The Fascinating Cross-Paths of Pathogenic Bacteria, Human and Animal Faecal Sources in Water-Stressed Communities of Vhembe District, South Africa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site Description, Water Supply, and Storage
2.2. Ethical Clearance
2.3. Sample Collection at Different Points
2.4. Sample Concentration of Bacteria and DNA Extraction
2.5. Multiplex Real-Time PCR for the Detection of Enteric Pathogens
2.6. The Occurrence of Sources of Faecal Contamination in Different Water Sources
2.7. Statistical Analysis
3. Results
3.1. Parameters of qPCR Assay in Detection and Quantification of Enteric Pathogens
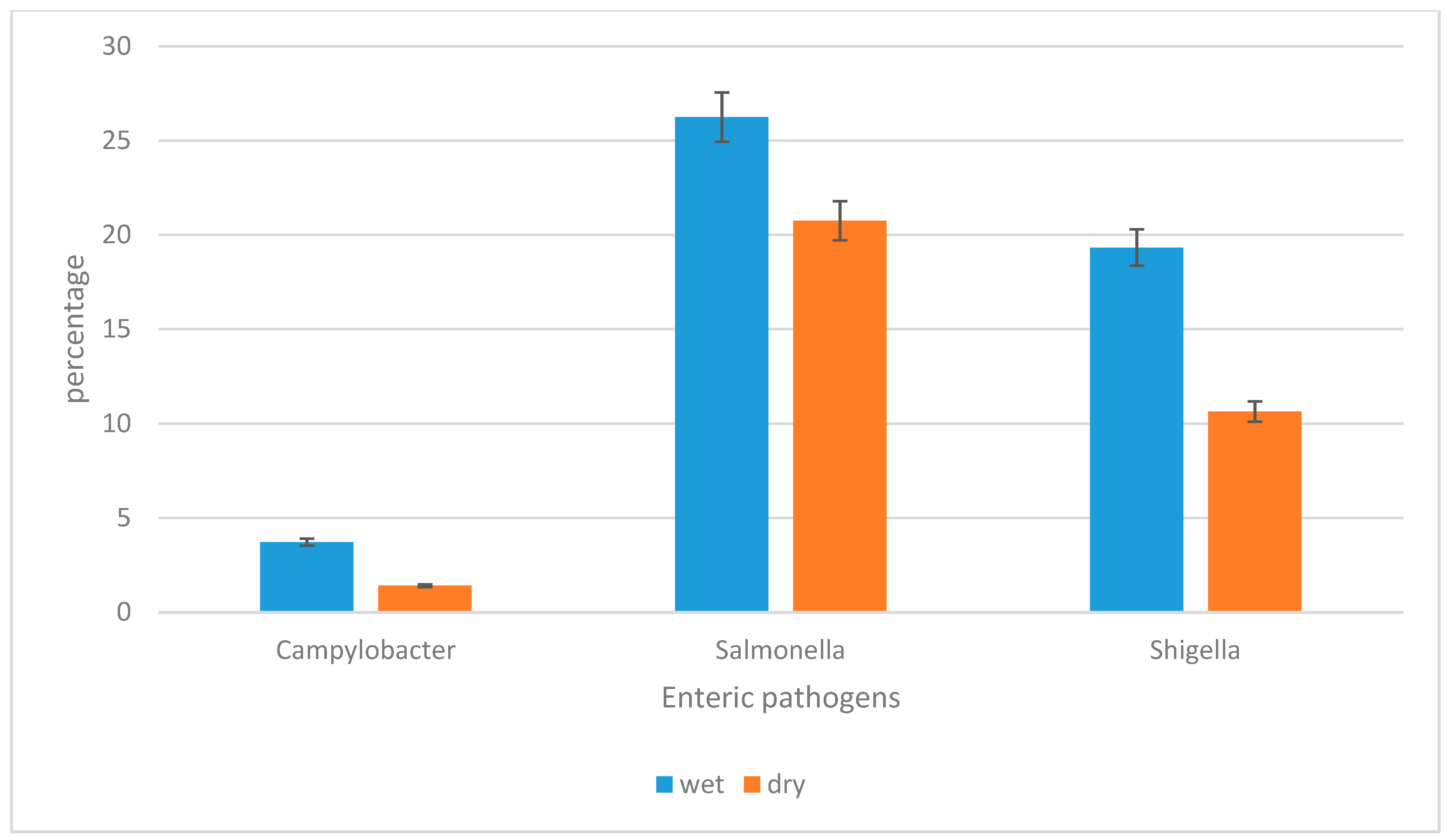
3.2. The Prevalence of Campylobacter jejuni subsp. jejuni, Shigella flexneri, and Salmonella enterica subsp. enterica Serovar Typhimurium str. LT2 in Water Sources per Season
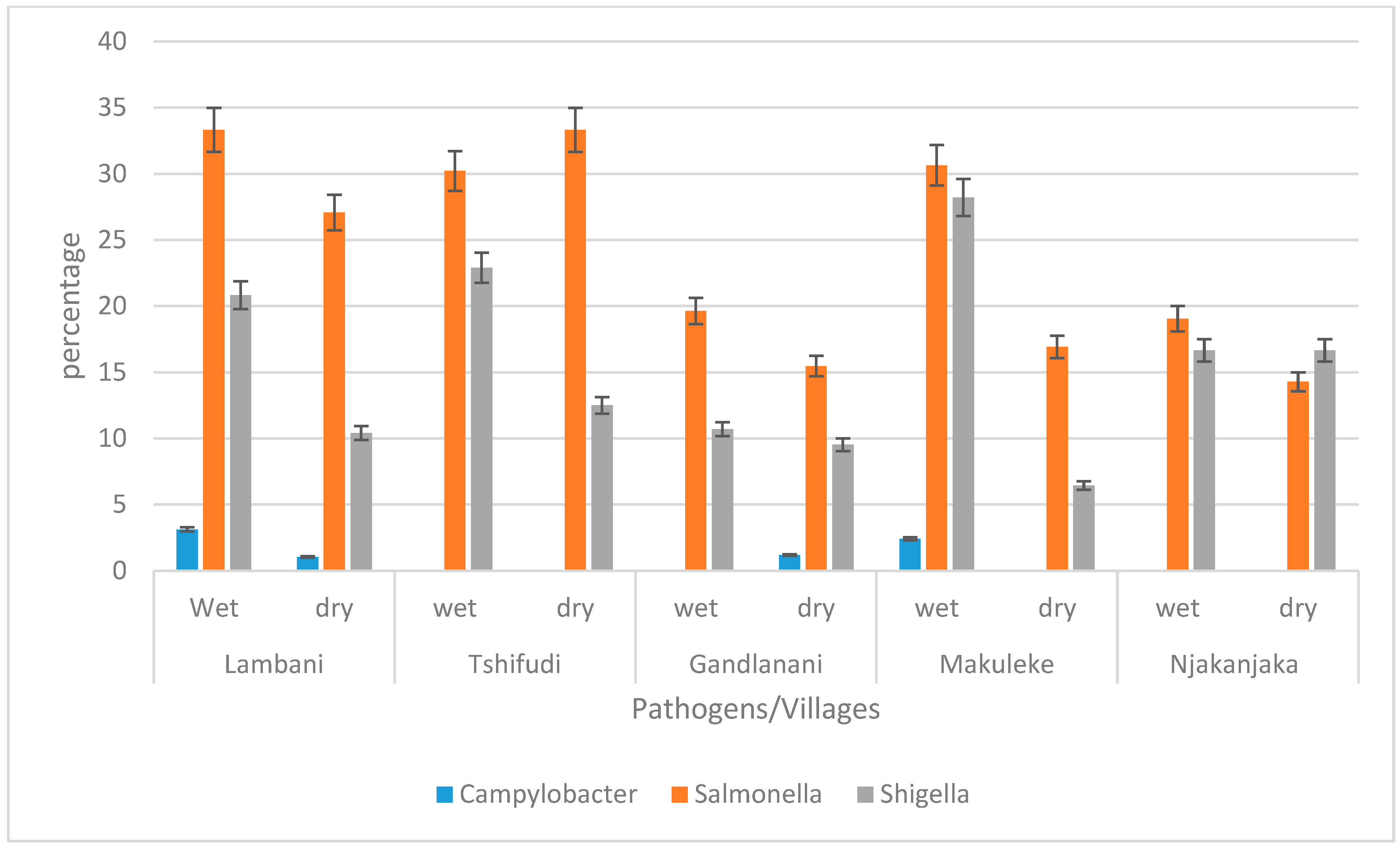
