The Relevance of the Predominant Clonal Evolution (PCE) Model for the Molecular Epidemiology and Subspecific Taxonomy of Trypanosoma cruzi
Abstract
:1. Introduction
2. Why Is Clonality vs. Sexuality Relevant for Both Molecular Epidemiology and Taxonomy?
3. Clones and Clonality: Multiple Meanings
4. Misconceptions Regarding the PCE Model Being “Challenged”
4.1. Total Absence of Genetic Recombination/Sexuality
4.2. Genetic Recombination Has No or Little Evolutionary/Epidemiological Significance
4.3. Use of Inadequate Genetic Tools
4.4. Improper Sampling
4.5. Inappropriate Evolutionary Scale
5. Main Features of the PCE Model and the Case of Trypanosoma cruzi
5.1. Mandatory Analysis of Multiple Genetic Loci
5.2. Widespread Propagation of Unchanged MLGs Across Space and Time
5.3. Linkage Disequilibrium (LD)
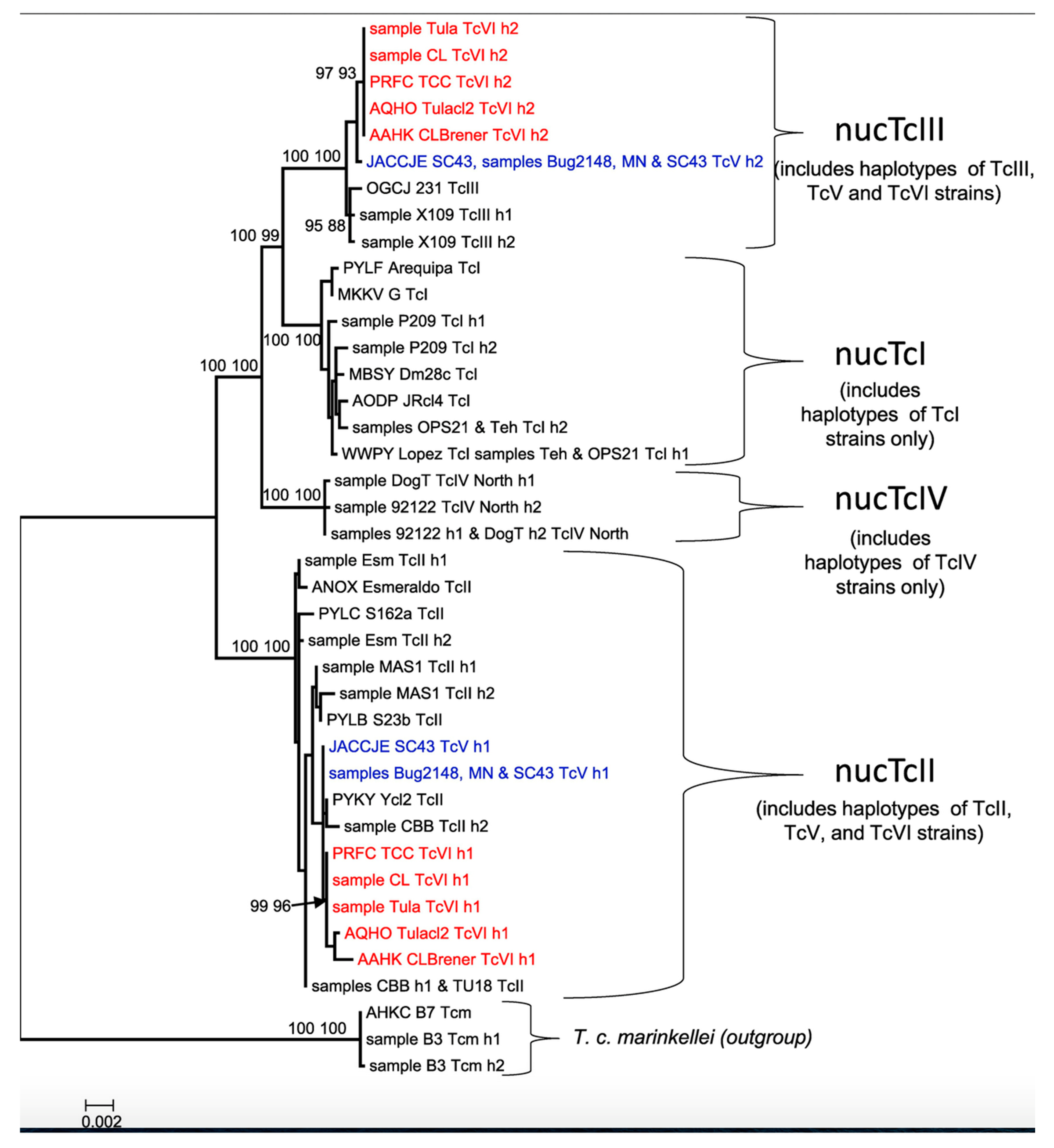
5.4. Multigene Bifurcating Trees (MGBTs)
6. Additional Key Concepts Related to the PCE Model
6.1. Clonets
6.2. Discrete Typing Units (DTUs)
6.3. Near-Clades
6.4. Russian Doll Evolution; Russian Doll Patterns (RDPs)
6.5. Is Apparent Clonality in T. cruzi Attributable to a Lack of Mating Opportunity? The “Starving Sex” Hypothesis
7. Conclusions and Future Direction
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Glossary of Specialized Terms
| Amplification fragment length polymorphism (AFLP) | Selective amplification of genomic restriction fragments (obtained by RFLP) by PCR*. |
| Barcoding | The DNA barcode. Some DNA fragments are highly conserved within the same species and variable between species. These are genetic markers or barcodes. cf. metabarcoding. |
| Clade | An evolutionary lineage defined by cladistic analysis. A clade is monophyletic (it has a single common ancestor) and genetically isolated (it evolves independently) from other clades, with no genetic exchange. See cladistic analysis. |
| Cladistic analysis | A method of phylogenetic analysis that relies on the polarization of characters, distinguishing between ancestral (plesiomorphic) and derived (apomorphic) traits. Only apomorphic characters shared by all members of a given clade (synapomorphic characters) are considered phylogenetically informative. |
| Concatenated phylogenetic tree | Method of constructing a phylogenetic tree by concatenating different gene sequences that have been aligned into a supergene matrix. |
| Discrete typing unit (DTU) | A group of genetic stocks that are more closely related to each other than to any other stock, remain stable over time and space, and can be identified by specific genetic, molecular, or immunological markers called tags. DTUs serve as reliable analytical units in studies considering pathogen genetic diversity and are ideal targets for molecular epidemiology surveys. |
| Genetic recombination | The exchange of genetic material between different individuals at two or more genetic loci, resulting in offspring with genetic combinations distinct from those of either parent. |
| Genomics | While genetics focuses on the study of individual genes, genomics examines the entire genome as a whole. |
| Homogamy | The tendency of an organism to mate with individuals that are genetically very similar or identical to itself. |
| Inbreeding | cf. homogamy. |
| Isoenzymes | Different electrophoretic variants of a given enzyme reflecting the genetic variability of the gene encoding that enzyme within the surveyed population. Variations in migration patterns among isoenzymes of the same enzyme arise from differences in their overall electric charge, which is determined by the specific electric charges of the amino acids composing the enzyme. Consequently, electrophoretic differences correspond to variations in the amino acid sequence and, ultimately, differences in the upstream gene sequence. |
| Linkage disequilibrium | The nonrandom association of genotypes at different genetic loci. In a population with no linkage disequilibrium (i.e., random genetic recombination), knowing an individual’s genotype at one locus does not provide information about their genotype at another locus. For example, in a randomly mating human population, knowing an individual’s ABO blood group does not indicate their Rhesus blood group. Conversely, if such an association is observed, it suggests linkage disequilibrium, indicating limited genetic recombination, which can be quantified using various statistical methods. |
