Establishment of a Simple and Rapid Nucleic Acid Detection Method for Hookworm Identification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Parasites and DNA Samples
2.2. Target Sequence Screening and Primer Design
2.3. Establishment of an RAA Reaction System
2.4. Sensitivity and Specificity Evaluation
2.5. Comparison of Detection Efficacy for Human Fecal Samples Using Different Methods
2.6. Ethics Statement
2.7. Statistical Analysis
3. Results
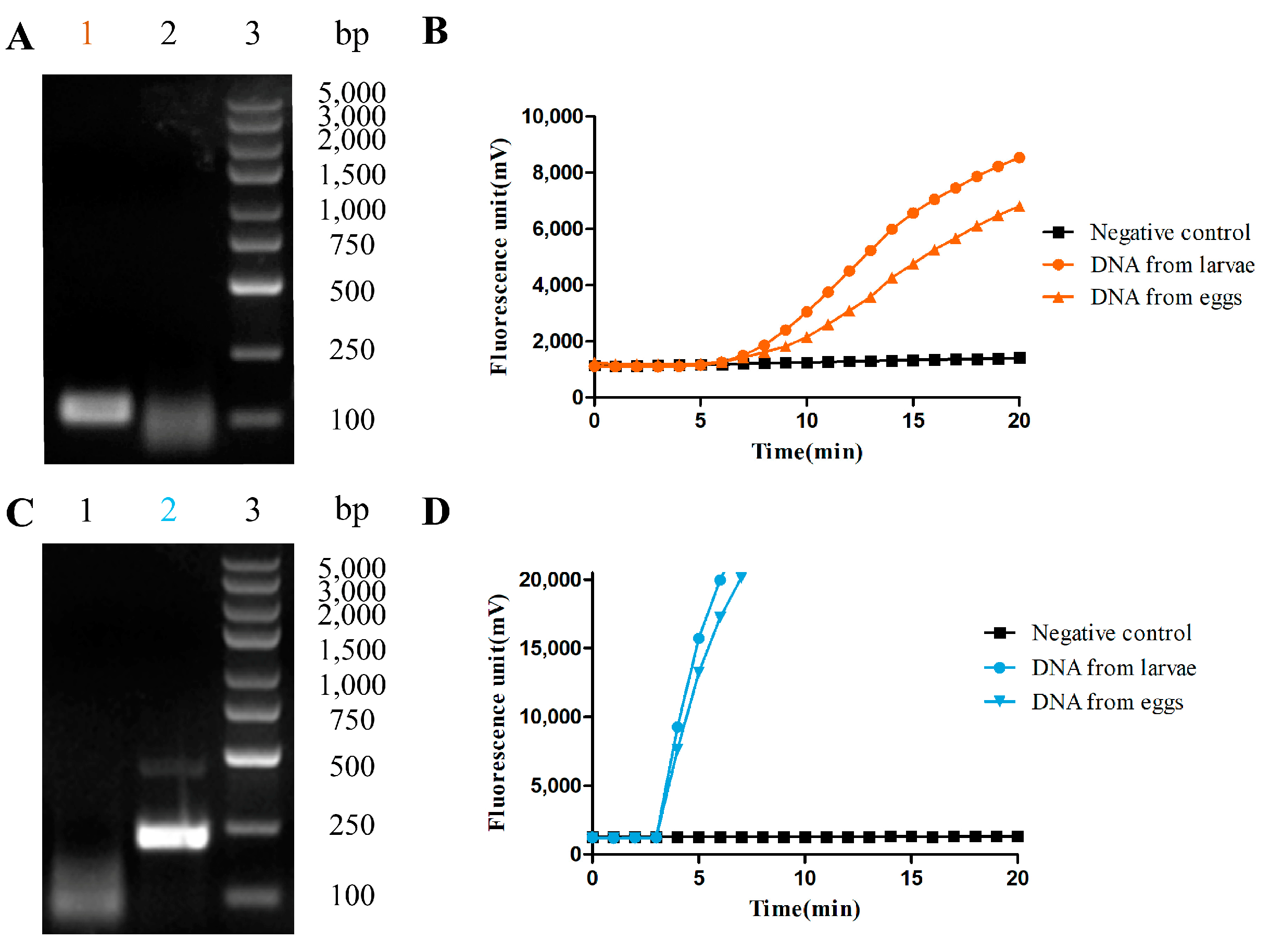
3.1. The RAA-Based Method Was Successfully Established for Hookworm Detection
3.2. Thr Fluorescence RAA-Based Method Showed High Sensitivity and Specificity for Hookworm Detection
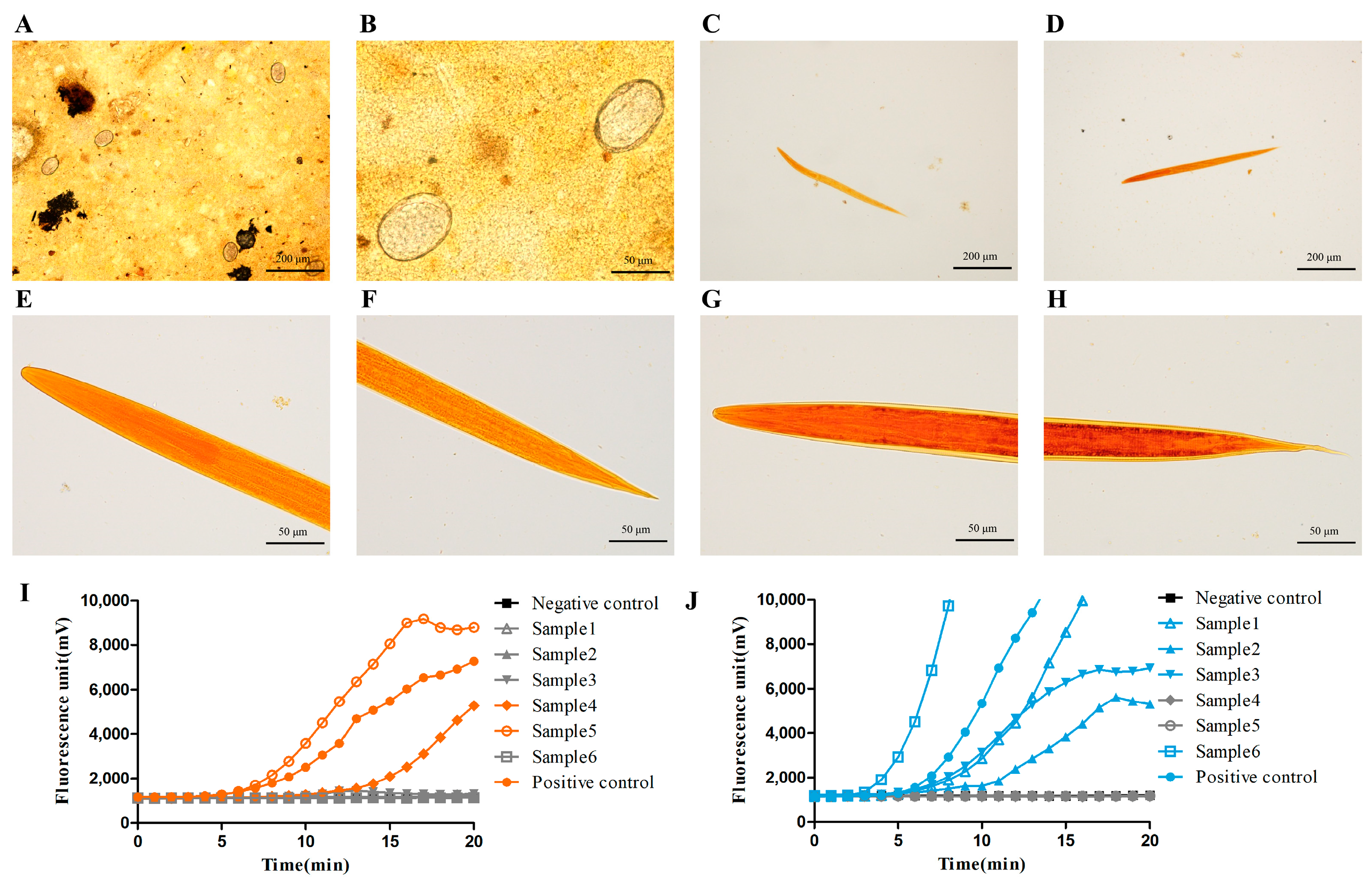
3.3. The Fluorescence RAA-Based Method Showed a High Detection Capacity of Hookworm Infection for Human Fecal Samples
3.4. The Fluorescence RAA-Based Method Could Be Applied to Hookworm Species Identification in Human Fecal Samples
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Álvarez-Hernández, D.-A.; Rivero-Zambrano, L.; Martínez-Juárez, L.-A.; García-Rodríguez-Arana, R. Overcoming the global burden of neglected tropical diseases. Ther. Adv. Infect. Dis. 2020, 7, 2049936120966449. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Chen, Y.; Zhang, H.-B.; Chen, J.-X.; Zhou, X.-N. The control of hookworm infection in China. Parasites Vectors 2009, 2, 44. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, S.M.; Hotez, P.J.; Asti, L.; Zapf, K.M.; Bottazzi, M.E.; Diemert, D.J.; Lee, B.Y. The global economic and health burden of human hookworm infection. PLoS Negl. Trop. Dis. 2016, 10, e0004922. [Google Scholar] [CrossRef]
- Chen, Y.D.; Zhou, C.H.; Zhu, H.H.; Huang, J.L.; Duan, L.; Zhu, T.G.; Qian, M.B.; Li, S.Z.; Chen, H.G.; Cai, L.; et al. National survey on the current status of important human parasitic diseases in China in 2015. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi 2020, 38, 5–16. [Google Scholar] [CrossRef]
- Dai, Y.; Xu, X.; Liu, J.; Jin, X.; Shen, M.; Wang, X.; Cao, J.; Yang, H. Prevalence of intestinal helminth infections in Jiangsu Province, eastern China; a cross-sectional survey conducted in 2015. BMC Infect. Dis. 2019, 19, 1–9. [Google Scholar] [CrossRef]
