The Production of Pyruvate in Biological Technology: A Critical Review
Abstract
:1. Introduction
2. General Methods of Pyruvic Acid Production
2.1. Chemical Method
2.2. Biotechnological Methods
2.3. Enzymatic Processes
2.4. Resting Cell Processes
2.5. Fermentation Processes
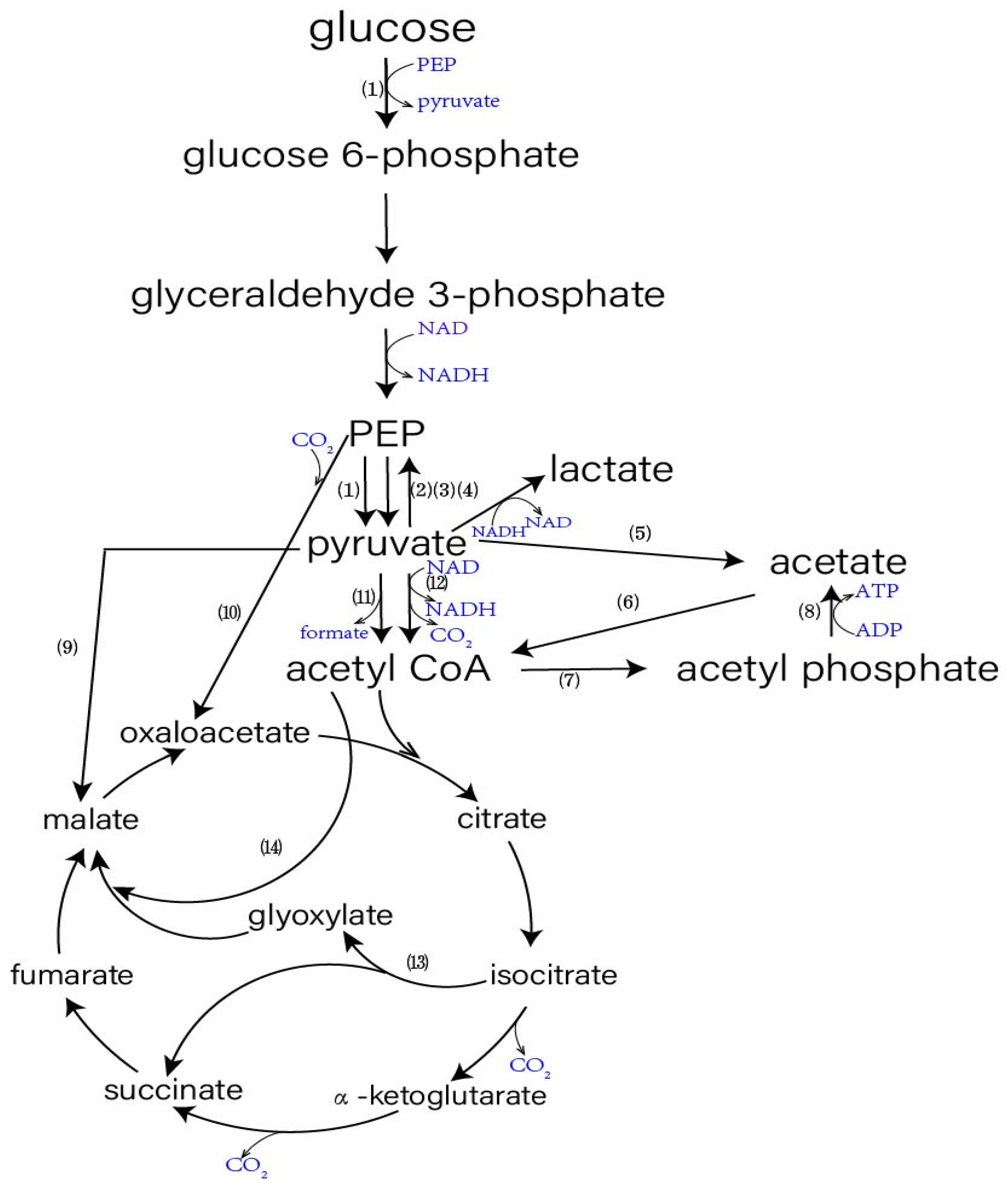
3. Biochemical Pathways Involved in Pyruvate
3.1. Biochemical Pathways Directly Impacting Pyruvate
3.1.1. Pyruvate Dehydrogenase (PDH)
3.1.2. Pyruvate Formate-Lyase (PFL)
3.1.3. Pyruvate Oxidase (POXB)
3.1.4. Lactate Dehydrogenase (LDH)
3.1.5. PEP Synthase (PPS)
3.2. Biochemical Pathways Indirectly Impacting Pyruvate Formation
3.2.1. PEP Carboxylase (PPC or PEPC. EC:4.1.1.31)
3.2.2. Energy and (F1F0) H+-ATP Synthase Complex
3.3. By-Products in the Production of Pyruvic Acid
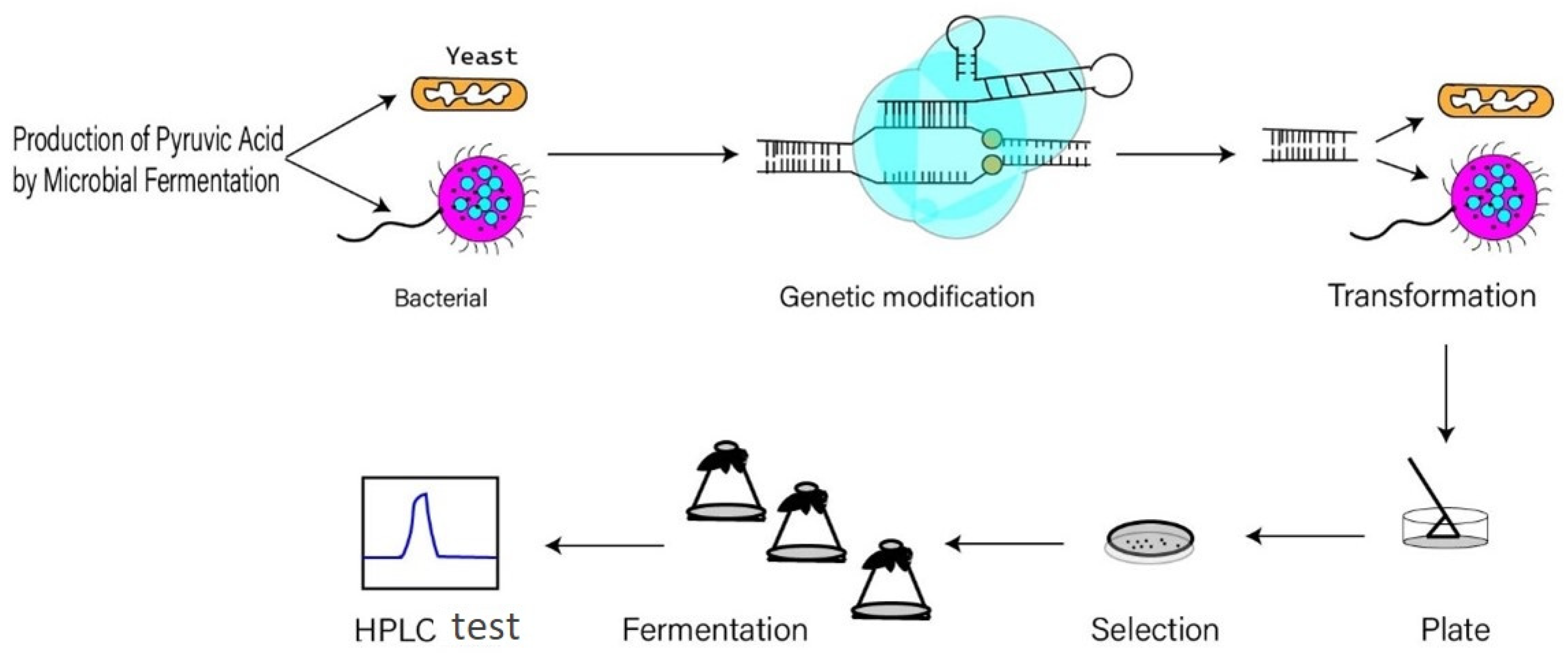
4. Production of Pyruvate by Recombinant Microbes
4.1. Production of Pyruvate by Recombinant E. coli
| Strain | Mutations | Auxotroph | Yield (g/g) | Pyruvate (g/L) | Productivity (g/L·h) | Media | Reference |
|---|---|---|---|---|---|---|---|
| W1485lip2 | Lipoic acid | 0.51 | 25.5 | 0.64 | Complex | [46] | |
| TBLA-1 | atpA401 | Lipoic acid | 0.60 | 30.0 | 1.25 | Complex | [47] |
| CGSC6162 | aceF | 0.72 | >30.0 | 1.50 | Complex | [22] | |
| CGSC7916 | aceF, ppc | 0.78 | >30.0 | 1.20 | Complex | [22] | |
| E. coliTC44 | 0.75 | 65.96 | [49] | ||||
| E. coli CGSC6162 Deltappc | 0.78 | 35 | [22] | ||||
| E. coli ALS929 | 0.68 | 90 | [50] | ||||
| E. coli YYC202 ldhA::Kan | 0.87 | 110.0 | [32] | ||||
| YYC202 ldhA::Kan | aceEF, ldhA, pps, pfl, poxB | 0.56 | 62.0 | 1.75 | [48] | ||
| TC44 | adhE, atpFH, ackA, ldhA, frdBC, pfl, poxB, sucA | 0.75 | 67.4 | 1.20 | [49] |
4.2. Production of Pyruvate by Recombinant Yeast
| Strain | Substrate | Pyruvate (g/L) | Yield (g/g) | Details and References |
|---|---|---|---|---|
| Bacillus megaterium | glucose | 27.8 | 0.38 | [61] |
| Blastobotrys adeninivorans VKM Y-2677 | glucose | 43.2 | 0.77 | [53] |
| S. cerevisiae | glucose | 135 | 0.54 | [51] |
| Trichosporoncutaneum PD70 | glucose | 34.6 | 0.429 | [62] |
| Torulopsis glabrata | glucose | 94.3 | 0.635 | [63] |
| Y. lipolyticaa 374/4 | glycerol | 61.3 | 0.71 | Y. lipolytica [64] |
| Y. lipolyticaa WSH-Z06 | glycerol | 39.3 | 0.71 | [65] |
| Y. lipolyticaa | glycerol | 48.1 | 0.48 | [66] |
| Y. lipolyticaa | glycerol | 97.2 | 0.795 | [52] |
5. Summary
Funding
Data Availability Statement
Conflicts of Interest
References