| Locus | The physical location of a gene on a chromosome. By extension, in genetic terminology, the term locus may also refer to the gene itself (plural: loci). |
| Metabarcoding | Use of Next Generation Sequencing. Metabarcoding is an extension of barcoding through the use of NGS technology, which allows blind and one-time identification of all species present in a sample. cf. barcoding. |
| Microsatellite | A short DNA sequence, typically 1–4 base pairs long, repeated in tandem along the DNA molecule. In many species, including pathogens, the number of repeats varies significantly between individuals and across populations and pathogen strains. The number of repeats at a specific locus defines microsatellite alleles. Microsatellites are present at hundreds of locations in most species. These markers evolve rapidly and offer high-resolution analysis. |
| Mitosis | Equational cell division that gives rise to genetically identical cells. |
| Multilocus | Referring to a trait involving multiple loci. |
| Multilocus enzyme electrophoresis (MLEE) | Isoenzyme* analysis involving a wide range of enzyme systems, each corresponding to one or more genetic loci. Multilocus enzyme electrophoresis (MLEE) has been extensively applied in population genetics across a diverse array of living organisms, including pathogens. |
| Multilocus genotype | A combined genotype determined by multiple genetic loci. |
| Multilocus sequence typing (MLST) | A method for characterizing pathogens based on sequencing several housekeeping genes. |
| Panmixia: panmictic, panmictic expectations | A genetic structure in which genetic exchange occurs randomly within a given population. Panmictic expectations are the confirmation of this state through various population genetics tests. |
| Parthenogenesis | A mode of reproduction observed in certain metazoans (e.g., insects, amphibians, fish, reptiles) that occurs without the genetic contribution of a mating partner. |
| Phylogenetics | A branch of genetics dedicated to reconstructing the evolutionary history and relationships of taxa or distinct evolutionary lineages. |
| Polymerase chain reaction (PCR) | A technique that amplifies the complementary strands of a target DNA sequence through a series of cycles until the desired amount of DNA is produced. PCR employs synthesized primers whose nucleotide sequences are complementary to the DNA flanking the target region. The DNA is heated to denature and separate the complementary strands and then cooled to allow the primers to bind to the flanking sequences. Taq DNA polymerase is added, and the reaction undergoes the necessary number of replication cycles to achieve amplification. |
| Population genetics | The study of genetic variation across space and time within and among populations. This field emphasizes the population or species as a whole rather than individual organisms (see also population genomics). |
| Population genomics | The study of genomic variation across space and time within and among populations. |
| Pulse field gel electrophoresis (PFGE) | The separation of large DNA fragments achieved through a specialized electrophoresis technique that uses alternately pulsed, perpendicularly oriented electrical fields. Strains sharing the same pulsed-field gel electrophoresis (PFGE) pattern are referred to as pulse types. In bacteria, the large DNA fragments are produced by the action of a low-frequency restriction enzyme (a bacterial endonuclease that cuts at a low frequency) on the bacterial chromosome. As such, PFGE is a specific form of restriction fragment length polymorphism (RFLP). In the case of parasitic protozoa (such as Trypanosoma and Leishmania) and yeasts, the large DNA fragments represent entire chromosomes, reflecting the organism’s molecular karyotype. |
