Cell-Free Supernatants (CFSs) from the Culture of Bacillus subtilis Inhibit Pseudomonas sp. Biofilm Formation
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain Selection
2.2. In Vitro Antagonistic Test
2.3. Biofilm-Formation Activity
2.4. Inhibition of Biofilm Formation by CFSs
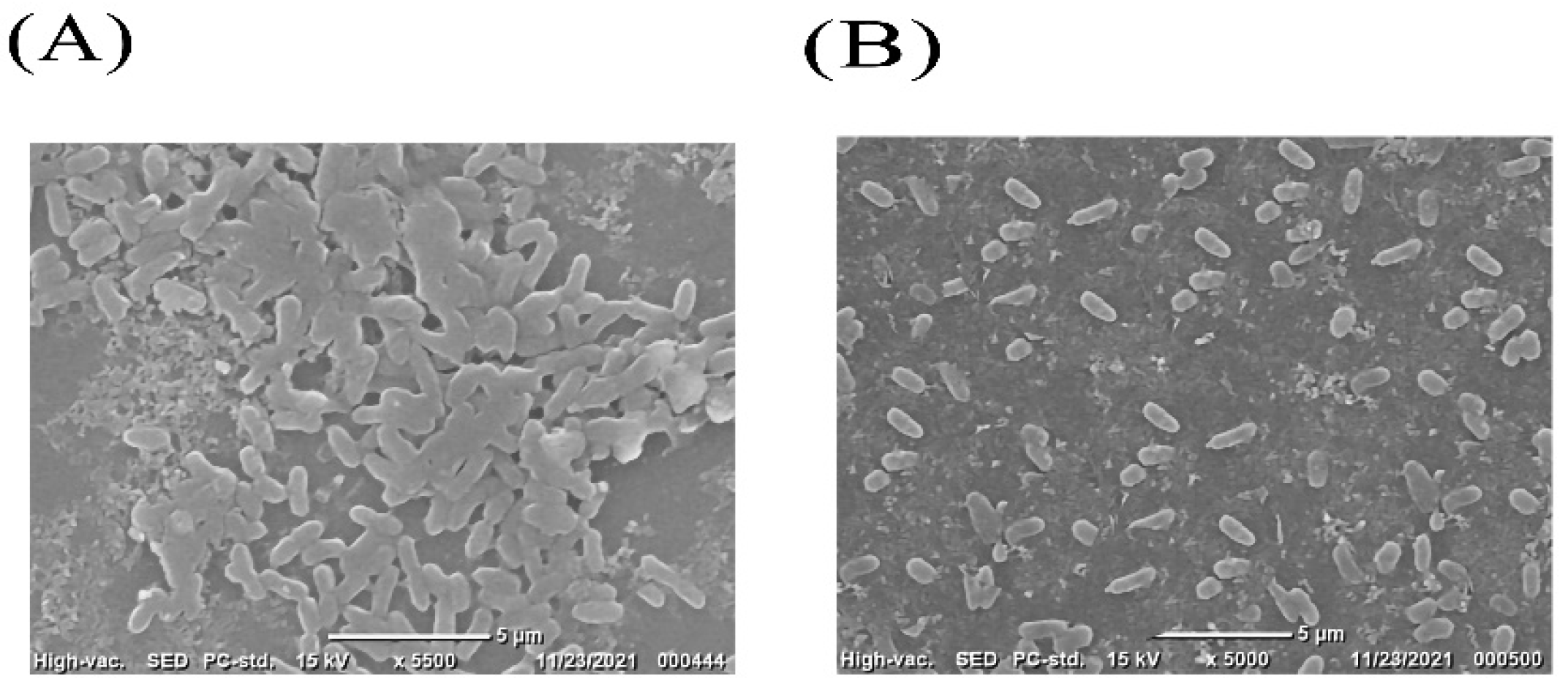
2.5. Scanning Electronic Microscope (SEM)
2.6. Volatile Compound Analysis through GC-MS
2.7. Molecular Identification of Anti-Biofilm Compounds
2.8. Pharmacokinetic Properties Analysis
2.9. Molecular Dynamics Simulation
2.10. Statistical Analysis
3. Results
3.1. In Vitro Antagonistic Test
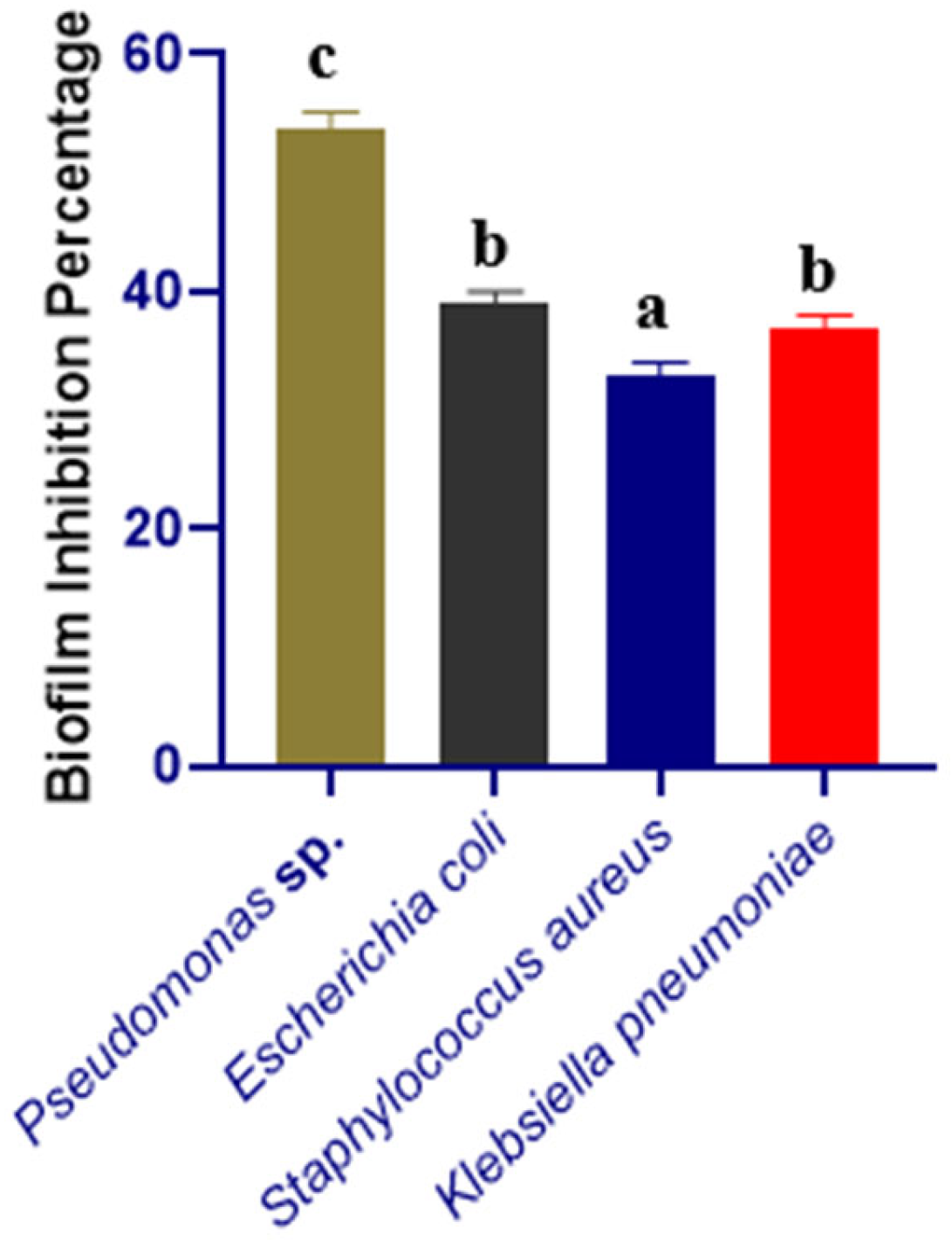
3.2. Biofilm-Inhibition Assay
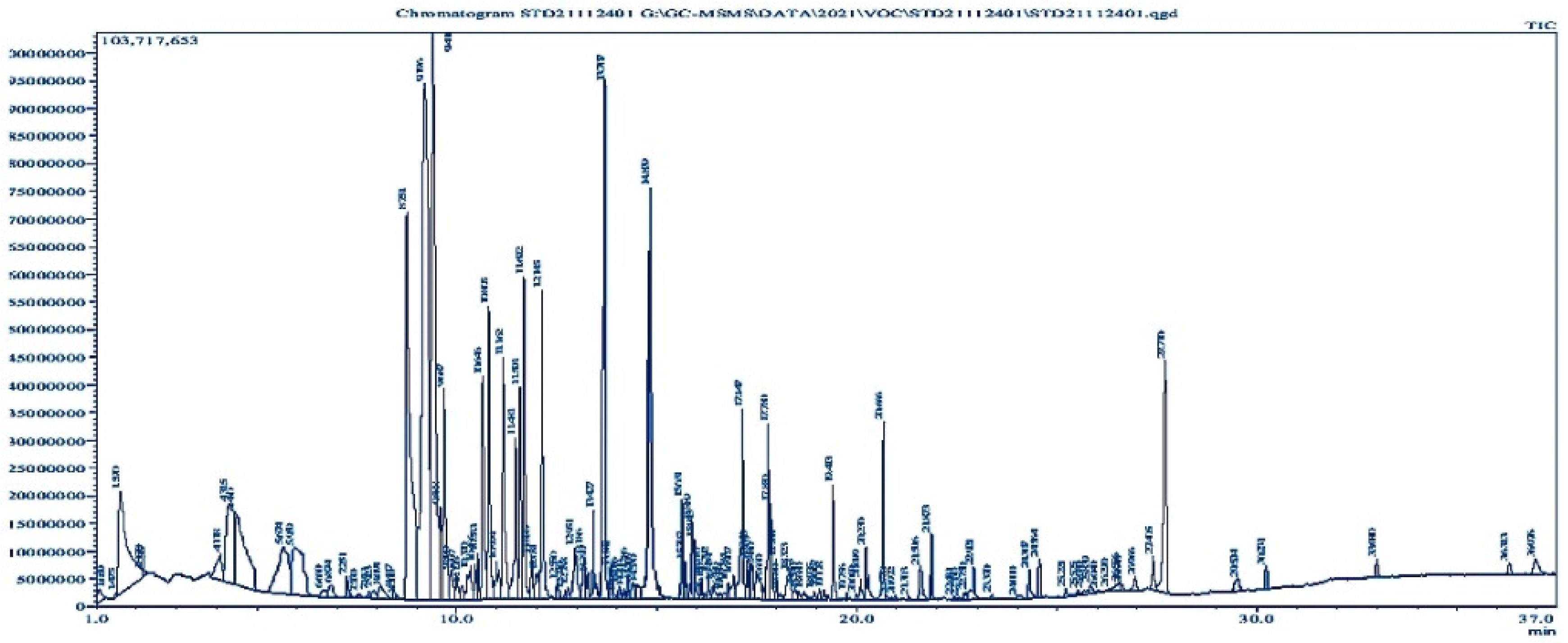
3.3. Volatile Compound Analysis by GC-MS
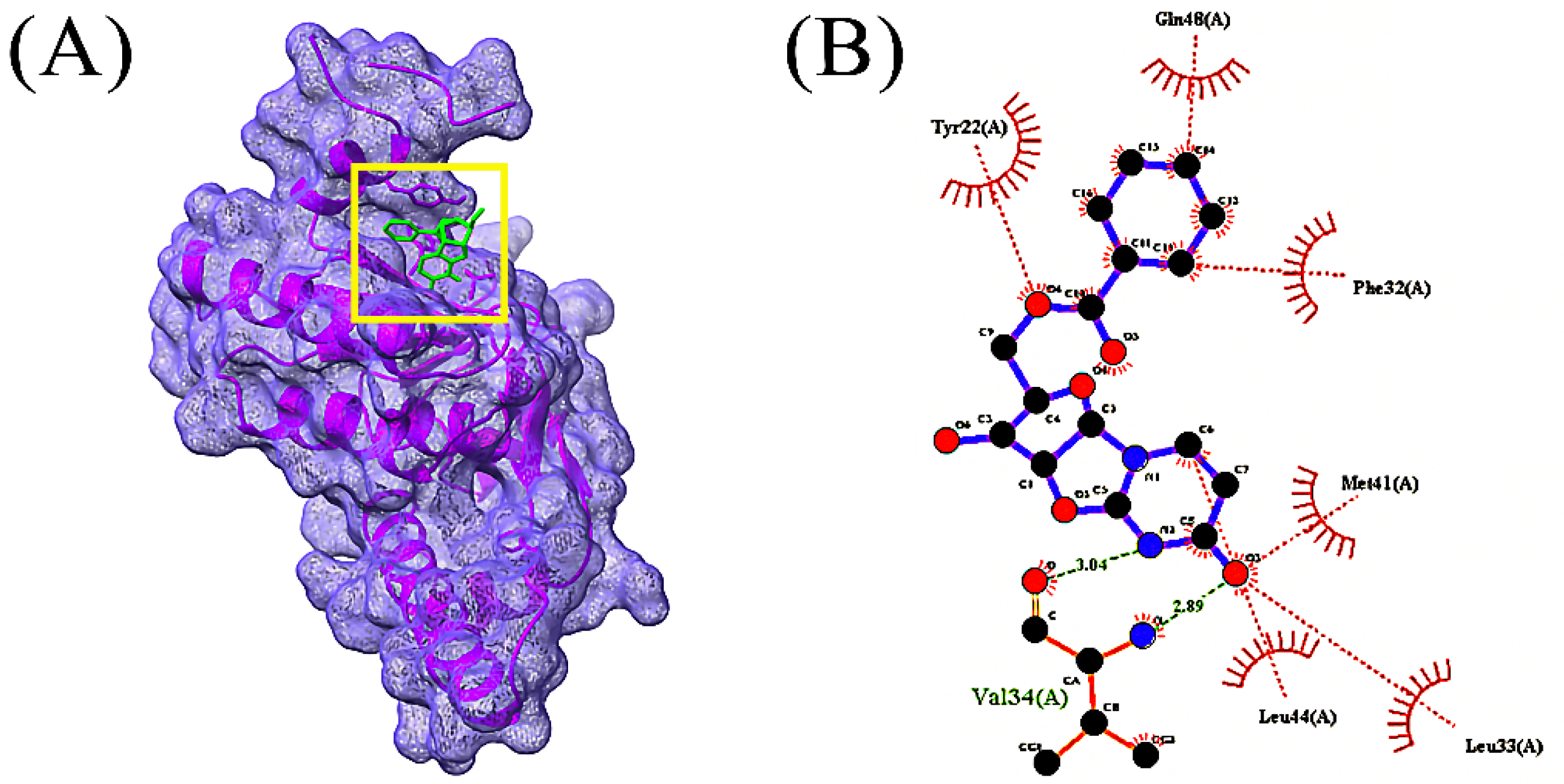
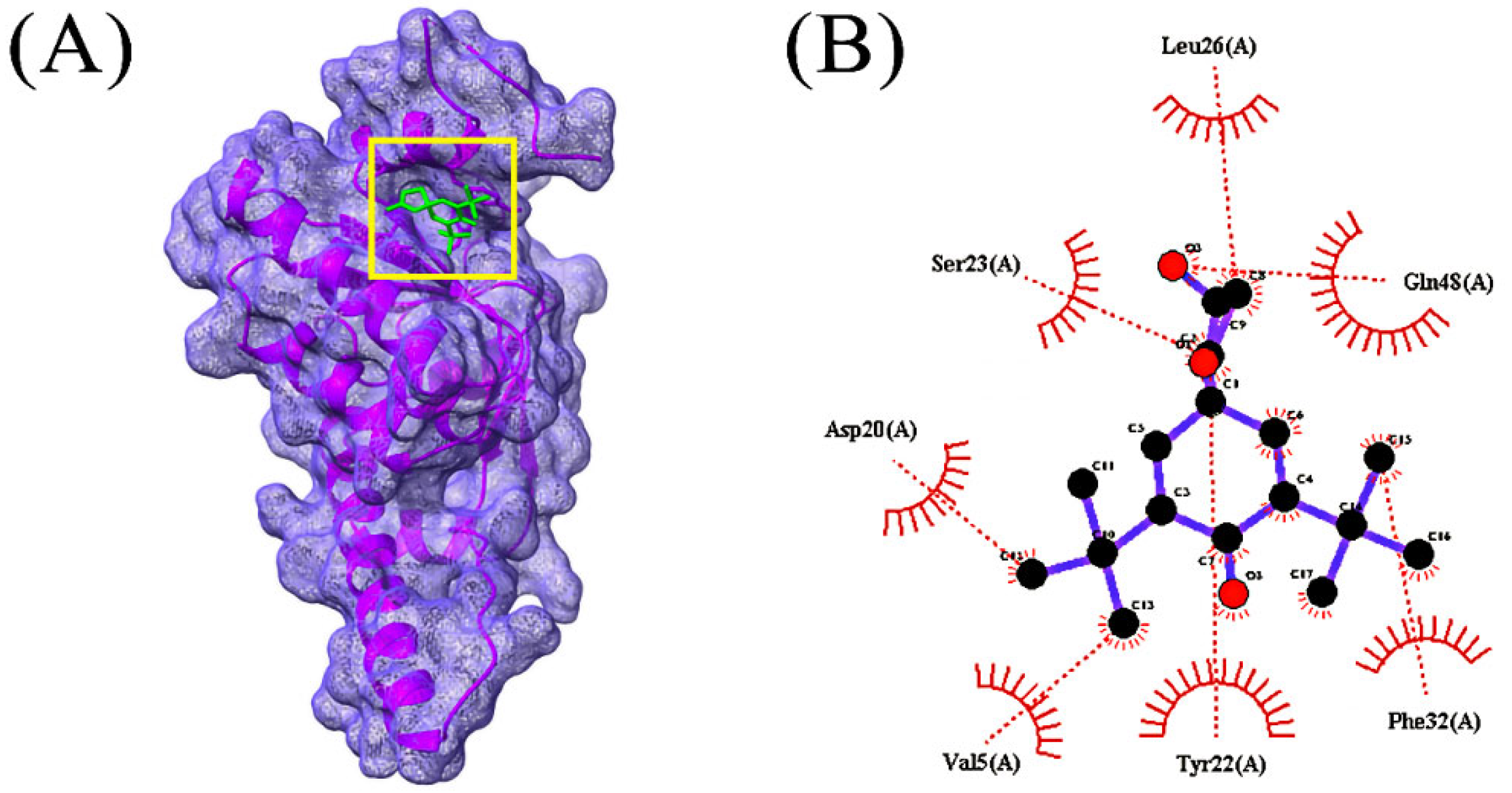
3.4. Molecular Docking Analysis
3.5. ADMET Prediction
3.6. Molecular Dynamics Simulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hopkins, A. Antibiotics at the Crossroads. Nature 2004, 43, 899–902. [Google Scholar]
- Qais, F.A.; Khan, M.S.; Ahmad, I.; Husain, F.M.; Khan, R.A.; Hassan, I.; Shahzad, S.A.; AlHarbi, W. Coumarin Exhibits Broad-Spectrum Antibiofilm and Antiquorum Sensing Activity against Gram-Negative Bacteria: In Vitro and In Silico Investigation. ACS Omega 2021, 6, 18823–18835. [Google Scholar] [CrossRef] [PubMed]
- Rather, I.A.; Kim, B.-C.; Bajpai, V.K.; Park, Y.-H. Self-medication and antibiotic resistance: Crisis, current challenges, and prevention. Saudi J. Biol. Sci. 2017, 24, 808–812. [Google Scholar] [CrossRef]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamr, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Zamani, H.; Rahbar, S.; Garakoui, S.R.; Sahebi, A.A.; Jafari, H. Antibiofilm potential of Lactobacillus plantarum spp. cell free supernatant (CFS) against multidrug resistant bacterial pathogens. Pharm. Biomed. Res. 2017, 3, 39–44. [Google Scholar] [CrossRef]