- McCommis, K.S.; Finck, B.N. Mitochondrial pyruvate transport: A historical perspective and future research directions. Biochem. J. 2015, 466, 443–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Chen, J.; Lun, S.Y. Biotechnological production of pyruvic acid. Appl. Microbiol. Biotechnol. 2001, 57, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Soma, Y.; Tsuruno, K.; Wada, M.; Yokota, A.; Hanai, T. Metabolic flux redirection from a central metabolic pathway toward a synthetic pathway using a metabolic toggle switch. Metab. Eng. 2014, 23, 175–184. [Google Scholar] [CrossRef]
- Jager, R.; Metzger, J.; Lautmann, K.; Shushakov, V.; Purpura, M.; Geiss, K.R.; Maassen, N. The effects of creatine pyruvate and creatine citrate on performance during high intensity exercise. J. Int. Soc. Sport. Nutr. 2008, 5, 4. [Google Scholar] [CrossRef] [Green Version]
- Han, H.S.; Kang, G.; Kim, J.S.; Choi, B.H.; Koo, S.H. Regulation of glucose metabolism from a liver-centric perspective. Exp. Mol. Med. 2016, 48, e218. [Google Scholar] [CrossRef] [Green Version]
- Ju, K.D.; Shin, E.K.; Cho, E.J.; Yoon, H.B.; Kim, H.S.; Kim, H.; Yang, J.; Hwang, Y.-H.; Ahn, C.; Oh, K.-H. Ethyl pyruvate ameliorates albuminuria and glomerular injury in the animal model of diabetic nephropathy. Am. J. Physiol.-Ren. Physiol. 2012, 302, F606–F613. [Google Scholar] [CrossRef] [Green Version]
- Plotnikov, E.; Losenkov, I.; Epimakhova, E.; Bohan, N. Protective effects of pyruvic acid salt against lithium toxicity and oxidative damage in human blood mononuclear cells. Adv. Pharm. Bull. 2019, 9, 302–306. [Google Scholar] [CrossRef]
- Cybulski, K.; Tomaszewska-Hetman, L.; Rakicka, M.; Juszczyk, P.; Rywińska, A. Production of pyruvic acid from glycerol by Yarrowia lipolytica. Folia Microbiol. 2019, 64, 809–820. [Google Scholar] [CrossRef]
- Maleki, N.; Eiteman, M.A. Recent progress in the microbial production of pyruvic acid. Fermentation 2017, 3, 8. [Google Scholar] [CrossRef]
- Descotes, J. The popliteal lymph node assay: A tool for studying the mechanisms of drug-induced autoimmune disorders. Toxicol. Lett. 1992, 64–65, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Yang, q.; Yi, X.; Guo, l. Synthesis of Pyruvic Acid from Lactic Acid by Oxidation with Oxygen. Dep. Chem. 2002, 43, 307–309. [Google Scholar]
- Gao, C.; Ma, C.; Xu, P. Progress in biotransformation of bio-based lactic acid. Sheng Wu Gong Cheng Xue Bao Chin. J. Biotechnol. 2013, 29, 1411–1420. [Google Scholar]
- Rui, L. Energy metabolism in the liver. Compr. Physiol. 2014, 4, 177–197. [Google Scholar] [CrossRef] [Green Version]