| Random primer amplified polymorphic DNA (RAPD) | In the classical polymerase chain reaction (PCR) method, the primers used are known DNA sequences, whereas the RAPD (random amplified polymorphic DNA) technique relies on primers with arbitrarily determined sequences. RAPD primers are typically 10 base pairs long, and the possible combinations are virtually unlimited. For a given genotype of an individual or strain, different primers will reveal different polymorphisms. RAPDs are an extremely powerful method for exploring the genetic variability of organisms. However, their use in routine strain identification is limited due to their lack of reproducibility. |
| Recombination (genetic) | cf. Genetic recombination. |
| Restriction fragment length polymorphism (RFLP) | DNA variability in a given organism can be detected using bacterial restriction endonucleases. These enzymes cut DNA at specific restriction sites defined by particular DNA sequences. The resulting polymorphism in DNA fragments can be visualized on gels, either directly through ethidium bromide staining or via Southern blot hybridization using specific probes. |
| Selfing: self-fertilization | Fertilization of an organism by itself, hence by a genotype that is identical to itself. |
| Sexuality | In this context, sexuality is used broadly to encompass all forms of genetic exchange between two distinct cells or individuals. |
| Single nucleotide polymorphism | Polymorphisms resulting from single nucleotide variations in the DNA sequence, known as single nucleotide polymorphisms (SNPs), contributing to differences among individuals, populations, and pathogen strains. SNPs are commonly used as high-resolution population markers. |
| Wahlund effect | In population genetics, the Wahlund effect traditionally refers to a heterozygote deficiency arising when two genetically distinct populations with different allele frequencies are mistakenly considered a single population, despite being separated by physical barriers (e.g., time or space). Here, we extend the term to refer to any apparent deviation from panmixia that results solely from physical obstacles (e.g., time or space) impeding genetic exchange. |
References
- de Sousa, A.S.; Vermeij, D.; Novaes Ramos, A., Jr.; Luquetti, A.O. Chagas Disease. Lancet 2024, 403, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Roman, F.; Iñiguez, A.M.; Yeo, M.; Jansen, A.M. Multilocus sequence typing: Genetic diversity in Trypanosoma cruzi I (TcI) isolates from Brazilian didelphids. Parasit. Vectors 2018, 11, 107. [Google Scholar] [CrossRef]
- Barnabé, C.; Sempéré, G.; Manzanilla, V.; Millan, J.M.; Amblard-Rambert, A.; Waleckx, E. mbctools: A User-Friendly Metabarcoding and Cross-Platform Pipeline for Analyzing Multiple Amplicon Sequencing Data across a Large Diversity of Organisms. Peer Community J. 2024, 4, e114. [Google Scholar] [CrossRef]
- Tavares de Oliveira, M.; Sulleiro, E.; da Silva, M.C.; Silgado, A.; de Lana, M.; da Silva, J.S.; Molina, I.; Marin-Neto, A. Intra-Discrete Typing Unit TcV Genetic Variability of Trypanosoma cruzi in Chronic Chagas’ Disease Bolivian Immigrant Patients in Barcelona, Spain. Front. Cardiovasc. Med. 2021, 8, 665624. [Google Scholar]
- Schwabl, P.; Sánchez, J.M.; Costales, J.A.; Sofıá Ocaña-Mayorga, S.; Segovia, M.; Carrasco, H.J.; Hernández, C.; Ramírez, J.D.; Lewis, M.D.; Grijalva, M.J.; et al. Culture-free genome-wide locus sequence typing (GLST) provides new perspectives on Trypanosoma cruzi dispersal and infection complexity. PLoS Genet. 2020, 16, e1009170. [Google Scholar] [CrossRef] [PubMed]
- Tibayrenc, M.; Cariou, M.L.; Solignac, M.; Carlier, Y. Arguments génétiques contre l’existence d’une sexualité actuelle chez Trypanosoma cruzi; Implications taxinomiques. C. R. Acad. Sci. 1981, 293, 207–209. [Google Scholar]
- Tibayrenc, M.; Ward, P.; Moya, A.; Ayala, F.J. Natural populations of Trypanosoma cruzi, the agent of Chagas’disease, have a complex multiclonal structure. Proc. Nat. Acad. Sci. USA 1986, 83, 115–119. [Google Scholar] [CrossRef]