- Bowler, P.G. Antibiotic resistance and biofilm tolerance: A combined threat in the treatment of chronic infections. J. Wound Care 2018, 27, 273–277. [Google Scholar] [CrossRef]
- Kalpana, B.J.; Aarthy, S.; Pandian, S.K. Antibiofilm Activity of α-Amylase from Bacillus subtilis S8-18 against Biofilm Forming Human Bacterial Pathogens. Appl. Biochem. Biotechnol. 2012, 167, 1778–1794. [Google Scholar] [CrossRef]
- Park, S.-C.; Park, Y.; Hahm, K.-S. The Role of Antimicrobial Peptides in Preventing Multidrug-Resistant Bacterial Infections and Biofilm Formation. Int. J. Mol. Sci. 2011, 12, 5971–5992. [Google Scholar] [CrossRef]
- Lee, H.-W.; Koh, Y.; Kim, J.; Lee, J.-C.; Lee, Y.-C.; Seol, S.-Y.; Cho, D.-T. Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin. Microbiol. Infect. 2008, 14, 49–54. [Google Scholar] [CrossRef]
- Zhang, J.; Liang, X.; Zhang, S.; Song, Z.; Wang, C.; Xu, Y. Maipomycin A, a Novel Natural Compound with Promising Anti-biofilm Activity against Gram-Negative Pathogenic Bacteria. Front. Microbiol. 2021, 11, 598024. [Google Scholar] [CrossRef] [PubMed]
- Ahammad, I.; Lira, S.S. Designing a novel mRNA vaccine against SARS-CoV-2: An immunoinformatics approach. Int. J. Biol. Macromol. 2020, 162, 820–837. [Google Scholar] [CrossRef] [PubMed]
- Garrido, A.; Atencio, L.A.; Bethancourt, R.; Bethancourt, A.; Guzmán, H.; Gutiérrez, M.; Durant-Archibold, A.A. Antibacterial Activity of Volatile Organic Compounds Produced by the Octocoral-Associated Bacteria Bacillus sp. BO53 and Pseudoalteromonas sp. GA327. Antibiotics 2020, 9, 923. [Google Scholar] [CrossRef] [PubMed]
- Mawla, G.D.; Hall, B.M.; Cárcamo-Oyarce, G.; Grant, R.A.; Zhang, J.J.; Kardon, J.R.; Ribbeck, K.; Sauer, R.T.; Baker, T.A. ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa. Mol. Microbiol. 2020, 115, 1094–1109. [Google Scholar] [CrossRef] [PubMed]