- Meza, E.; Becker, J.; Bolivar, F.; Gosset, G.; Wittmann, C. Consequences of phosphoenolpyruvate:sugar phosphotranferase system and pyruvate kinase isozymes inactivation in central carbon metabolism flux distribution in Escherichia coli. Microb. Cell Factories 2012, 11, 127. [Google Scholar] [CrossRef] [Green Version]
- Postma, P.W.; Lengeler, J.W.; Jacobson, G.R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 1993, 57, 543–594. [Google Scholar] [CrossRef]
- Dimitrova, M.N.; Peterkofsky, A.; Ginsburg, A. Opposing effects of phosphoenolpyruvate and pyruvate with Mg(2+) on the conformational stability and dimerization of phosphotransferase enzyme I from Escherichia coli. Protein Sci. A Publ. Protein Soc. 2003, 12, 2047–2056. [Google Scholar] [CrossRef] [Green Version]
- Ginsburg, A.; Peterkofsky, A. Enzyme I: The gateway to the bacterial phosphoenolpyruvate: Sugar phosphotransferase system. Arch. Biochem. Biophys. 2002, 397, 273–278. [Google Scholar] [CrossRef]
- Mattevi, A.; Valentini, G.; Rizzi, M.; Speranza, M.L.; Bolognesi, M.; Coda, A. Crystal structure of Escherichia coli pyruvate kinase type I: Molecular basis of the allosteric transition. Structure 1995, 3, 729–741. [Google Scholar] [CrossRef] [Green Version]
- Ponce, E.; Flores, N.; Martinez, A.; Valle, F.; Bolivar, F. Cloning of the two pyruvate kinase isoenzyme structural genes from Escherichia coli: The relative roles of these enzymes in pyruvate biosynthesis. J. Bacteriol. 1995, 177, 5719–5722. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; St Leger, R.J.; Fang, W. Pyruvate Accumulation Is the First Line of Cell Defense against Heat Stress in a Fungus. mBio 2017, 8, e01284-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomar, A.; Eiteman, M.A.; Altman, E. The effect of acetate pathway mutations on the production of pyruvate in Escherichia coli. Appl. Microbiol. Biotechnol. 2003, 62, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Yonehara, T.; Miyata, R. Fermentative production of pyruvate from glucose by Torulopsis glabrata. J. Ferment. Bioeng. 1994, 78, 155–159. [Google Scholar] [CrossRef]
- Lehtio, L.; Leppanen, V.M.; Kozarich, J.W.; Goldman, A. Structure of Escherichia coli pyruvate formate-lyase with pyruvate. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002, 58, 2209–2212. [Google Scholar] [CrossRef]
- Sawers, G.; Bock, A. Anaerobic regulation of pyruvate formate-lyase from Escherichia coli K-12. J. Bacteriol. 1988, 170, 5330–5336. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Hamid, A.M.; Attwood, M.M.; Guest, J.R. Pyruvate oxidase contributes to the aerobic growth efficiency of Escherichia coli. Microbiology 2001, 147, 1483–1498. [Google Scholar] [CrossRef] [Green Version]