- Tibayrenc, M.; Kjellberg, F.; Ayala, F.J. A clonal theory of parasitic protozoa: The population structure of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas and Trypanosoma, and its medical and taxonomical consequences. Proc. Nat. Acad. Sci. USA 1990, 87, 2414–2418. [Google Scholar] [CrossRef]
- Tibayrenc, M.; Ayala, F.J. Relevant units of analysis for applied and basic research dealing with neglected transmissible diseases: The predominant clonal evolution model of pathogenic microorganisms. PLoS Neglect. Trop. Dis. 2017, 11, e0005293. [Google Scholar] [CrossRef]
- Ochman, H.; Selander, R.K. Evidence for clonal population structure in Escherichia coli. Proc. Nat. Acad. Sci. USA 1984, 81, 198–201. [Google Scholar] [CrossRef]
- Ørskov, F.; Ørskov, I. Summary of a workshop on the clone concept in the epidemiology, taxonomy, and evolution of the Enterobacteriaceae and other Bacteria. J. Infect. Dis. 1983, 148, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Rougeron, V.; De Meeûs, T.; Hide, M.; Waleckx, E.; Bermudez, H.; Arevalo, J.; Llanos-Cuentas, A.; Dujardin, J.C.; De Doncker, S.; Le Ray, D.; et al. Extreme inbreeding in Leishmania braziliensis. Proc. Nat. Acad. Sci. USA 2009, 106, 10224–10229. [Google Scholar] [CrossRef] [PubMed]
- Rougeron, V.; De Meeûs, T.; Hide, M.; Le Falher, G.; Bucheton, B.; Dereure, J.; El-Safi, S.H.; Dessein, A.; Bañuls, A. Multifaceted Population Structure and Reproductive Strategy in Leishmania donovani Complex in One Sudanese Village. PLoS Neglect. Trop. Dis. 2011, 5, e1448. [Google Scholar] [CrossRef] [PubMed]
- Sturm, N.R.; Campbell, D.A. Alternative lifestyles: The population structure of Trypanosoma cruzi. Acta Trop. 2010, 115, 35–43. [Google Scholar] [CrossRef]
- Avise, J.C. Evolutionary perspectives on clonal reproduction in vertebrate animals. Proc. Nat. Acad. Sci. USA 2015, 29, 8867–8873. [Google Scholar] [CrossRef]
- Tibayrenc, M. Molecular Epidemiology of Pathogenic Microorganisms and the Predominant Clonal Evolution (PCE) model. In Genetics and Evolution of Infectious Diseases, 3rd ed.; Tibayrenc, M., Ed.; Elsevier: Amsterdam, The Netherlands; Academic Press: Cambridge, MA, USA, 2024. [Google Scholar]
- Calo, S.; Billmyre, B.B.; Heitman, J. Generators of Phenotypic Diversity in the Evolution of Pathogenic Microorganisms. PLoS Pathog. 2013, 9, e1003181. [Google Scholar] [CrossRef]
- Feretzaki, F.; Heitman, J. Unisexual Reproduction Drives Evolution of Eukaryotic Microbial Pathogens. PLoS Pathog. 2013, 9, e1003674. [Google Scholar] [CrossRef]
- Gibson, W.; Lewis, M.D.; Yeo, M.; Miles, M. Genetic Exchange in Trypanosomatids and its Relevance to Epidemiology. In Genetics and Evolution of Infectious Diseases; Tibayrenc, M., Ed.; Elsevier: Amsterdam, The Netherlands, 2024. [Google Scholar]
- Gutiérrez-Corbo, C.; Domínguez-Asenjo, B.; Martínez-Valladares, M.; Pérez-Pertejo, Y.; García-Estrada, C.; Balaña-Fouce, R.; Reguera, R.M. Reproduction in Trypanosomatids. Past Present. Biol. 2021, 10, 471. [Google Scholar]
- Miles, M.A.; Llewellyn, M.S.; Lewis, M.D.; Yeo, M.; Baleela, R.; Fitzpatrick, S.; Gaunt, M.W.; Mauricio, I.L. The molecular epidemiology and phylogeography of Trypanosoma cruzi and parallel research on Leishmania: Looking back and to the future. J. Parasitol. 2009, 136, 1509–1526. [Google Scholar] [CrossRef]
- Rougeron, V.; De Meeûs, T.; Kako Ouraga, S.; Hide, M.; Bañuls, A. ‘‘Everything You Always Wanted to Know about Sex (but Were Afraid to Ask)’’ in Leishmania after Two Decades of Laboratory and Field Analyses. PLoS Pathog. 2010, 6, e1001004. [Google Scholar] [CrossRef]
- Tomasini, N. Introgression of the Kinetoplast DNA: An Unusual Evolutionary Journey in Trypanosoma cruzi. Curr. Genom. 2018, 19, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Gaunt, M.W.; Yeo, M.; Frame, I.A.; Tothard, J.R.; Carrasco, H.J.; Taylor, M.C.; Mena, S.S.; Veazey, P.; Miles, G.A.; Acosta, N.; et al. Mechanism of genetic exchange in American trypanosomes. Nature 2003, 421, 936–939. [Google Scholar] [CrossRef] [PubMed]