- Al-Dulaimi, M.; Algburi, A.; Abdelhameed, A.; Mazanko, M.S.; Rudoy, D.V.; Ermakov, A.M.; Chikindas, M.L. Antimicrobial and Anti-Biofilm Activity of Polymyxin E Alone and in Combination with Probiotic Strains of Bacillus subtilis KATMIRA1933 and Bacillus amyloliquefaciens B-1895 against Clinical Isolates of Selected Acinetobacter spp.: A Preliminary Study. Pathogens 2021, 10, 1574. [Google Scholar] [CrossRef] [PubMed]
- Lahiri, D.; Nag, M.; Dutta, B.; Mukherjee, I.; Ghosh, S.; Dey, A.; Banerjee, R.; Ray, R.R. Catechin as the Most Efficient Bioactive Compound from Azadirachta indica with Antibiofilm and Anti-quorum Sensing Activities against Dental Biofilm: An In Vitro and In Silico Study. Appl. Biochem. Biotechnol. 2021, 193, 1617–1630. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2011, 47, e2437. [Google Scholar] [CrossRef]
- Ray, S.; Jin, J.-O.; Choi, I.; Kim, M. Cell-Free Supernatant of Bacillus thuringiensis Displays Anti-Biofilm Activity against Staphylococcus aureus. Appl. Biochem. Biotechnol. 2022, 1–15. [Google Scholar] [CrossRef]
- Ansari, M.J.; Al-Ghamdi, A.; Usmani, S.; Al-Waili, N.S.; Sharma, D.; Nuru, A.; Al-Attal, Y. Effect of Jujube Honey on Candida albicans Growth and Biofilm Formation. Arch. Med. Res. 2013, 44, 352–360. [Google Scholar] [CrossRef]
- Lin, J.; Wu, M.; Wu, H.; Zhang, T.; Wu, C.; Li, F. Epidemiological Characteristics of Coronavirus Disease 2019 in Zhejiang Province. J. Prev. Med. 2020, 12, 217–223. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Kawsar, S.M.A.; Kumer, A. Computational Investigation of Methyl α-d-Glucopyranoside Derivatives as Inhibitor against Bacteria, Fungi and COVID-19 (Sars-2). J. Chil. Chem. Soc. 2021, 66, 5206–5214. [Google Scholar] [CrossRef]
- Punjabi, M.; Bharadvaja, N.; Sachdev, A.; Krishnan, V. Molecular characterization, modeling, and docking analysis of late phytic acid biosynthesis pathway gene, inositol polyphosphate 6-/3-/5-kinase, a potential candidate for developing low phytate crops. 3 Biotech 2018, 8, 344. [Google Scholar] [CrossRef]