- Loukas, A.; Hotez, P.J.; Diemert, D.; Yazdanbakhsh, M.; McCarthy, J.S.; Correa-Oliveira, R.; Croese, J.; Bethony, J.M. Hookworm infection. Nat. Rev. Dis. Prim. 2016, 2, 16088. [Google Scholar] [CrossRef] [PubMed]
- de Silva, N.R.; Brooker, S.; Hotez, P.; Montresor, A.; Engels, D.; Savioli, L. Soil-transmitted helminth infections: Updating the global picture. Trends Parasitol. 2003, 19, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Roche, M.; Layrisse, M. The nature and causes of “hookworm anemia”. Am. J. Trop. Med. Hyg. 1966, 15, 1029–1102. [Google Scholar] [CrossRef]
- Knopp, S.; Mgeni, A.F.; Khamis, I.S.; Steinmann, P.; Stothard, R.; Rollinson, D.; Marti, H.; Utzinger, J. Diagnosis of soil-transmitted helminths in the era of preventive chemotherapy: Effect of multiple stool sampling and use of different diagnostic techniques. PLoS Negl. Trop. Dis. 2008, 2, e331. [Google Scholar] [CrossRef]
- Bärenbold, O.; Raso, G.; Coulibaly, J.T.; N’goran, E.K.; Utzinger, J.; Vounatsou, P. Estimating sensitivity of the Kato-Katz technique for the diagnosis of Schistosoma mansoni and hookworm in relation to infection intensity. PLoS Negl. Trop. Dis. 2017, 11, e0005953. [Google Scholar] [CrossRef]
- Tarafder, M.; Carabin, H.; Joseph, L.; Balolong, E.; Olveda, R.; McGarvey, S. Estimating the sensitivity and specificity of Kato-Katz stool examination technique for detection of hookworms, Ascaris lumbricoides and Trichuris trichiura infections in humans in the absence of a ‘gold standard’. Int. J. Parasitol. 2010, 40, 399–404. [Google Scholar] [CrossRef]
- Dacombe, R.; Crampin, A.; Floyd, S.; Randall, A.; Ndhlovu, R.; Bickle, Q.; Fine, P. Time delays between patient and laboratory selectively affect accuracy of helminth diagnosis. Trans. R. Soc. Trop. Med. Hyg. 2007, 101, 140–145. [Google Scholar] [CrossRef]
- Jonker, F.A.M.; Calis, J.C.J.; Phiri, K.; Brienen, E.A.T.; Khoffi, H.; Brabin, B.; Verweij, J.J.; Van Hensbroek, M.B.; Van Lieshout, L. Real-time PCR demonstrates Ancylostoma duodenale is a key factor in the etiology of severe anemia and iron deficiency in Malawian pre-school children. PLoS Negl. Trop. Dis. 2012, 6, e1555. [Google Scholar] [CrossRef] [PubMed]