- Jiang, G.R.; Nikolova, S.; Clark, D.P. Regulation of the ldhA gene, encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 2001, 147, 2437–2446. [Google Scholar] [CrossRef] [Green Version]
- Tarmy, E.M.; Kaplan, N.O. Chemical characterization of D-lactate dehydrogenase from Escherichia coli B. J. Biol. Chem. 1968, 243, 2579–2586. [Google Scholar] [CrossRef]
- Patnaik, R.; Roof, W.D.; Young, R.F.; Liao, J.C. Stimulation of glucose catabolism in Escherichia coli by a potential futile cycle. J. Bacteriol. 1992, 174, 7527–7532. [Google Scholar] [CrossRef] [Green Version]
- Narindrasorasak, S.; Bridger, W.A. Phosphoenolypyruvate synthetase of Escherichia coli: Molecular weight, subunit composition, and identification of phosphohistidine in phosphoenzyme intermediate. J. Biol. Chem. 1977, 252, 3121–3127. [Google Scholar] [CrossRef]
- Gulick, A.M.; Starai, V.J.; Horswill, A.R.; Homick, K.M.; Escalante-Semerena, J.C. The 1.75 A crystal structure of acetyl-CoA synthetase bound to adenosine-5′-propylphosphate and coenzyme A. Biochemistry 2003, 42, 2866–2873. [Google Scholar] [CrossRef] [PubMed]
- Zelic, B.; Gostovic, S.; Vuorilehto, K.; Vasic-Racki, D.; Takors, R. Process strategies to enhance pyruvate production with recombinant Escherichia coli: From repetitive fed-batch to in situ product recovery with fully integrated electrodialysis. Biotechnol. Bioeng. 2004, 85, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Izui, K.; Matsumura, H.; Furumoto, T.; Kai, Y. Phosphoenolpyruvate carboxylase: A new era of structural biology. Annu. Rev. Plant Biol. 2004, 55, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Schaaff, I.; Heinisch, J.; Zimmermann, F.K. Overproduction of glycolytic enzymes in yeast. Yeast 1989, 5, 285–290. [Google Scholar] [CrossRef]
- Ruyter, G.J.; Postma, P.W.; van Dam, K. Control of glucose metabolism by enzyme IIGlc of the phosphoenolpyruvate-dependent phosphotransferase system in Escherichia coli. J. Bacteriol. 1991, 173, 6184–6191. [Google Scholar] [CrossRef] [Green Version]
- Koebmann, B.J.; Westerhoff, H.V.; Snoep, J.L.; Nilsson, D.; Jensen, P.R. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J. Bacteriol. 2002, 184, 3909–3916. [Google Scholar] [CrossRef] [Green Version]
- Kotlarz, D.; Garreau, H.; Buc, H. Regulation of the amount and of the activity of phosphofructokinases and pyruvate kinases in Escherichia coli. Biochim. Biophys. Acta 1975, 381, 257–268. [Google Scholar] [CrossRef]