- Land, H.; Humble, M.S. ASARA: A Tool to Obtain Structural Guidance in Biocatalytic Investigations. Methods Mol. Biol. 2018, 1685, 43–67. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G.; Spronk, C. YASARA–Yet Another Scientific Artificial Reality Application. YASARA.org. 2013. Available online: http://www.yasara.org/ (accessed on 14 August 2022).
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, S.; Mita, M.A.; Biswas, S.; Paul, G.K.; Promi, M.M.; Afrose, S.; Hasan, R.; Shimu, M.S.S.; Zaman, S.; Uddin, S.; et al. Molecular docking and dynamics study to explore phytochemical ligand molecules against the main protease of SARS-CoV-2 from extensive phytochemical datasets. Expert Rev. Clin. Pharmacol. 2021, 14, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Harrach, M.F.; Drossel, B. Structure and dynamics of TIP3P, TIP4P, and TIP5P water near smooth and atomistic walls of different hydroaffinity. J. Chem. Phys. 2014, 140, 174501. [Google Scholar] [CrossRef]
- Mahmud, S.; Paul, G.K.; Biswas, S.; Afrose, S.; Mita, M.A.; Hasan, R.; Shimu, M.S.S.; Hossain, A.; Promi, M.M.; Ema, F.K.; et al. Prospective Role of Peptide-Based Antiviral Therapy against the Main Protease of SARS-CoV-2. Front. Mol. Biosci. 2021, 8, 628585. [Google Scholar] [CrossRef]
- Biswas, S.; Mahmud, S.; Mita, M.A.; Afrose, S.; Hasan, R.; Shimu, M.S.S.; Saleh, A.; Mostafa-Hedeab, G.; Alqarni, M.; Obaidullah, A.J.; et al. Molecular Docking and Dynamics Studies to Explore Effective Inhibitory Peptides against the Spike Receptor Binding Domain of SARS-CoV-2. Front. Mol. Biosci. 2022, 8, 791642. [Google Scholar] [CrossRef]
- Mahmud, S.; Hasan, R.; Biswas, S.; Paul, G.K.; Afrose, S.; Mita, M.A.; Shimu, M.S.S.; Promi, M.M.; Hani, U.; Rahamathulla, M.; et al. Screening of Potent Phytochemical Inhibitors against SARS-CoV-2 Main Protease: An Integrative Computational Approach. Front. Bioinform. 2021, 1, 717141. [Google Scholar] [CrossRef]