- Nongmaithem, O.; Shantikumar, T.; Dutta, S. Evaluation of various culture techniques for identification of hookworm species from stool samples of children. Indian J. Med. Res. 2019, 149, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Verweij, J.J.; Brienen, E.A.T.; Ziem, J.; Yelifari, L.; Polderman, A.M.; Van Lieshout, L. Simultaneous detection and quantification of Ancylostoma duodenale, Necator americanus, and Oesophagostomum bifurcum in fecal samples using Multiplex Real-Time PCR. Am. J. Trop. Med. Hyg. 2007, 77, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Vasoo, S.; Pritt, B.S. Molecular diagnostics and parasitic disease. Clin. Lab. Med. 2013, 33, 461–503. [Google Scholar] [CrossRef] [PubMed]
- Barda, B.; Schindler, C.; Wampfler, R.; Ame, S.; Ali, S.M.; Keiser, J. Comparison of real-time PCR and the Kato-Katz method for the diagnosis of soil-transmitted helminthiasis and assessment of cure in a randomized controlled trial. BMC Microbiol. 2020, 20, 1–8. [Google Scholar] [CrossRef]
- Benjamin-Chung, J.; Pilotte, N.; Ercumen, A.; Grant, J.R.; Maasch, J.R.M.A.; Gonzalez, A.M.; Ester, A.C.; Arnold, B.F.; Rahman, M.; Haque, R.; et al. Comparison of multi-parallel qPCR and double-slide Kato-Katz for detection of soil-transmitted helminth infection among children in rural Bangladesh. PLoS Negl. Trop. Dis. 2020, 14, e0008087. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef]
- Yan, T.-F.; Li, X.-N.; Wang, L.; Chen, C.; Duan, S.-X.; Qi, J.-J.; Li, L.-X.; Ma, X.-J. Development of a reverse transcription recombinase-aided amplification assay for the detection of coxsackievirus A10 and coxsackievirus A6 RNA. Arch. Virol. 2018, 163, 1455–1461. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, L.; Ma, R.; Cong, L.; Wu, Z.; Wei, Y.; Xue, S.; Zheng, W.; Tang, S. Rapid detection of Salmonella with Recombinase Aided Amplification. J. Microbiol. Methods 2017, 139, 202–204. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, X.-N.; Li, G.-X.; Zhao, L.; Duan, S.-X.; Yan, T.-F.; Feng, Z.-S.; Ma, X.