- Kotlarz, D.; Buc, H. Phosphofructokinases from Escherichia coli. Methods Enzymol. 1982, 90 Pt E, 60–70. [Google Scholar] [CrossRef]
- Sorgen, P.L.; Caviston, T.L.; Perry, R.C.; Cain, B.D. Deletions in the second stalk of F1F0-ATP synthase in Escherichia coli. J. Biol. Chem. 1998, 273, 27873–27878. [Google Scholar] [CrossRef] [Green Version]
- Weber, J.; Senior, A.E. ATP synthesis driven by proton transport in F1F0-ATP synthase. FEBS Lett. 2003, 545, 61–70. [Google Scholar] [CrossRef] [Green Version]
- Awasthi, D.; Wang, L.; Rhee, M.S.; Wang, Q.; Chauliac, D.; Ingram, L.O.; Shanmugam, K.T. Metabolic engineering of Bacillus subtilis for production of D-lactic acid. Biotechnol. Bioeng. 2018, 115, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; He, P.; Lu, D.; Shen, A.; Jiang, N.J.E.; Technology, M. Metabolic engineering of Torulopsis glabrata for improved pyruvate production. Enzym. Microb. Technol. 2005, 36, 832–839. [Google Scholar] [CrossRef]
- Wieschalka, S.; Blombach, B.; Eikmanns, B.J. Engineering Corynebacterium glutamicum for the production of pyruvate. Appl. Microbiol. Biotechnol. 2012, 94, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Hofvendahl, K.; Hahn-Hagerdal, B. Factors affecting the fermentative lactic acid production from renewable resources (1). Enzym. Microb. Technol. 2000, 26, 87–107. [Google Scholar] [CrossRef]
- Chang, D.E.; Jung, H.C.; Rhee, J.S.; Pan, J.G. Homofermentative production of D- or L-lactate in metabolically engineered Escherichia coli RR1. Appl. Environ. Microbiol. 1999, 65, 1384–1389. [Google Scholar] [CrossRef] [Green Version]
- Yokota, A.; Shimizu, H.; Terasawa, Y.; Takaoka, N.; Tomita, F. Pyruvic acid production by a lipoic acid auxotroph of Escherichia coli W1485. Appl. Microbiol. Biotechnol. 1994, 41, 638–646. [Google Scholar] [CrossRef]
- Yokota, A.; Terasawa, Y.; Takaoka, N.; Shimizu, H.; Tomita, F. Pyruvic acid production by an F1-ATPase-defective mutant of Escherichia coli W1485lip2. Biosci. Biotechnol. Biochem. 1994, 58, 2164–2167. [Google Scholar] [CrossRef] [Green Version]
- Zelic, B.; Gerharz, T.; Bott, M.; Vasic-Racki, D.; Wandrey, C.; Takors, R. Fed-batch process for pyruvate production by recombinant Escherichia coli YYC202 strain. Eng. Life Sci. 2003, 3, 299–305. [Google Scholar] [CrossRef]