- Mahmud, S.; Biswas, S.; Paul, G.; Mita, M.; Promi, M.; Afrose, S.; Hasan, R.; Zaman, S.; Uddin, S.; Dhama, K.; et al. Plant-Based Phytochemical Screening by Targeting Main Protease of SARS-CoV-2 to Design Effective Potent Inhibitors. Biology 2021, 10, 589. [Google Scholar] [CrossRef] [PubMed]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Krieger, E.; Nielsen, J.E.; Spronk, C.A.; Vriend, G. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model. 2006, 25, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, S.; Paul, G.; Afroze, M.; Islam, S.; Gupt, S.; Razu, M.; Biswas, S.; Zaman, S.; Uddin, S.; Khan, M.; et al. Efficacy of Phytochemicals Derived from Avicennia officinalis for the Management of COVID-19: A Combined In Silico and Biochemical Study. Molecules 2021, 26, 2210. [Google Scholar] [CrossRef] [PubMed]
- Paul, G.K.; Mahmud, S.; Aldahish, A.A.; Afroze, M.; Biswas, S.; Gupta, S.B.R.; Razu, M.H.; Zaman, S.; Uddin, S.; Nahari, M.H.; et al. Computational screening and biochemical analysis of Pistacia integerrima and Pandanus odorifer plants to find effective inhibitors against Receptor-Binding domain (RBD) of the spike protein of SARS-Cov-2. Arab. J. Chem. 2021, 15, 103600. [Google Scholar] [CrossRef]
- Mahmud, S.; Rafi, O.; Paul, G.K.; Promi, M.M.; Shimu, M.S.S.; Biswas, S.; Bin Emran, T.; Dhama, K.; Alyami, S.A.; Moni, M.A.; et al. Designing a multi-epitope vaccine candidate to combat MERS-CoV by employing an immunoinformatics approach. Sci. Rep. 2021, 11, 15431. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, S.; Afrose, S.; Biswas, S.; Nagata, A.; Paul, G.K.; Mita, M.A.; Hasan, R.; Shimu, M.S.S.; Zaman, S.; Uddin, S.; et al. Plant-derived compounds effectively inhibit the main protease of SARS-CoV-2: An in silico approach. PLoS ONE 2022, 17, e0273341. [Google Scholar] [CrossRef]