3.3. The Prevalence of Campylobacter jejuni subsp. jejuni, Shigella flexneri, and Salmonella enterica subsp. enterica Serovar Typhimurium str. LT2 in Water Sources per Village during Wet and Dry Seasons
3.4. The Occurrence of Campylobacter jejuni subsp. Jejuni, Shigella flexneri, and Salmonella enterica subsp. enterica Serovar Typhimurium str. LT2 in Different Water Sources during Wet and Dry Seasons
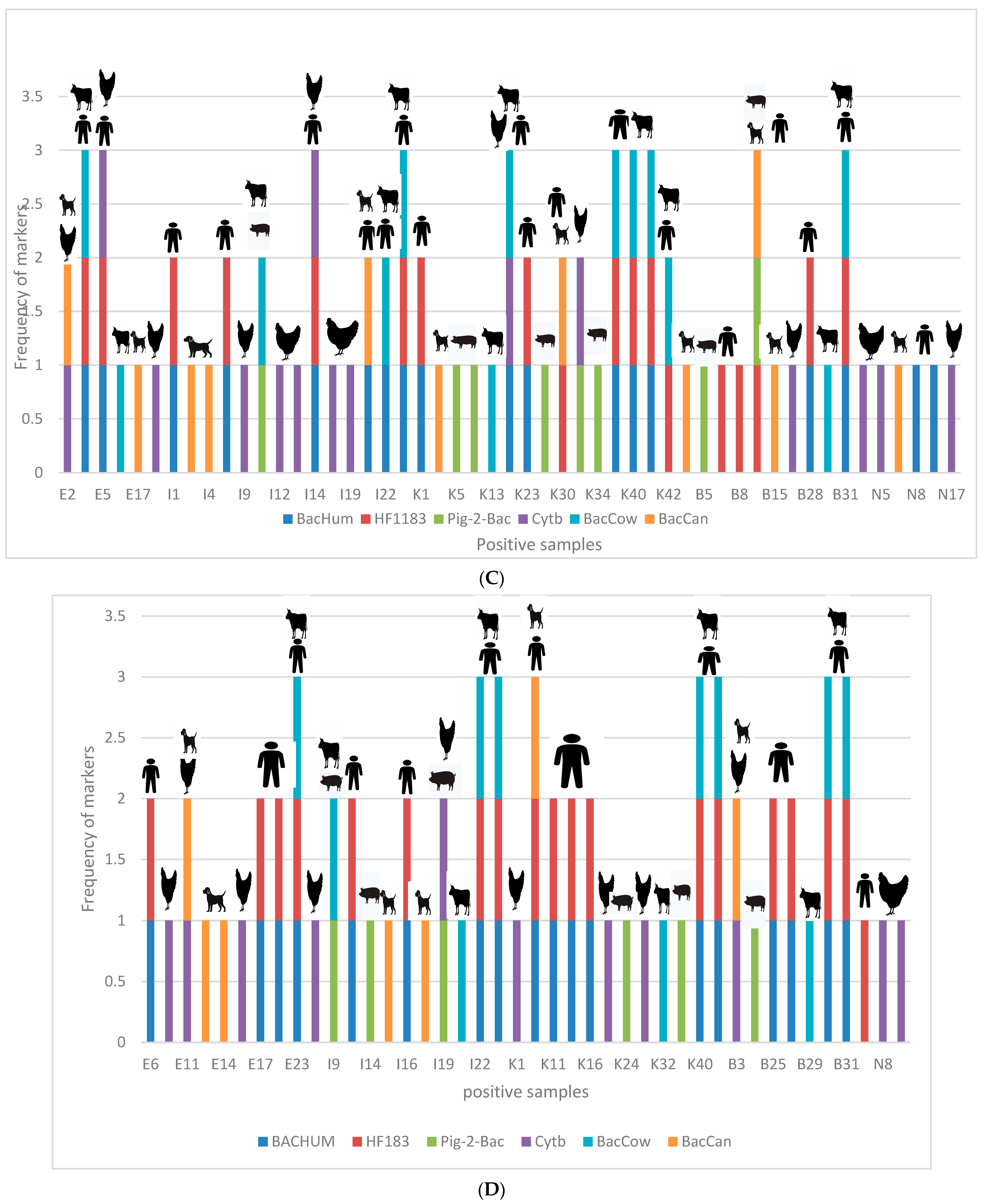
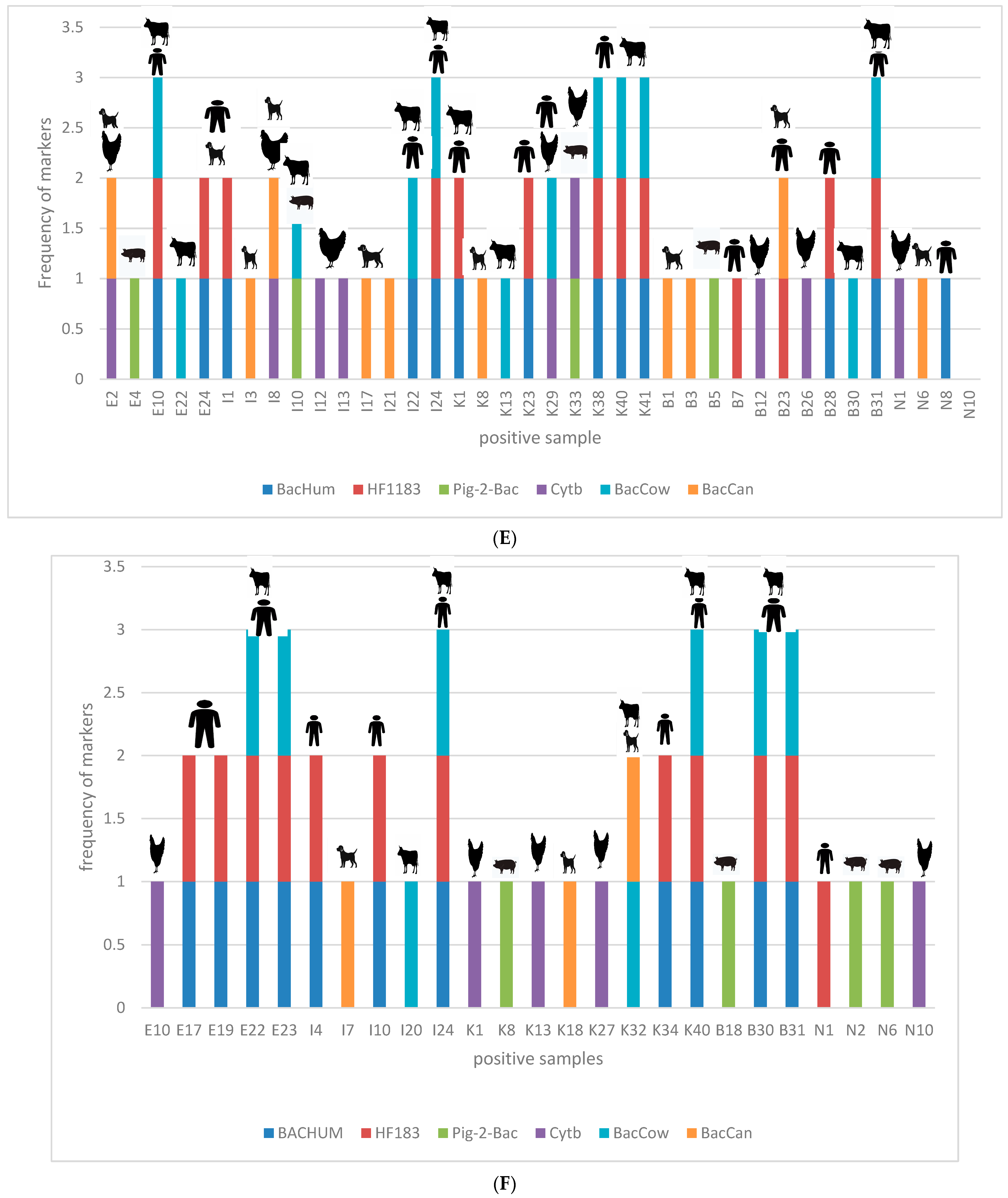
3.5. Correlation between Occurrence of Enteric Pathogens and Different Sources of Contamination Using Host-Specific Markers
4. Discussion
5. Conclusions
6. Study Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saylan, Y.; Erdem, Ö.; Cihangir, N.; Denizli, A. Detecting fingerprints of waterborne bacteria on a sensor. Chemosensors 2019, 7, 33. [Google Scholar] [CrossRef]
- Mosisa, D.; Aboma, M.; Girma, T.; Shibru, A. Determinants of diarrheal diseases among under five children in Jimma Geneti District, Oromia region, Ethiopia, 2020: A case-control study. BMC Pediatr. 2021, 21, 532. [Google Scholar] [CrossRef]