- Lewis, M.D.; Llewellyn, M.S.; Yeo, M.; Messenger, L.A.; Miles, M.A. Experimental and natural recombination in Trypanosoma cruzi. In American Trypanosomiasis: Chagas Disease. One Hundred Years of Research, 2nd ed.; Telleria, J., Tibayrenc, M., Eds.; Elsevier: Amsterdam, The Netherlands; Academic Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Messenger, L.A.; Miles, M. Evidence and importance of genetic exchange among field populations of Trypanosoma cruzi. Acta Trop. 2015, 151, 150–155. [Google Scholar] [CrossRef]
- Ramírez, J.D.; Llewellyn, M.S. Reproductive clonality in protozoan pathogens—Truth or artefact? Mol. Ecol. 2014, 23, 4195–4202. [Google Scholar] [CrossRef] [PubMed]
- Weedall, G.D.; Hall, N. Sexual reproduction and genetic exchange in parasitic protists. J. Parasitol. 2015, 142, S120–S127. [Google Scholar] [CrossRef]
- Zingales, B.; Macedo, A.M. Fifteen Years after the Definition of Trypanosoma cruzi DTUs: What have we learned? Life 2023, 13, 2339. [Google Scholar] [CrossRef]
- Gelanew, T.; Kuhls, K.; Hurissa, Z.; Weldegebreal, T.; Hailu, W.; Kassahun, A.; Abebe, T.; Hailu, A.; Schönian, G. Inference of Population Structure of Leishmania donovani Strains Isolated from Different Ethiopian Visceral Leishmaniasis Endemic Areas. PLoS Negl. Trop. Dis. 2010, 4, e889. [Google Scholar] [CrossRef]
- Mauricio, I.L.; Yeo, M.; Baghaei, M.; Doto, D.; Pratlong, F.; Zemanova, E.; Dedet, J.P.; Lukes, J.; Miles, M.A. Towards multilocus sequence typing of the Leishmania donovani complex: Resolving genotypes and haplotypes for five polymorphic metabolic enzymes (ASAT, GPI, NH1, NH2, PGD). Int. J. Parasitol. 2006, 36, 757–769. [Google Scholar] [CrossRef]
- Ramírez, J.D.; Llewellyn, M.S. Response to Tibayrenc and Ayala: Reproductive clonality in protozoan pathogens—Truth or artefact? Mol. Ecol. 2015, 24, 5782–5784. [Google Scholar] [CrossRef]
- Shaik, J.S.; Dobson, D.E.; Sacks, D.L.; Beverley, S.M. Leishmania Sexual Reproductive Strategies as Resolved through Computational Methods Designed for Aneuploid Genomes. Genes 2021, 12, 167. [Google Scholar] [CrossRef]
- Tibayrenc, M.; Ayala, F.J. Reproductive clonality of pathogens: A perspective on pathogenic viruses, bacteria, fungi, and parasitic protozoa. Proc. Nat. Acad. Sci. USA 2012, 109, E3305–E3313. [Google Scholar] [CrossRef] [PubMed]
- Morrison, L.J.; Tweedie, A.; Black, A.; Pinchbeck, G.L.; Christley, R.M.; Schoenefeld, A.; Hertz-Fowler, C.; MacLeod, A.; Turner, C.M.R.; Tait, A. Discovery of Mating in the Major African Livestock Pathogen Trypanos. Congolense. PLoS ONE 2009, 4, e5564. [Google Scholar] [CrossRef]
- Dumonteil, E.; Desale, H.; Tu, W.; Hernandez-Cuevas, N.; Shroyer, M.; Goff, K.; Marx, P.A.; Herrera, C. Intra-host Trypanosoma cruzi strain dynamics shape disease progression: The missing link in Chagas disease pathogenesis. Microbiol. Spectr. 2023, 11, e04236-22. [Google Scholar] [CrossRef]
- Cura, C.I.; Mejía-Jaramillo, A.M.; Duffy, T.; Burgos, J.M.; Rodriguero, M.; Cardinal, M.V.; Kjos, S.; Gurgel-Gonçalves, R.; Blanchet, D.; De Pablos, L.M.; et al. Trypanosoma cruzi I genotypes in different geographical regions and transmission cycles based on a microsatellite motif of the intergenic spacer of spliced-leader genes. Int. J. Parasitol. 2010, 40, 1599–1607. [Google Scholar] [CrossRef] [PubMed]
- Miles, M.A.; Souza, A.; Povoa, M.; Shaw, J.J.; Lainson, R.; Toyé, P.J. Isozymic heterogeneity of Trypanosoma cruzi in the first autochtonous patients with Chagas’disease in Amazonian Brazil. Nature 1978, 272, 819–821. [Google Scholar] [CrossRef]
- Tibayrenc, M.; Ayala, F. Isozyme variability of Trypanosoma cruzi, the agent of Chagas’ disease: Genetical, taxonomical and epidemiological significance. Evolution 1988, 42, 277–292. [Google Scholar] [CrossRef]
- Barnabé, C.; Brisse, S.; Tibayrenc, M. Population structure and genetic typing of Trypanosoma cruzi, the agent of Chagas’disease: A multilocus enzyme electrophoresis approach. Parasitology 2000, 150, 513–526. [Google Scholar] [CrossRef] [PubMed]
- Messenger, L.A.; Garcia, L.; Vanhove, M.; Huaranca, C.; Bustamante, M.; Torrico, M.; Torrico, F.; Miles, M.A.; Llewellyn, M.S. Ecological host fitting of Trypanosoma cruzi TcI in Bolivia: Mosaic population structure, hybridization and a role for humans in Andean parasite dispersal. Mol. Ecol. 2015, 245, 2406–2422. [Google Scholar] [CrossRef]