- Mahmud, S.; Biswas, S.; Paul, G.K.; Mita, M.A.; Afrose, S.; Hasan, R.; Shimu, M.S.S.; Uddin, M.A.R.; Uddin, S.; Zaman, S.; et al. Antiviral peptides against the main protease of SARS-CoV-2: A molecular docking and dynamics study. Arab. J. Chem. 2021, 14, 103315. [Google Scholar] [CrossRef]
- Mahfuz, A.M.U.B.; Khan, A.; Biswas, S.; Afrose, S.; Mahmud, S.; Bahadur, N.M.; Ahmed, F. In search of novel inhibitors of anti-cancer drug target fibroblast growth factor receptors: Insights from virtual screening, molecular docking, and molecular dynamics. Arab. J. Chem. 2022, 15, 103882. [Google Scholar] [CrossRef]
- Jagannathan, R. Characterization of Drug-like Chemical Space for Cytotoxic Marine Metabolites Using Multivariate Methods. ACS Omega 2019, 4, 5402–5411. [Google Scholar] [CrossRef] [PubMed]
- Azam, M.W.; Khan, A.U. Updates on the pathogenicity status of Pseudomonas aeruginosa. Drug Discov. Today 2018, 24, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules 2020, 25, 1340. [Google Scholar] [CrossRef] [PubMed]
- Rohde, C.; Wittmann, J.; Kutter, E. Bacteriophages: A Therapy Concept against Multi-Drug–Resistant Bacteria. Surg. Infect. 2018, 19, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.R.; Isabella, V.M.; Lewis, K. Pseudomonas aeruginosa Biofilms in Disease. Microb. Ecol. 2013, 68, 1–12. [Google Scholar] [CrossRef]
- Wang, T.; Liang, Y.; Wu, M.; Chen, Z.; Lin, J.; Yang, L. Natural products from Bacillus subtilis with antimicrobial properties. Chin. J. Chem. Eng. 2015, 23, 744–754. [Google Scholar] [CrossRef]
- Ozabor, T.P.; Fadahunsi, I.F. Antimicrobial Activity of Bacillus Subtilis against Some Selected Food Borne Pathogens. Acta Sci. Microbiol. 2019, 2, 89–95. [Google Scholar]
- A Kadhum, H.; Hasan2, T.H. The Study of Bacillus subtils Antimicrobial Activity on Some of the Pathological Isolates. Int. J. Drug Deliv. Technol. 2019, 9, 193–196. [Google Scholar] [CrossRef]
- Ramachandran, R.; Chalasani, A.G.; Lal, R.; Roy, U. A Broad-Spectrum Antimicrobial Activity of Bacillus subtilis RLID 12.1. Sci. World J. 2014, 2014, 968487. [Google Scholar] [CrossRef][Green Version]