-J. Use of a rapid reverse-transcription recombinase aided amplification assay for respiratory syncytial virus detection. Diagn. Microbiol. Infect. Dis. 2018, 90, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Liu, Y.H.; Ni, B.X.; Wang, X.T.; Xu, X.Z.; Ying, Q.J.; Dai, Y.; Cao, J. Establishment of a nucleic acid assay for detection of Echinococcus granulosus based on recombinase-aided isothermal amplification assay. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi 2020, 32, 340–344. [Google Scholar] [PubMed]
- Zhang, Q.; Ding, X.; Wu, X.M.; Liu, Y.H.; Liu, J.F.; Xu, X.Z.; Ying, Q.J.; Cao, J.; Dai, Y. Establishment and preliminary evaluation of recombinase aided isothermal amplification (RAA) assay for specific nucleic acid detection of Clonorchis sinensis. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi 2019, 31, 468–473. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ding, X.; Liu, Y.H.; Liu, J.F.; Xu, X.Z.; Ying, Q.J.; Dai, Y.; Cao, J. Establishment of a recombinase-aided isothermal amplification assay for nucleic acid detection of Angiostrongylus cantonensis. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi 2020, 32, 350–354. [Google Scholar] [CrossRef]
- Wang, J.-X.; Pan, C.-S.; Cui, L.-W. Application of a real-time PCR method for detecting and monitoring hookworm Necator americanus infections in Southern China. Asian Pac. J. Trop. Biomed. 2012, 2, 925–929. [Google Scholar] [CrossRef]
- Ngui, R.; Ching, L.S.; Kai, T.T.; Roslan, M.A.; Lim, Y.A. Molecular identification of human hookworm infections in economically disadvantaged communities in Peninsular Malaysia. Am. J. Trop. Med. Hyg. 2012, 86, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Katz, N.; Chaves, A.; Pellegrino, J. A simple device for quantitative stool thick-smear technique in Schistosomiasis mansoni. Rev. Inst. Med. Trop. Sao Paulo 1972, 14, 397–400. [Google Scholar]
- Zendejas-Heredia, P.A.; Colella, V.; Hii, S.F.; Traub, R.J. Comparison of the egg recovery rates and limit of detection for soil-transmitted helminths using the Kato-Katz thick smear, faecal flotation and quantitative real-time PCR in human stool. PLoS Negl. Trop. Dis. 2021, 15, e0009395. [Google Scholar] [CrossRef]