- Ramírez, J.D.; Hernández, C. Trypanosoma cruzi I: Towards the need of genetic subdivision? Part II. Acta Tropica 2018, 184, 53–58. [Google Scholar] [CrossRef]
- Rivas-Garcıá, L.; Carballo-Amador, M.A.; Flores-López, C.A. Design of a AFLP-PCR and PCR-RFLP test that identify the majority of discrete typing units of Trypanosoma cruzi. PLoS ONE 2020, 15, e0237180. [Google Scholar] [CrossRef]
- Cosentino, R.O.; Agüero, F. A Simple Strain Typing Assay for Trypanosoma cruzi: Discrimination of Major Evolutionary Lineages from a Single Amplification Product. PLoS Neglect. Trop. Dis. 2012, 6, e1777. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, P.B.; Lewis, M.D.; Cruickshank, C.; Gaunt, M.W.; Yeo, M.; Llewellyn, M.S.; Valente, S.A.; Maia da Silva, F.; Stevens, J.R.; Miles, M.A.; et al. Identification and lineage genotyping of South American trypanosomes using fluorescent fragment length barcoding. Infect. Genet. Evol. 2011, 11, 44–51. [Google Scholar] [CrossRef]
- Tibayrenc, M.; Neubauer, K.; Barnabé, C.; Guerrini, F.; Sarkeski, D.; Ayala, F.J. Genetic characterization of six parasitic protozoa: Parity of random-primer DNA typing and multilocus isoenzyme electrophoresis. Proc. Natl. Acad. Sci. USA 1993, 90, 1335–1339. [Google Scholar] [CrossRef] [PubMed]
- Brisse, S.; Barnabé, C.; Tibayrenc, M. Identification of six Trypanosoma cruzi phylogenetic lineages by random amplified polymorphic DNA and multilocus enzyme electrophoresis. Int. J. Parasitol. 2000, 30, 35–44. [Google Scholar] [CrossRef]
- Carrasco, H.J.; Segovia, M.; Llewellyn, M.S.; Morocoima, A.; Urdaneta-Morales, S.; Martínez, C.; Martínez, C.E.; Garcia, C.; Rodríguez, M.; Espinosa, R.; et al. Geographical Distribution of Trypanosoma cruzi Genotypes in Venezuela. PLoS Neglect. Trop. Dis. 2012, 6, e1707. [Google Scholar] [CrossRef]
- Rozas, M.; De Doncker, S.; Adaui, V.; Coronado, X.; Barnabé, C.; Tibayrenc, M.; Solari, A.; Dujardin, J.C. Multilocus Polymerase Chain Reaction Restriction Fragment–Length Polymorphism Genotyping of Trypanosoma cruzi (Chagas Disease): Taxonomic and Clinical Applications. J. Infect. Dis. 2007, 195, 1381–1388. [Google Scholar] [CrossRef] [PubMed]
- Hanage, W.P. Not So Simple After All: Bacteria, Their Population Genetics, and Recombination. Cold Spring Harb. Perspect. Biol. 2016, 8, a018069. [Google Scholar] [CrossRef]
- Maiden, M.C.J. Multilocus Sequence Typing of Bacteria. Ann. Rev. Microbiol. 2006, 60, 561–588. [Google Scholar] [CrossRef]
- Pearson, T.; Giffard, P.; Beckstrom-Sternberg, S.; Auerbach, R.; Hornstra, H.; Tuanyok, A.; Erin, P.; Price, E.P.; Glass, M.B.; Leadem, B.; et al. Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol. 2009, 7, 78. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Browne, E.B.; Madsen, A.; Wirth, T.; Viscidi, R.P.; Crandall, K.A. Population genetics of microbial pathogens estimated from multilocus sequence typing (MLST) data. Infect. Genet. Evol. 2006, 6, 97–112. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A.; Holmes, E.C. Recombination in evolutionary genomics. Ann. Rev. Genet. 2002, 36, 75–97. [Google Scholar] [CrossRef] [PubMed]
- Tibayrenc, M.; Kjellberg, F.; Ayala, F.J. The clonal theory of parasitic protozoa: A taxonomic proposal applicable to other clonal organisms. Bioscience 1991, 41, 767–774. [Google Scholar] [CrossRef]
- Pratlong, F.; Lami, P.; Ravel, C.; Balard, Y.; Dereure, J.; Serres, G.; El Baidouri, F.; Dedet, J.P. Geographical distribution and epidemiological features of Old World Leishmania infantum and Leishmania donovani foci, based on the isoenzyme analysis of 2277 strains. J. Parasitol. 2013, 40, 423–434. [Google Scholar] [CrossRef]
- Kuhls, K.; Chicharro, C.; Cañavate, C.; Cortes, S.; Campino, L.; Haralambous, C.; Soteriadou, K.; Pratlong, F.; Dedet, J.P.; Mauricio, I.; et al. Differentiation and Gene Flow among European Populations of Leishmania infantum MON-1. PLoS Negl. Trop. Dis. 2008, 2, e261. [Google Scholar] [CrossRef]
- Tibayrenc, M. Genetic epidemiology of parasitic protozoa and other infectious agents: The need for an integrated approach. Int. J. Parasitol. 1998, 28, 85–104. [Google Scholar] [CrossRef] [PubMed]