- World Health Organization. Water, Sanitation, Hygiene and Health: A Primer for Health Professionals; (No. WHO/CED/PHE/WSH/19.149); World Health Organization: Geneva, Switzerland, 2019; Available online: https://apps.who.int/iris/bitstream/handle/10665/330100/WHO-CED-PHE-WSH-19.149-eng.pdf (accessed on 12 December 2019).
- Ramírez-Castillo, F.Y.; Loera-Muro, A.; Jacques, M.; Garneau, P.; Avelar-González, F.J.; Harel, J.; Guerrero-Barrera, A.L. Waterborne pathogens: Detection methods and challenges. Pathogens 2015, 4, 307–334. [Google Scholar] [CrossRef] [PubMed]
- Mahagamage, M.G.Y.L.; Pathirage, M.V.S.C.; Manage, P.M. Contamination status of Salmonella spp., Shigella spp.; Campylobacter spp. in surface and groundwater of the Kelani River Basin, Sri Lanka. Water 2020, 12, 2187. [Google Scholar] [CrossRef]
- Edokpayi, J.N.; Rogawski, E.T.; Kahler, D.M.; Hill, C.L.; Reynolds, C.; Nyathi, E.; Smith, J.A.; Odiyo, J.O.; Samie, A.; Bessong, P.; et al. Challenges to sustainable safe drinking water: A case study of water quality and use across seasons in rural communities in Limpopo Province, South Africa. Water 2018, 10, 159. [Google Scholar] [CrossRef] [PubMed]
- Obi, C.L.; Potgieter, N.; Bessong, P.O.; Matsaung, G. Assessment of the microbial quality of river water sources in rural Venda communities in South Africa. Water 2002, 28, 287–292. [Google Scholar] [CrossRef]
- Chouhan, S. Recovery of Salmonella and Shigella isolates from drinking water. Eur. J. Exp. Biol. 2015, 5, 49–61. [Google Scholar]
- Bardhan, P.; Faruque, A.S.G.; Naheed, A.; Sack, D.A. Decreasing shigellosis-related deaths without Shigella spp.–specific interventions, Asia. Emerg. Infect. Dis. 2010, 16, 1718. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Campylobacter; Fact sheet; World Health Organization: Geneva, Switzerland, 2020; Available online: https://www.who.int/news-room/fact-sheets/detail/campylobacter (accessed on 1 May 2020).