- Causey, T.B.; Shanmugam, K.T.; Yomano, L.P.; Ingram, L.O. Engineering Escherichia coli for efficient conversion of glucose to pyruvate. Proc. Natl. Acad. Sci. USA 2004, 101, 2235–2240. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Eiteman, M.A.; Altman, R.; Altman, E. High glycolytic flux improves pyruvate production by a metabolically engineered Escherichia coli strain. Appl. Environ. Microbiol. 2008, 74, 6649–6655. [Google Scholar] [CrossRef] [Green Version]
- van Maris, A.J.; Geertman, J.M.; Vermeulen, A.; Groothuizen, M.K.; Winkler, A.A.; Piper, M.D.; van Dijken, J.P.; Pronk, J.T. Directed evolution of pyruvate decarboxylase-negative Saccharomyces cerevisiae, yielding a C2-independent, glucose-tolerant, and pyruvate-hyperproducing yeast. Appl. Environ. Microbiol. 2004, 70, 159–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, W.; Lin, X.; Zhong, S.; Chen, J.; Wang, Z.; Sun, J. Enhanced pyruvic acid yield in an osmotic stress-resistant mutant of Yarrowia lipolytica. Electron. J. Biotechnol. 2020, 44, 19–24. [Google Scholar] [CrossRef]
- Kamzolova, S.V.; Morgunov, I.G. Biosynthesis of pyruvic acid from glucose by Blastobotrys adeninivorans. Appl. Microbiol. Biotechnol. 2016, 100, 7689–7697. [Google Scholar] [CrossRef]
- Aßkamp, M.R.; Klein, M.; Nevoigt, E. Saccharomyces cerevisiae exhibiting a modified route for uptake and catabolism of glycerol forms significant amounts of ethanol from this carbon source considered as ‘non-fermentable’. Biotechnol. Biofuels 2019, 12, 257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavlik, P.; Simon, M.; Schuster, T.; Ruis, H. The glycerol kinase (GUT1) gene of Saccharomyces cerevisiae: Cloning and characterization. Curr. Genet. 1993, 24, 21–25. [Google Scholar] [CrossRef]
- Ronnow, B.; Kiellandbrandt, M.C. GUT2, a gene for mitochondrial glycerol 3-phosphate dehydrogenase of Saccharomyces cerevisiae. Yeast 1993, 9, 1121–1130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tang, Y.; Guo, Z.; Shi, G. Engineering of the glycerol decomposition pathway and cofactor regulation in an industrial yeast improves ethanol production. J. Ind. Microbiol. Biotechnol. 2013, 40, 1153–1160. [Google Scholar] [CrossRef] [Green Version]
- Miyata, R.; Yonehara, T. Improvement of fermentative production of pyruvate from glucose by Torulopsis glabrata IFO 0005. J. Ferment. Bioeng. 1996, 82, 475–479. [Google Scholar] [CrossRef]
- Maaheimo, H.; Fiaux, J.; Cakar, Z.P.; Bailey, J.E.; Sauer, U.; Szyperski, T. Central carbon metabolism of Saccharomyces cerevisiae explored by biosynthetic fractional (13)C labeling of common amino acids. Eur. J. Biochem. 2001, 268, 2464–2479. [Google Scholar] [CrossRef] [Green Version]