- Zingales, B.; Miles, M.A.; Campbell, D.; Tibayrenc, M.; Macedo, A.M.; Teixeira, M.M.; Schijman, A.; Llewellyn, M.S.; Lages-Silva, E.; Machado, C.R.; et al. The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications. Infect. Genet. Evol. 2012, 12, 240–253. [Google Scholar] [CrossRef]
- Marcili, A.; Lima, L.; Cavazzsana, M.; Junqueira, A.C.V.; Veludo, H.H.; Maia da Silva, F.; Campaner, M.; Paiva, F.; Nunes, V.L.B.; Teixeira, M.M.G. A new genotype of Trypanosoma cruzi associated with bats evidenced by phylogenetic analyses using SSU rDNA, cytochrome b and histone H2B genes and genotyping. J. Parasitol. 2009, 136, 641–655. [Google Scholar] [CrossRef]
- Brisse, S.; Henriksson, J.; Barnabé, C.; Douzery, E.J.P.; Berkvens, D.; Serrano, M.; De Carvalho, M.R.C.; Buck, G.A.; Dujardin, J.C.; Tibayrenc, M. Evidence for genetic exchange and hybridization in Trypanosoma cruzi based on nucleotide sequences and molecular karyotype. Infect. Genet. Evol. 2003, 2, 173–183. [Google Scholar] [CrossRef]
- Sturm, N.R.; Vargas, N.S.; Westenberger, S.J.; Zingales, B.; Campbell, D.A. Evidence for multiple hybrid groups in Trypanosoma cruzi. Int. J. Parasitol. 2003, 33, 269–279. [Google Scholar] [CrossRef]
- Tibayrenc, M.; Ayala, F.J. How clonal are Trypanosoma and Leishmania? Trends Parasitol. 2013, 29, 264–269. [Google Scholar] [CrossRef]
- Costales, J.A.; Jara-Palacio, M.A.; Llewellyn, M.S.; Messenger, L.A.; Ocaña-Mayorgaa, S.; Villacísa, A.G.; Tibayrenc, M.; Grijalva, M.J. Trypanosoma cruzi population dynamics in the Central Ecuadorian Coast. Acta Tropica 2015, 151, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Llewellyn, M.S.; Miles, M.A.; Carrasco, H.J.; Lewis, M.D.; Yeo, M.; Vargas, J.; Torrico, F.; Diosque, P.; Valente, V.; Valente, S.A.; et al. Genome-Scale Multilocus Microsatellite Typing of Trypanosoma cruzi Discrete Typing Unit I Reveals Phylogeographic Structure and Specific Genotypes Linked to Human Infection. PLoS Pathog. 2009, 5, e1000410. [Google Scholar] [CrossRef] [PubMed]
- Llewellyn, M.S.; Lewis, M.D.; Acosta, N.; Yeo, M.; Carrasco, H.J.; Segovia, M.; Vargas, J.; Torrico, F.; Miles, M.A.; Gaunt, M. Trypanosoma cruzi IIc: Phylogenetic and phylogeographic insights from sequence and microsatellite analysis and potential impact on emergent Chagas disease. PLoS Negl. Trop. Dis. 2009, 3, e510. [Google Scholar] [CrossRef] [PubMed]
- Barnabé, C.; Brenière, S.F.; Santillán-Guayasamín, S.; Douzery, E.J.P.; Waleckx, E. Revisiting gene typing and phylogeny of Trypanosoma cruzi reference strains: Comparison of the relevance of mitochondrial DNA, single-copy nuclear DNA, and the intergenic region of mini-exon gene. Infect. Genet. Evol. 2023, 115, 105504. [Google Scholar] [CrossRef]
- Majeau, A.; Murphy, L.; Herrera, C.; Dumonteil, E. Assessing Trypanosoma cruzi Parasite Diversity through Comparative Genomics: Implications for Disease Epidemiology and Diagnostics. Pathogens 2021, 10, 212. [Google Scholar] [CrossRef]
- Cruz-Saavedra, L.; Schwabl, P.; Vallejo, G.A.; Carranza, J.C.; Muñoz, M.; Patino, L.H.; Paniz-Mondolfi, A.; Llewellyn, M.S.; Ramírez, J.D. Genome plasticity driven by aneuploidy and loss of heterozygosity in Trypanosoma cruzi. Microb. Genom. 2022, 8, 843. [Google Scholar] [CrossRef]
- Flores-López, C.; Mitchell, E.A.; Reisenman, C.E.; Sarkar, S.; Williamson, P.C.; Machado, C.A. Phylogenetic diversity of two common Trypanosoma cruzi lineages in the Southwestern United States. Infect. Genet. Evol. 2022, 99, 105251. [Google Scholar] [CrossRef] [PubMed]
- Tibayrenc, M.; Ayala, F.J. New insights into clonality and panmixia in Plasmodium and Toxoplasma. Adv. Parasitol. 2014, 84, 253–268. [Google Scholar]
- Cibulskis, R.E. The microdistribution of Trypanosoma cruzi. Trans. R. Soc. Trop. Med. Hyg. 1985, 79, 138–139. [Google Scholar] [CrossRef]
- Brénière, S.F.; Tibayrenc, M.; Antezana, G.; Pabon, J.; Carrasco, R.; Selaès, H.; Desjeux, P. Résultats préliminaires en faveur d’une relation faible ou inexistante entre les formes cliniques de la maladie de Chagas et les souches isoenzymatiques de Trypanosoma cruzi. C. R. Acad. Sci. 1985, 300, 555–558. [Google Scholar]