- Fennell, J.; Geris, J.; Wilkinson, M.E.; Daalmans, R.; Soulsby, C. Lessons from the 2018 drought for management of local water supplies in upland areas: A tracer-based assessment. Hydrol. Process. 2020, 34, 4190–4210. [Google Scholar] [CrossRef]
- Stats, S.A. The State of Basic Service Delivery in South Africa: In-Depth Analysis of the Community Survey 2016 Data, Report 03-01-22; Statistics South Africa: Pretoria, South Africa, 2016.
- Mudau, M. Application of Microbial Source Tracking Markers for the Identification of Predominant Faecal Pollution Sources in Water-Stressed Rural, Communities of South Africa. Master’s Dissertation, Tshwane University of Technology, Pretoria, South Africa, 2023. [Google Scholar]
- Silva, N.; Taniwaki, M.; Junqueira, V.; Silveira, N.; Nascimento, M.; Gomes, R. Plate Count Method APHA 2001 for Total Coliforms in Foods: Microbiological Examination Methods of Food and Water a Laboratory Manual; Institute of Food Technology: Campinas, Brazil, 2016. [Google Scholar]
- Haramoto, E.; Osada, R. Assessment and application of host-specific Bacteroidales genetic markers for microbial source tracking of river water in Japan. PLoS ONE 2018, 13, e0207727. [Google Scholar] [CrossRef]
- Wiemer, D.; Loderstaedt, U.; Von Wulffen, H.; Priesnitz, S.; Fischer, M.; Tannich, E.; Hagen, R.M. Real-time multiplex PCR for simultaneous detection of Campylobacter jejuni, Salmonella, Shigella and Yersinia species in faecal samples. Int. J. Med. Microbiol. 2011, 301, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Ashbolt, N.J. Microbial contamination of drinking water and human health from community water systems. Curr. Environ. Health Rep. 2015, 2, 95–106. [Google Scholar] [CrossRef]
- Prüss, A.; Kay, D.; Fewtrell, L.; Bartram, J. Estimating the burden of disease from water, sanitation, and hygiene at a global level. Environ. Health Perspect. 2002, 110, 537–542. [Google Scholar] [PubMed]
- Troeger, C.; Forouzanfar, M.; Rao, P.C.; Khalil, I.; Brown, A.; Reiner, R.C.; Fullman, N.; Thompson, R.L.; Abajobir, A.; Ahmed, M.; et al. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: A systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect. Dis. 2017, 17, 909–948. [Google Scholar] [CrossRef]
- Mulatu, G.; Beyene, G.; Zeynudin, A. Prevalence of Shigella, Salmonella and Cmpylobacter species and their susceptibility patters among under five children with diarrhea in Hawassa Town, South Ethiopia. Ethiop. J. Health Sci. 2014, 24, 101. [Google Scholar] [PubMed]
- Hagedorn, C.; Benham, B.L.; Zeckoski, S.C. Microbial Source Tracking and The TMDL (Total Maximum Daily Loads) Process. Virginia Cooperative Extension. 2009. Available online: http://hdl.handle.net/10919/47483 (accessed on 1 May 2009).