- Hamza, F.; Kumar, A.R.; Zinjarde, S. Antibiofilm potential of a tropical marine Bacillus licheniformis isolate: Role in disruption of aquaculture associated biofilms. Aquac. Res. 2015, 47, 2661–2669. [Google Scholar] [CrossRef]
- Ghosh, S.; Lahiri, D.; Nag, M.; Dey, A.; Sarkar, T.; Biswas, R.; Dutta, B.; Mukherjee, D.; Pati, S.; Pattanaik, S.; et al. Analysis of Antibiofilm Activities of Bioactive Compounds from Honeyweed (Leonurus sibiricus) against P. aeruginosa: An In Vitro and In Silico Approach. Appl. Biochem. Biotechnol. 2022, 1–17. [Google Scholar] [CrossRef] [PubMed]



| Name of the Strains | Zone of Inhibition (mm) | Status |
|---|---|---|
| Klebsiella pneumoniae | 07.00 ± 0.33 a | No inhibitory activity (−) |
| Pseudomonas sp. | 18.33 ± 1.00 c | Moderate/average inhibition (++) |
| Escherichia coli | 08.33 ± 0.57 ab | No inhibitory activity (−) |
| Staphylococcus aureus | 09.00 ± 1.00 a | No inhibitory activity (−) |
| Compounds | CID | Docking Score (kcal/mol) | |
|---|---|---|---|
| 1 | 1-(9H-Fluoren-2-yl)-2-(1-phenyl-1H-tetrazol-5-ylsulfanyl)- ethanone | 606333 | −7.0 |
| 2 | Oxalic acid, cyclohexylmethyl tridecyl ester | 6421725 | −6.4 |
| 3 | Bis(pentamethylcyclotrisiloxy)tetramethyldisiloxane | 553163 | −6.2 |
| 4 | 2,2’-Anhydro-1-arabinofuranosyluracil | 569411 | −6.0 |
| 5 | Pyridine, 2,3,6-trimethyl- | 15100 | −5.8 |
| 6 | D-Limonene | 440917 | −5.8 |
| 7 | Azulene | 9231 | −5.8 |
| 8 | Benzene, 1,3-bis(1,1-dimethylethyl)- | 71343282 | −5.8 |
| 9 | Propionic acid, (3,6,7,8-tetrahydro-3,7-methano-2,4,6- trimethyl-2H-oxocin-7-yl)methyl ester | 583617 | −5.7 |
| 10 | 2,5-di-tert-Butyl-1,4-benzoquinone | 17161 | −5.6 |
| Compounds Name and CID | MW g/mol | H. Ac | H. Do | Log Po/w | Log S | Number of Lipinski Violation | TPSA (Å2) | Human Intestinal Absorption) | BBB (+ve/−ve) |
|---|---|---|---|---|---|---|---|---|---|
