murray, P.A.; Thompson, A.; Rigden, D.J.; Rees, H.H. Characterisation of a Desmosterol Reductase Involved in Phytosterol Dealkylation in the Silkworm, Bombyx mori. PLoS ONE 2011, 6, e21316. [Google Scholar] [CrossRef]
- Fujimori, H.; Zhou, Y.J.; Fukumura, K.; Matsumoto, S.; Tukamoto, Y.; Nagata, S. Specific Distribution of Expression and Enzymatic Activity of Cholesterol Biosynthetic Enzyme DHCR24 Orthologs in the Phytophagous Insect. Biosci. Biotechnol. Biochem. 2020, 84, 126–133. [Google Scholar] [CrossRef]
- Mushegian, A.R.; Koonin, E.V. A Putative FAD-Binding Domain in a Distinct Group of Oxidases Including a Protein Involved in Plant Development. Protein Sci. 1995, 4, 1243–1244. [Google Scholar] [CrossRef] [PubMed]
- Shamsuzzama; Lebedev, R.; Trabelcy, B.; Goncalves, I.L.; Gerchman, Y.; Sapir, A. Metabolic Reconfiguration in C. elegans Suggests a Pathway for Widespread Sterol Auxotrophy in the Animal Kingdom. Curr. Biol. 2020, 30, 3031–3038.e7. [Google Scholar] [CrossRef] [PubMed]
- Counsell, R.E.; Klimstra, P.D.; Ranney, R.E. Hypocholesterolemic Agents. III.1 N-Methyl-N-(dialkylamino)alkyl-17β- aminoandrost-5-en-3β-ol Derivatives. J. Med. Pharm. Chem. 1962, 5, 1224–1233. [Google Scholar] [CrossRef]
- Liebrich, W.; Hoffmann, K. Ecdysone 20-Monooxygenase in a Cricket, Gryllus Bimaculatus (Ensifera, Gryllidae)—Characterization Microsomal Midgut Steroid Hydroxylase Adult Females. J. Comp. Physiol. B 1991, 161, 93–99. [Google Scholar] [CrossRef]
- Fukumura, K.; Konuma, T.; Tsukamoto, Y.; Nagata, S. Adipokinetic Hormone Signaling Determines Dietary Fatty Acid Preference through Maintenance of Hemolymph Fatty Acid Composition in the Cricket Gryllus Bimaculatus. Sci. Rep. 2018, 8, 4737. [Google Scholar] [CrossRef]
- Finn, R.D.; Tate, J.; Mistry, J.; Coggill, P.C.; Sammut, S.J.; Hotz, H.R.; Ceric, G.; Forslund, K.; Eddy, S.R.; Sonnhammer, E.L.L.; et al. The Pfam Protein Families Database. Nucleic Acids Res. 2007, 36, D281–D288. [Google Scholar] [CrossRef]
- Serra, M.; Matabosch, X.; Ying, L.; Watson, G.; Shackleton, C. Hair and Skin Sterols in Normal Mice and those with Deficient Dehydrosterol Reductase (DHCR7), the Enzyme Associated with Smith-Lemli-Opitz Syndrome. J. Steroid Biochem. Mol. Biol. 2010, 122, 318–325. [Google Scholar] [CrossRef][Green Version]
- Waterham, H.R.; Koster, J.; Romeijn, G.J.; Hennekam, R.C.; Vreken, P.; Andersson, H.C.; FitzPatrick, D.R.; Kelley, R.I.; Wanders, R.J. Mutations in the 3β-Hydroxysterol Δ24-Reductase Gene Cause Desmosterolosis, an Autosomal Recessive Disorder of Cholesterol Biosynthesis. Am. J. Hum. Genet. 2001, 69, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Nagahashi, J.; Hiraike, K. Effects of Centrifugal Force and Centrifugation Time on the Sedimentation of Plant Organelles. Plant Physiol. 1982, 69, 546–548. [Google Scholar] [CrossRef]
- Graham, J. Preparation of Crude Subcellular Fractions by Differential Centrifugation. Sci. World J. 2002, 2, 1638–1642. [Google Scholar] [CrossRef][Green Version]
- Greeve, I.; Hermans-Borgmeyer, I.; Brellinger, C.; Kasper, D.; Gomez-Isla, T.; Behl, C.; Levkau, B.; Nitsch, R.M. The Human DIMINUTO/DWARF1 Homolog Seladin-1 Confers Resistance to Alzheimer’s Disease-Associated Neurodegeneration and Oxidative Stress. J. Neurosci. 2000, 20, 7345. [Google Scholar] [CrossRef]
- Zerenturk, E.J.; Sharpe, L.J.; Brown, A.J. DHCR24 Associates Strongly with the Endoplasmic Reticulum beyond Predicted Membrane Domains: Implications for the Activities of this Multi-Functional Enzyme. Biosci. Rep. 2014, 34. [Google Scholar] [CrossRef]
- Luu, W.; Zerenturk, E.J.; Kristiana, I.; Bucknall, M.P.; Sharpe, L.J.; Brown, A.J. Signaling Regulates Activity of DHCR24, the Final Enzyme in Cholesterol Synthesis. J. Lipid Res. 2014, 55, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Blom, N.; Gammeltoft, S.; Brunak, S. Sequence and Structure-Based Prediction of Eukaryotic Protein Phosphorylation Sites. J. Mol. Biol. 1999, 294, 1351–1362. [Google Scholar] [CrossRef] [PubMed]