- Nybo, K. qPCR efficiency calculations. Biotechniques 2011, 51, 401–402. [Google Scholar] [CrossRef] [PubMed]
- Potgieter, N.; Karambwe, S.; Mudau, L.S.; Barnard, T.; Traore, A. Human enteric pathogens in eight rivers used as rural household drinking water sources in the northern region of South Africa. Int. J. Environ. Res. Public Health 2020, 17, 2079. [Google Scholar] [CrossRef]
- Bessong, P.O.; Odiyo, J.O.; Musekene, J.N.; Tessema, A. Spatial distribution of diarrhoea and microbial quality of domestic water during an outbreak of diarrhoea in the Tshikuwi community in Venda, South Africa. J. Health Popul. Nutr. 2009, 27, 652. [Google Scholar] [CrossRef]
- Samie, A.; Mashao, M.B.; Bessong, P.O.; NKgau, T.F.; Momba, M.N.B.; Obi, C.L. Diversity and antibiograms of bacterial organisms isolated from samples of household drinking-water consumed by HIV-positive individuals in rural settings, South Africa. J. Health Popul. Nutr. 2012, 30, 241. [Google Scholar] [CrossRef]
- Lipp, E.K.; Kurz, R.; Vincent, R.; Rodriguez-Palacios, C.; Farrah, S.R.; Rose, J.B. The effects of seasonal variability and weather on microbial faecal pollution and enteric pathogens in a subtropical estuary. Estuaries 2001, 24, 266–276. [Google Scholar] [CrossRef]
- Sadik, N.J.; Uprety, S.; Nalweyiso, A.; Kiggundu, N.; Banadda, N.E.; Shisler, J.L.; Nguyen, T.H. Quantification of multiple waterborne pathogens in drinking water, drainage channels, and surface water in Kampala, Uganda, during seasonal variation. GeoHealth 2017, 1, 258–269. [Google Scholar] [CrossRef]
- Shah, M.; Kathiiko, C.; Wada, A.; Odoyo, E.; Bundi, M.; Miringu, G.; Guyo, S.; Karama, M.; Ichinose, Y. Prevalence, seasonal variation, and antibiotic resistance pattern of enteric bacterial pathogens among hospitalized diarrheic children in suburban regions of central Kenya. Trop. Med. Health 2016, 44, 39. [Google Scholar] [CrossRef] [PubMed]
- Egberongbe, H.O.; Bankole, M.O.; Popoola, T.O.S.; Olowofeso, O. Seasonal variation of enteric bacteria population in surface water sources among rural communities of Ijebu North, Ogun State, Nigeria. Agro-Science 2021, 20, 81–85. [Google Scholar] [CrossRef]
- Momba, M.N.B.; Binda, M.A. Combining chlorination and chloramination processes for the inhibition of biofilm formation in drinking surface water system models. J. Appl. Microbiol. 2002, 92, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Momba, M.N.; Notshe, T.L. The microbiological quality of groundwater-derived drinking water after long storage in household containers in a rural community of South Africa. J. Water Supply: Res. Technol.—AQUA 2003, 52, 67–77. [Google Scholar] [CrossRef]
- Khabo-Mmekoa, C.M.; Genthe, B.; Momba, M.N.B. Enteric pathogens risk factors associated with household drinking water: A case study in Ugu District Kwa-Zulu Natal Province, South Africa. Int. J. Environ. Res. Public Health 2022, 19, 4431. [Google Scholar] [CrossRef]
- Muringani, B.N.; Obi, C.L.; Apalata, T.; Vasaikar, S.D. Prevalence of potential enteric pathogens in treated and untreated water sources around Eastern Cape region. J. Bacteriol. Mycol. 2015, 1, 14–19. [Google Scholar] [CrossRef]
- Ghasemzadeh, I.; Namazi, S.H. Review of bacterial and viral zoonotic infections transmitted by dogs. J. Med. Life 2015, 8, 1. [Google Scholar] [PubMed]
- Boyle, S.F.; Corrigan, V.K.; Buechner-Maxwell, V.; Pierce, B.J. Evaluation of risk of zoonotic pathogen transmission in a university-based animal assisted intervention (AAI) program. Front. Vet. Sci. 2019, 6, 167. [Google Scholar] [CrossRef]
- Santaniello, A.; Varriale, L.; Dipineto, L.; Borrelli, L.; Pace, A.; Fioretti, A.; Menna, L.F. Presence of Campylobacter jejuni and C. coli in dogs under training for animal-assisted therapies. Int. J. Environ. Res. Public Health 2021, 18, 3717. [Google Scholar] [CrossRef]