- Kumar, S.; Gupta, S.; Mohmad, A.; Fular, A.; Parthasarathi, B.C.; Chaubey, A.K. Molecular tools-advances, opportunities and prospects for the control of parasites of veterinary importance. Int. J. Trop. Insect Sci. 2020, 41, 33–42. [Google Scholar] [CrossRef]
- Ngui, R.; Lim, Y.A.L.; Chua, K.H. Rapid detection and identification of human hookworm infections through High Resolution Melting (HRM) analysis. PLoS ONE 2012, 7, e41996. [Google Scholar] [CrossRef] [PubMed]
- van Mens, S.P.; Aryeetey, Y.; Yazdanbakhsh, M.; van Lieshout, L.; Boakye, D.; Verweij, J.J. Comparison of real-time PCR and Kato smear microscopy for the detection of hookworm infections in three consecutive faecal samples from schoolchildren in Ghana. Trans. R. Soc. Trop. Med. Hyg. 2013, 107, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Papaiakovou, M.; Pilotte, N.; Grant, J.R.; Traub, R.J.; Llewellyn, S.; McCarthy, J.S.; Krolewiecki, A.J.; Cimino, R.; Mejia, R.; Williams, S.A. A novel, species-specific, real-time PCR assay for the detection of the emerging zoonotic parasite Ancylostoma ceylanicum in human stool. PLoS Negl. Trop. Dis. 2017, 11, e0005734. [Google Scholar] [CrossRef] [PubMed]
- Vercruysse, J.; Albonico, M.; Behnke, J.M.; Kotze, A.C.; Prichard, R.K.; McCarthy, J.S.; Montresor, A.; Levecke, B. Is anthelmintic resistance a concern for the control of human soil-transmitted helminths? Int. J. Parasitol. Drugs Drug Resist. 2011, 1, 14–27. [Google Scholar] [CrossRef]
- Gasser, R.; Cantacessi, C.; Loukas, A. DNA technological progress toward advanced diagnostic tools to support human hookworm control. Biotechnol. Adv. 2008, 26, 35–45. [Google Scholar] [CrossRef]
- Chidambaram, M.; Parija, S.C.; Toi, P.C.; Mandal, J.; Sankaramoorthy, D.; George, S.; Natarajan, M.; Padukone, S. Evaluation of the utility of conventional polymerase chain reaction for detection and species differentiation in human hookworm infections. Trop. Parasitol. 2017, 7, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Basuni, M.; Rahumatullah, A.; Miswan, N.; Ahmad, M.; Noordin, R.; Verweij, J.J.; Zainudin, N.S.; Aziz, F.A.; Othman, N.; Muhi, J. A Pentaplex Real-Time Polymerase Chain reaction assay for detection of four species of soil-transmitted helminths. Am. J. Trop. Med. Hyg. 2011, 84, 338–343. [Google Scholar] [CrossRef]