- Fiaux, J.; Cakar, Z.P.; Sonderegger, M.; Wüthrich, K.; Szyperski, T.; Sauer, U. Metabolic-flux profiling of the yeasts Saccharomyces cerevisiae and Pichia stipitis. Eukaryot. Cell 2003, 2, 170–180. [Google Scholar] [CrossRef] [Green Version]
- Hollmann, R.; Deckwer, W.D. Pyruvate formation and suppression in recombinant Bacillus megaterium cultivation. J. Biotechnol. 2004, 111, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; He, P.; Lu, D.; Shen, A.; Jiang, N. Screening of pyruvate-producing yeast and effect of nutritional conditions on pyruvate production. Lett. Appl. Microbiol. 2002, 35, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xu, Q.; Li, Y.; Shi, Z.; Zhu, Y.; Du, G.; Chen, J. Enhancement of pyruvate production by osmotic-tolerant mutant of Torulopsis glabrata. Biotechnol. Bioeng. 2007, 97, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Morgunov, I.G.; Kamzolova, S.V.; Perevoznikova, O.A.; Shishkanova, N.V.; Finogenova, T.V. Pyruvic acid production by a thiamine auxotroph of Yarrowia lipolytica. Process Biochem. 2004, 39, 1469–1474. [Google Scholar] [CrossRef]
- Zeng, W.; Zhang, H.; Xua, S.; Fang, F.; Zhou, J. Biosynthesis of keto acids by fed-batch culture of Yarrowia lipolytica WSH-Z06. Bioresour. Technol. 2017, 243, 1037–1043. [Google Scholar] [CrossRef] [PubMed]
- Krzysztof, C.; Ludwika, T.-H.; Magdalena, R.; Wojciech, Ł.; Waldemar, R.; Anita, R. The bioconversion of waste products from rapeseed processing into keto acids by Yarrowia lipolytica. Ind. Crops Prod. 2018, 119, 102–110. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, W.; Du, Y.; Yu, K.; Xu, S.; Liu, M.; Wang, S.; Yang, Y.; Zhang, Y.; Sun, J. The Production of Pyruvate in Biological Technology: A Critical Review. Microorganisms 2022, 10, 2454. https://doi.org/10.3390/microorganisms10122454
Yuan W, Du Y, Yu K, Xu S, Liu M, Wang S, Yang Y, Zhang Y, Sun J. The Production of Pyruvate in Biological Technology: A Critical Review. Microorganisms. 2022; 10(12):2454. https://doi.org/10.3390/microorganisms10122454
Chicago/Turabian StyleYuan, Wei, Yongbao Du, Kechen Yu, Shiyi Xu, Mengzhu Liu, Songmao Wang, Yuanyuan Yang, Yinjun Zhang, and Jie Sun. 2022. "The Production of Pyruvate in Biological Technology: A Critical Review" Microorganisms 10, no. 12: 2454. https://doi.org/10.3390/microorganisms10122454
APA StyleYuan, W., Du, Y., Yu, K., Xu, S., Liu, M., Wang, S., Yang, Y., Zhang, Y., & Sun, J. (2022). The Production of Pyruvate in Biological Technology: A Critical Review. Microorganisms, 10(12), 2454. https://doi.org/10.3390/microorganisms10122454