- Lemos, M.L.; Nunes, A.; Ancora, M.; Cammà, C.; Costa, P.M.D.; Oleastro, M. Campylobacter jejuni in different canine populations: Characteristics and zoonotic potential. Microorganisms 2021, 9, 2231. [Google Scholar] [CrossRef]
- Boehm, A.B.; Soller, J.A.; Shanks, O.C. Human-associated faecal quantitative polymerase chain reaction measurements and simulated risk of gastrointestinal illness in recreational waters contaminated with raw sewage. Environ. Sci. Technol. Lett. 2015, 2, 270–275. [Google Scholar] [CrossRef]
- Viau, E.J.; Lee, D.; Boehm, A.B. Swimmer risk of gastrointestinal illness from exposure to tropical coastal waters impacted by terrestrial dry-weather runoff. Environ. Sci. Technol. 2011, 45, 7158–7165. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.J.; Gabriel, E.; Leatherbarrow, A.J.; Cheesbrough, J.; Gee, S.; Bolton, E.; Fox, A.; Fearnhead, P.; Hart, C.A.; Diggle, P.J. Tracing the source of campylobacteriosis. PLoS Genet. 2008, 4, e1000203. [Google Scholar] [CrossRef] [PubMed]
- Savichtcheva, O.; Okayama, N.; Okabe, S. Relationships between Bacteroides 16S rRNA genetic markers and presence of bacterial enteric pathogens and conventional faecal indicators. Water Res. 2007, 41, 3615–3628. [Google Scholar] [CrossRef] [PubMed]
- Mughini-Gras, L.; Enserink, R.; Friesema, I.; Heck, M.; van Duynhoven, Y.; van Pelt, W. Risk factors for human salmonellosis originating from pigs, cattle, broiler chickens and egg laying hens: A combined case-control and source attribution analysis. PLoS ONE 2014, 9, e87933. [Google Scholar] [CrossRef] [PubMed]
- Heredia, N.; García, S. Animals as sources of food-borne pathogens: A review. Anim. Nutr. 2018, 4, 250–255. [Google Scholar] [CrossRef]
- Bataller, E.; García-Romero, E.; Llobat, L.; Lizana, V.; Jiménez-Trigos, E. Dogs as a source of Salmonella spp. in apparently healthy dogs in the Valencia Region. Could it be related with intestinal lactic acid bacteria? BMC Vet. Res. 2020, 16, 268. [Google Scholar] [CrossRef]
- Askari, N.; Rafiee, M.; Amini, K. A case control study of Salmonella spp. infection in stray dogs in Tehran shelters and the correlation between paraclinical tests results and clinical findings. Arch. Razi Inst. 2020, 75, 93. [Google Scholar]
- Shi, R.; Yang, X.; Chen, L.; Chang, H.T.; Liu, H.Y.; Zhao, J.; Wang, X.W.; Wang, C.Q. Pathogenicity of Shigella in chickens. PLoS ONE 2014, 9, e100264. [Google Scholar] [CrossRef]



| Sources of Water | Villages | |||||
|---|---|---|---|---|---|---|
| Lambani | Tshifudi | Gandlanani | Makuleke | Njhakanjhaka | Total | |
| WPPB | 0 | 0 | 0 | 0 | 112 | 112 |
| River | 16 | 16 | 16 | 16 | 0 | 64 |
| Treatment plant | 16 | 16 | 16 | 16 | 0 | 64 |
| Household storage | 24 | 8 | 152 | 72 | 48 | 304 |
| Household/communal tap | 136 | 160 | 144 | 144 | 0 | 584 |
| Total | 192 | 200 | 328 | 248 | 160 | 1128 |
| Pathogen | Target Gene | Primer/Probe | Sequence 5–3 | Conditions | Reference |
|---|---|---|---|---|---|
| Campylobacter jejuni subsp. jejuni | gyrA | 366F | CTA TAA CAA CTG CAC CTA CTA AT |  | Wiemer et al. (2011) [16] |
| 614R | ATG AAA TTT TTG CCA GTG GTG | ||||
| 409P | Fam-CTT AAT AGC CGT CAC CCC AC-Tam. | ||||
| Shigella flexneri | ipaH | 1635F | CAG AAG AGC AGA AGT ATG AG | ||
| 1804R | CAG TAC CTC GTC AGT CAG | ||||
| 1747P | ROX-ACA GGT GAT GCG TGA GAC TG-BHQ2 | ||||
| Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | ttrC | 4136F | AAT TAG CCA TGT TGT AAT CTC | ||
| 4315R | ATT GTT GAT TCA GGT ACA AAC | ||||
| 4163P | JOE-CAA GTT CAA CGC GCA ATT TA-BHQ1a |
| Sources of Contamination | Water Source Categories | |||||||
|---|---|---|---|---|---|---|---|---|
| Wet Season | ||||||||
| Household | River | Treatment Plant | Total | |||||
| Frequency | Percent | Frequency | Percent | Frequency | Percent | Frequency | Percent | |
| BacHum | 20 | 10.86 | 3 | 18.75 | 5 | 35.71 | 28 | 13.08 |
| HF183 | 23 | 12.5 | 7 | 43.75 | 5 | 35.71 | 35 | 16.35 |
| Cytb | 44 | 23.91 | 0 | 0 | 0 | 0 | 44 | 20.56 |