| Sequences | AD (5′-3′) | NA (5′-3′) |
|---|---|---|
| Forward primer | CGATACGCGAATCGACCGATCCATCGCTGAAG | CTGTTTGTCGAACGGTACTTGCTCTGTACTACG |
| Reverse primer | ATCTGCTAACGCGGACGCCAGTACAGCAATAAC | TCCGTTCAACCACGCTCATAAGTCGCGAGAGC |
| Fluorescence probe ∗ | TCGACCGATCCATCGCTGAAGCTAGTCGA(FAM)TT(THF)(BHQ)TGACATAAAGTCACG | TGCAACATGTGCACGCTGTTATTCACTACGT(FAM)TA(THF)(BHQ)TTRGCTAGTTTACTAAC |
| Results | Method | ||
|---|---|---|---|
| RAA | Kato-Katz | Larvae Culture | |
| Negative | 129 | 133 | 158 |
| Positive | 77 | 73 | 48 |
| Kappa value | - | 0.168 | 9.658 |
| (vs. RAA) | (>0.05) | (<0.01) | |
| EPG | Method | ||
|---|---|---|---|
| RAA | Kato-Katz | Larvae Culture | |
| 1–100 | 21 | 18 | 7 |
| 100–500 | 18 | 17 | 11 |
| 500–1000 | 15 | 15 | 11 |
| 1000–2000 | 11 | 11 | 8 |
| 2000–4000 | 7 | 7 | 6 |
| >4000 | 5 | 5 | 5 |
| Total | 77 | 73 | 48 |
| Hookworm Species | Method | |
|---|---|---|
| RAA | Larvae Culture | |
| AD | 5 | 2 |
| NA | 65 | 44 |
| Mix | 7 | 2 |
| Total | 77 | 48 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, X.; Yang, Y.; Zhang, Y.; Zhang, Q.; Mao, F.; Dai, Y. Establishment of a Simple and Rapid Nucleic Acid Detection Method for Hookworm Identification. Pathogens 2023, 12, 630. https://doi.org/10.3390/pathogens12040630
Ding X, Yang Y, Zhang Y, Zhang Q, Mao F, Dai Y. Establishment of a Simple and Rapid Nucleic Acid Detection Method for Hookworm Identification. Pathogens. 2023; 12(4):630. https://doi.org/10.3390/pathogens12040630
Chicago/Turabian StyleDing, Xin, Yougui Yang, Yingshu Zhang, Qiang Zhang, Fanzhen Mao, and Yang Dai. 2023. "Establishment of a Simple and Rapid Nucleic Acid Detection Method for Hookworm Identification" Pathogens 12, no. 4: 630. https://doi.org/10.3390/pathogens12040630
APA StyleDing, X., Yang, Y., Zhang, Y., Zhang, Q., Mao, F., & Dai, Y. (2023). Establishment of a Simple and Rapid Nucleic Acid Detection Method for Hookworm Identification. Pathogens, 12(4), 630. https://doi.org/10.3390/pathogens12040630