- Gibon, Y.; Usadel, B.; Blaesing, O.E.; Kamlage, B.; Hoehne, M.; Trethewey, R.; Stitt, M. ntegration of Metabolite with Transcript and Enzyme Activity Profiling during Diurnal Cycles in Arabidopsis rosettes. Genome Biol. 2006, 7, R76. [Google Scholar] [CrossRef] [PubMed]
- Glanemann, C.; Loos, A.; Gorret, N.; Willis, L.B.; O’Brien, X.M.; Lessard, P.A.; Sinskey, A.J. Disparity between Changes in mRNA Abundance and Enzyme Activity in Corynebacterium glutamicum: Implications for DNA Microarray Analysis. Appl. Microbiol. Biotechnol. 2003, 61, 61–68. [Google Scholar] [CrossRef]
- Svoboda, J.A.; Robbins, W.E. The Inhibitive Effects of Azasterols on Sterol Metabolism and Growth and Development in Insects with Special Reference to the Tobacco hornworm. Lipids 1971, 6, 113–119. [Google Scholar] [CrossRef]
- Jansen, M.; Pietiaïnen, V.M.; Pölönen, H.; Rasilainen, L.; Koivusalo, M.; Ruotsalainen, U.; Jokitalo, E.; Ikonen, E. Cholesterol Substitution Increases the Structural Heterogeneity of Caveolae. J. Biol. Chem. 2008, 283, 14610–14618. [Google Scholar] [CrossRef]
- Ahrens, R.A.; Dupont, J.; Thompson, M.J. Sterols in Brain and Liver of Young Rats Fed 20,25-Diaza-Cholesterol. Proc. Soc. Exp. Biol. Med. 1965, 118, 436–438. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, J.A.; Robbins, W.E. Desmosterol as a Common Intermediate in the Conversion of a Number of C28 and C29 Plant Sterols to Cholesterol by the Tobacco hornworm. Experientia 1968, 24, 1131–1132. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, J.A.; Robbins, W.E. Conversion of Beta Sitosterol to Cholesterol Blocked in an Insect by Hypocholesterolemic Agents. Science 1967, 156, 1637–1638. [Google Scholar] [CrossRef] [PubMed]
- Dahlin, P.; Srivastava, V.; Ekengren, S.; McKee, L.S.; Bulone, V. Comparative Analysis of Sterol Acquisition in the Oomycetes Saprolegnia parasitica and Phytophthora infestans. PLoS ONE 2017, 12, e0170873. [Google Scholar] [CrossRef]






| Sterols | Parent Ion | Peak for Identification | Relative Retention |
|---|---|---|---|
| (TMS Derivatives) | (m/z) | (m/z) | Time (RRT) |
| Cholesterol | 458 | 458 | 1.009 |
| Cholesterol-d6 | 464 | 464 | 1.013 |
| Cholesterol-d7 | 465 | 465 | 1.014 |
| Desmosterol | 456 | 343 | 0.978 |
| Desmosterol-d6 | 462 | 462 | 0.984 |
| 7-Dehydrocholesterol | 456 | 325 | 0.974 |
| 7-Dehydrodesmosterol | 454 | 349 | 0.943 |
| Experiment | Primer | Sequence (5′ to 3′) |
|---|---|---|
| qPCR | DHCR24-1 Forward | AACAATGATCCAGGCCTCGT |
| DHCR24-1 Reverse | AGGAACGCCGTATACACCAA | |
| DHCR24-2 Forward | ATGGGTGTTCGCGTCATTCT | |
| DHCR24-2 Reverse | TGAAGACCAACCAGTTGCGT | |
| -actin Forward | TTGACAATGGATCCGGAATGT | |
| -actin Reverse | AAAACTGCCCTGGGTGCAT | |
| RNAi | T7-DHCR24-1 Forward | GCTTCTAATACGACTCACTATAGCGATAAGAAGGTGCGCAACG |
| T7-DHCR24-1 Reverse | GCTTCTAATACGACTCACTATAGTAACGACACTGCCATCTGCC | |
| T7-DHCR24-2 Forward | GCTTCTAATACGACTCACTATAGATGCCTCCCAAGGTTTCGTT | |
| T7-DHCR24-2 Reverse | GCTTCTAATACGACTCACTATAGACACGTCAGGAAAAGCCTCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mack, Y.S.I.; Dehari, M.; Morooka, N.; Nagata, S. Identification and Characterization of 24-Dehydrocholesterol Reductase (DHCR24) in the Two-Spotted Cricket, Gryllus bimaculatus. Insects 2021, 12, 782. https://doi.org/10.3390/insects12090782
Mack YSI, Dehari M, Morooka N, Nagata S. Identification and Characterization of 24-Dehydrocholesterol Reductase (DHCR24) in the Two-Spotted Cricket, Gryllus bimaculatus. Insects. 2021; 12(9):782. https://doi.org/10.3390/insects12090782
Chicago/Turabian StyleMack, Yin Shan Isa, Masatoshi Dehari, Nobukatsu Morooka, and Shinji Nagata. 2021. "Identification and Characterization of 24-Dehydrocholesterol Reductase (DHCR24) in the Two-Spotted Cricket, Gryllus bimaculatus" Insects 12, no. 9: 782. https://doi.org/10.3390/insects12090782
APA StyleMack, Y. S. I., Dehari, M., Morooka, N., & Nagata, S. (2021). Identification and Characterization of 24-Dehydrocholesterol Reductase (DHCR24) in the Two-Spotted Cricket, Gryllus bimaculatus. Insects, 12(9), 782. https://doi.org/10.3390/insects12090782