- Schwabl, P.; Imamura, H.; Van den Broeck, F.; Costales, J.A.; Maiguashca-Sánchez, J.; Miles, M.A.; Andersson, B.; Grijalva, M.J.; Llewellyn, M.S. Meiotic sex in Chagas disease parasite Trypanosoma cruzi. Nat. Com. 2019, 10, 3972. [Google Scholar] [CrossRef] [PubMed]
- Rusman, F.; Floridia-Yapur, N.; Ragone, P.G.; Diosque, P.; Tomasini, N. Evidence of hybridization, mitochondrial introgression and biparental inheritance of the kDNA minicircles in Trypanosoma cruzi I. PLoS Negl. Trop. Dis. 2020, 14, e0007770. [Google Scholar] [CrossRef] [PubMed]
- Guhl, F.; Ramírez, J.D. Trypanosoma cruzi I diversity: Towards the need of genetic subdivision? Acta Tropica 2011, 119, 1–4. [Google Scholar] [CrossRef]
- Tibayrenc, M.; Ayala, F.J. Models in parasite and pathogen evolution: Genomic analysis reveals predominant clonality and progressive evolution at all evolutionary scales in parasitic protozoa, yeasts and bacteria. Adv. Parasitol. 2021, 111, 75–117. [Google Scholar] [PubMed]


| Viruses | Bacteria | Parasitic Protozoa | Fungi |
|---|---|---|---|
| clades | clades | assemblages | AFLP groups |
| clusters | clonal complexes | clades | clades |
| genogroups | clonal lineages | clonal haplogroups | clonal groups |
| genotypes | clonal subgroups | clonal haplotypes | clonal lineages |
| groups | clusters | clonal lineages | clusters |
| lineages | eBurst groups | clonal types | clonal groups |
| major genotypes | family strains | clones | genetically distinct subgroups |
| major lineages | genetic groups | clonotypes | genotypes |
| phylogenetic groups | genoclouds | clusters | genotypic groups |
| phylogroups | genogroups | core subgroups | groups |
| subclades | genome groups | discrete typing units (DTUs) | lineages |
| subgenotypes | genomospecies | divergent entities | major clades |
| subgenotype clusters | genospecies | genetic clades | minor clades |
| subgroups | groups | genetic groups | molecular genotypes |
| sublineages | haplotypes | genetic types | molecular types |
| substrains | lineages | genotypes | phylogenetic species |
| subtypes | major branches | groups | subclades |
| subvariants | major clusters | haplogroups | subclusters |
| types | main/major lineages | haplotypes | subgenotypes |
| variants | major phylogenetic groups | lesser subgroups | subgroups |
| phylogenetic clades | lineages | subpopulations | |
| phylogenetic groups | main haplogroups | varieties | |
| phylogenetic groupings | major clades | ||
| phylogroups | major clonal lineages | ||
| populations | major groups | ||
| primary clusters | major monophyletic groups | ||
| principal genetic groups | phylogenetic lineages | ||
| pulsotypes | populations | ||
| secondary clusters | subassemblages | ||
| semi discrete lineages | subclades | ||
| subclades | subclusters | ||
| subclones | subgroups | ||
| subclusters | sublineages | ||
| subgroups | subpopulations | ||
| sublineages | subgenotypes | ||
| subpopulations | subgroups | ||
| subspecies | subspecies | ||
| subspecies groups | subtypes | ||
| subtypes | subtype groups | ||
| types |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tibayrenc, M. The Relevance of the Predominant Clonal Evolution (PCE) Model for the Molecular Epidemiology and Subspecific Taxonomy of Trypanosoma cruzi. Pathogens 2025, 14, 407. https://doi.org/10.3390/pathogens14050407
Tibayrenc M. The Relevance of the Predominant Clonal Evolution (PCE) Model for the Molecular Epidemiology and Subspecific Taxonomy of Trypanosoma cruzi. Pathogens. 2025; 14(5):407. https://doi.org/10.3390/pathogens14050407
Chicago/Turabian StyleTibayrenc, Michel. 2025. "The Relevance of the Predominant Clonal Evolution (PCE) Model for the Molecular Epidemiology and Subspecific Taxonomy of Trypanosoma cruzi" Pathogens 14, no. 5: 407. https://doi.org/10.3390/pathogens14050407
APA StyleTibayrenc, M. (2025). The Relevance of the Predominant Clonal Evolution (PCE) Model for the Molecular Epidemiology and Subspecific Taxonomy of Trypanosoma cruzi. Pathogens, 14(5), 407. https://doi.org/10.3390/pathogens14050407