| BacCow | 26 | 14.13 | 5 | 31.25 | 4 | 28.57 | 35 | 16.35 |
| BacCan | 41 | 22.28 | 0 | 0 | 0 | 0 | 41 | 19.15 |
| Pig-2-Bac | 30 | 16.3 | 1 | 6.25 | 0 | 0 | 31 | 14.48 |
| Total | 184 | 100 | 16 | 100 | 14 | 100 | 214 | 100 |
| Dry Season | ||||||||
| BacHum | 34 | 23.94 | 3 | 21.43 | 3 | 18.75 | 40 | 23.26 |
| HF183 | 31 | 21.83 | 3 | 21.43 | 2 | 12.5 | 26 | 20.93 |
| Cytb | 23 | 16.2 | 1 | 7.14 | 2 | 12.5 | 26 | 15.12 |
| BacCow | 13 | 9.15 | 5 | 35.71 | 5 | 31.25 | 23 | 13.37 |
| BacCan | 21 | 14.79 | 2 | 14.29 | 2 | 12.5 | 25 | 14.53 |
| Pig-2-Bac | 20 | 14.08 | 0 | 0 | 2 | 12.5 | 22 | 12.79 |
| Total | 142 | 100 | 14 | 100 | 16 | 100 | 172 | 100 |
| Standard Curve Parameters for Enteric Pathogens | ||||||
|---|---|---|---|---|---|---|
| Target | Slope | y-Intercept | Efficiency | LLOQ (Ct) | Gene Copies per μL | Log10 Gene Copies per ng |
| Campylobacter jejuni subsp. jejuni | −3.882 | 39.212 | 81 | 27.5 | 4.00 × 1037 | 37.60 |
| Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | −3.493 | 38.882 | 93 | 28.39 | 3.32 × 1039 | 39.52 |
| Shigella flexneri | −3.799 | 39.604 | 83 | 28.58 | 1.01 × 1039 | 39.00 |
| Enteric Pathogens | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sources of Water | Campylobacter jejuni subsp. jejuni | Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | Shigella flexneri | p-Value | Pearson Correlation (r) | ||||
| River n = 28 per season | WET | 12 | 42.86 | 17 | 60.71 | 20 | 71.43 | 0.05 | 0.75 |
| DRY | 4 | 14.29 | 13 | 46.43 | 10 | 35.71 | |||
| Treatment plant (RW) n = 16 per season | WET | 7 | 43.75 | 14 | 87.5 | 7 | 43.75 | 0.01 | 1 |
| DRY | 4 | 25 | 10 | 62.5 | 4 | 25 | |||
| Treatment plant (FW) n = 16 per season | WET | 0 | 0 | 0 | 0 | 2 | 12.5 | 0.42 | 0 |
| DRY | 0 | 0.00 | 0 | 0 | 0 | 0.00 | |||
| Household (drinking water) n = 508 per season | WET | 2 | 0.39 | 117 | 23.03 | 82 | 16.14 | 0.17 | 0.97 |
| DRY | 0 | 0 | 94 | 18.50 | 46 | 9.06 | |||
| Enteric Pathogens (Correlation Coefficient (r)) | ||||||
|---|---|---|---|---|---|---|
| Sources of Contamination | WET SEASON | |||||
| Campylobacter jejuni subsp. jejuni | Salmonella enterica subsp. enterica Serovar Typhimurium str. LT2 | Shigella flexneri | ||||
| r | p | r | p | r | p | |
| BacHum | −0.19 | 0.05 | 0.6 | 0.01 | 0.05 | 0.01 |
| HF183 | −0.25 | 0.08 | 0.55 | 0.01 | 0.16 | 0.01 |
| Cytb | −0.08 | 0.01 | 0.72 | 0.01 | 0.24 | 0.02 |
| Pig-2-Bac | −0.01 | 0.01 | 0.62 | 0.01 | 0.04 | 0.01 |
| BacCow | −0.29 | 0.2 | 0.36 | 0.01 | −0.2 | 0.03 |
| BacCan | −0.02 | 0.02 | 0.74 | 0.01 | 0.2 | 0.02 |
| DRY SEASON | ||||||
| Campylobacter jejuni subsp. jejuni | Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | Shigella flexneri | ||||
| r | p | r | p | r | p | |
| BacHum | 0.34 | 0.03 | 0.92 | 0 | 0.08 | 0.3 |
| HF183 | 0.46 | 0.02 | 0.92 | 0 | 0.25 | 0.13 |
| Cytb | 0.65 | 0.02 | 0.45 | 0.01 | 0.54 | 0 |
| Pig-2-Bac | 0.25 | 0.02 | 0.78 | 0.01 | 0.25 | 0.01 |
| BacCow | −0.1 | 0.09 | 0.78 | 0.01 | −0.05 | 0.1 |
| BacCan | 0.84 | 0.02 | 0.66 | 0.01 | 0.5 | 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mudau, M.; Ngobeni-Nyambi, R.; Momba, M.N.B. The Fascinating Cross-Paths of Pathogenic Bacteria, Human and Animal Faecal Sources in Water-Stressed Communities of Vhembe District, South Africa. Pathogens 2023, 12, 1085. https://doi.org/10.3390/pathogens12091085
Mudau M, Ngobeni-Nyambi R, Momba MNB. The Fascinating Cross-Paths of Pathogenic Bacteria, Human and Animal Faecal Sources in Water-Stressed Communities of Vhembe District, South Africa. Pathogens. 2023; 12(9):1085. https://doi.org/10.3390/pathogens12091085
Chicago/Turabian StyleMudau, Mulalo, Renay Ngobeni-Nyambi, and Maggy Ndombo Benteke Momba. 2023. "The Fascinating Cross-Paths of Pathogenic Bacteria, Human and Animal Faecal Sources in Water-Stressed Communities of Vhembe District, South Africa" Pathogens 12, no. 9: 1085. https://doi.org/10.3390/pathogens12091085