| 1-(9H-Fluoren-2-yl)-2-(1-phenyl-1H-tetrazol-5-ylsulfanyl)- ethanone; 606333 | 384.5 | 4 | 0 | 3.20 | −5.70 | Yes; 0 violation | 85.97 | High | −ve |
| Oxalic acid, cyclohexylmethyl tridecyl ester; 6421725 | 368.5 | 4 | 0 | 5.03 | −6.61 | Yes; 4 violations | 52.60 | High | −ve |
| Bis(pentamethylcyclotrisiloxy)tetramethyldisiloxane; 553163 | 579.2 | 9 | 0 | 6.36 | −2.37 | Yes; 2 violations | 83.07 | High | −ve |
| 2,2’-Anhydro-1-arabinofuranosyluracil; 569411 | 330.29 | 7 | 1 | 2.11 | −1.50 | Yes; 0 violation | 99.88 | High | −ve |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, S.; Mahmud, M.L.; Almalki, W.H.; Biswas, S.; Islam, M.A.; Mortuza, M.G.; Hossain, M.A.; Ekram, M.A.-E.; Uddin, M.S.; Zaman, S.; et al. Cell-Free Supernatants (CFSs) from the Culture of Bacillus subtilis Inhibit Pseudomonas sp. Biofilm Formation. Microorganisms 2022, 10, 2105. https://doi.org/10.3390/microorganisms10112105
Islam S, Mahmud ML, Almalki WH, Biswas S, Islam MA, Mortuza MG, Hossain MA, Ekram MA-E, Uddin MS, Zaman S, et al. Cell-Free Supernatants (CFSs) from the Culture of Bacillus subtilis Inhibit Pseudomonas sp. Biofilm Formation. Microorganisms. 2022; 10(11):2105. https://doi.org/10.3390/microorganisms10112105
Chicago/Turabian StyleIslam, Shirmin, Md. Liton Mahmud, Waleed H. Almalki, Suvro Biswas, Md. Ariful Islam, Md. Golam Mortuza, Mohammad Akbar Hossain, Md. Akhtar-E Ekram, Md. Salah Uddin, Shahriar Zaman, and et al. 2022. "Cell-Free Supernatants (CFSs) from the Culture of Bacillus subtilis Inhibit Pseudomonas sp. Biofilm Formation" Microorganisms 10, no. 11: 2105. https://doi.org/10.3390/microorganisms10112105
APA StyleIslam, S., Mahmud, M. L., Almalki, W. H., Biswas, S., Islam, M. A., Mortuza, M. G., Hossain, M. A., Ekram, M. A.-E., Uddin, M. S., Zaman, S., & Saleh, M. A. (2022). Cell-Free Supernatants (CFSs) from the Culture of Bacillus subtilis Inhibit Pseudomonas sp. Biofilm Formation. Microorganisms, 10(11), 2105. https://doi.org/10.3390/microorganisms10112105







