Internalization of Polymeric Bacterial Peptidoglycan Occurs through Either Actin or Dynamin Dependent Pathways
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Analysis of Primary Human Peripheral Blood Mononuclear Cells
2.3. Bacteria Strain, Peptidoglycan Purification and Labeling
2.4. Sizing of PGN Polymers Purified from Bacterial Cell Wall
2.5. Monocyte Internalization of FITC-Labeled PGN and/or hkBa
2.6. Fluorescence Microscopy Analysis of FITC-PGN Uptake by Monocytes
2.7. Flow Cytometry Analysis of Procoagulant and Proinflammatory Responses to PGN
2.8. Data Analysis and Representation
3. Results
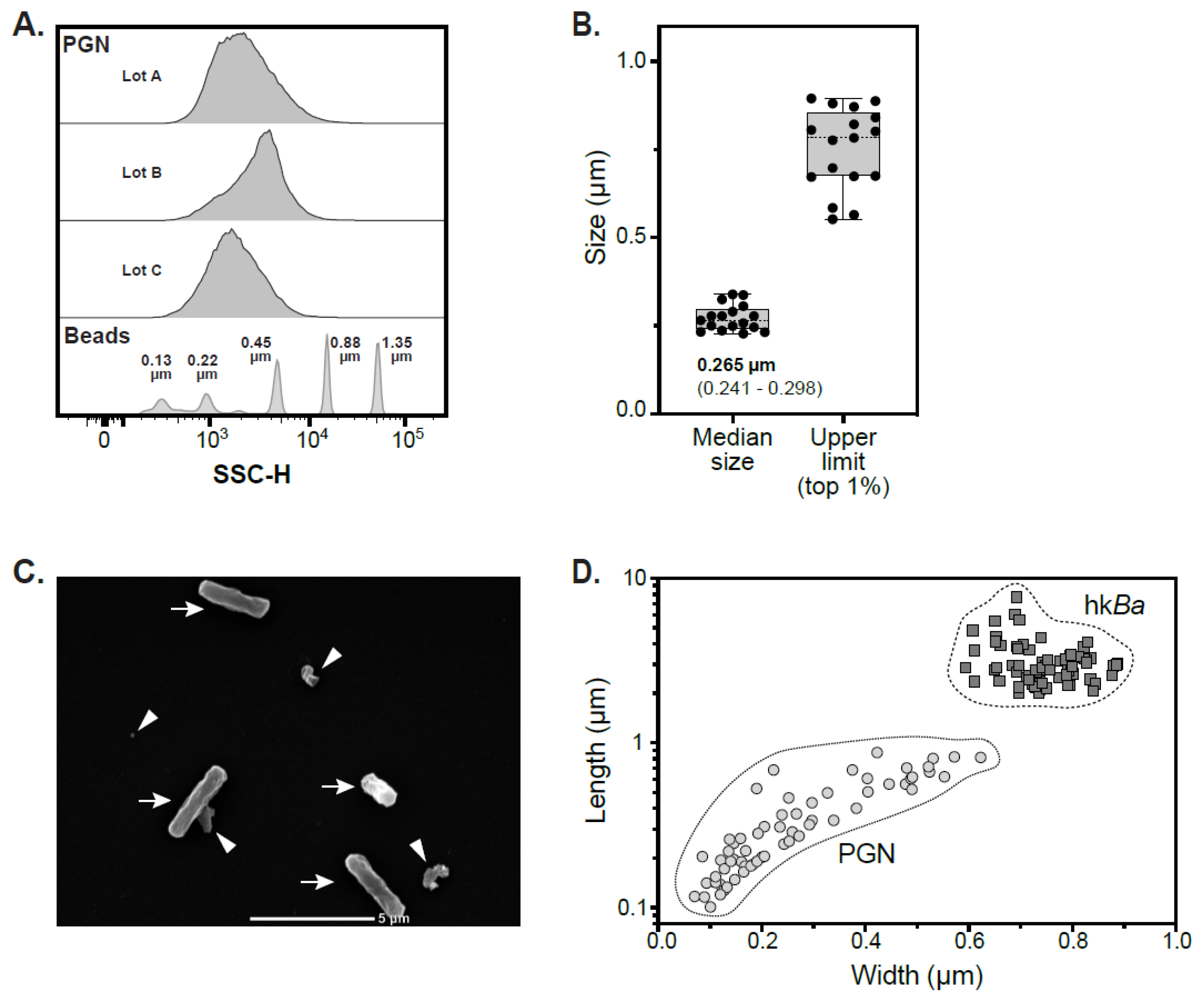
3.1. Sizing of PGN Macromolecules Purified from the Cell Wall of Bacillus Anthracis
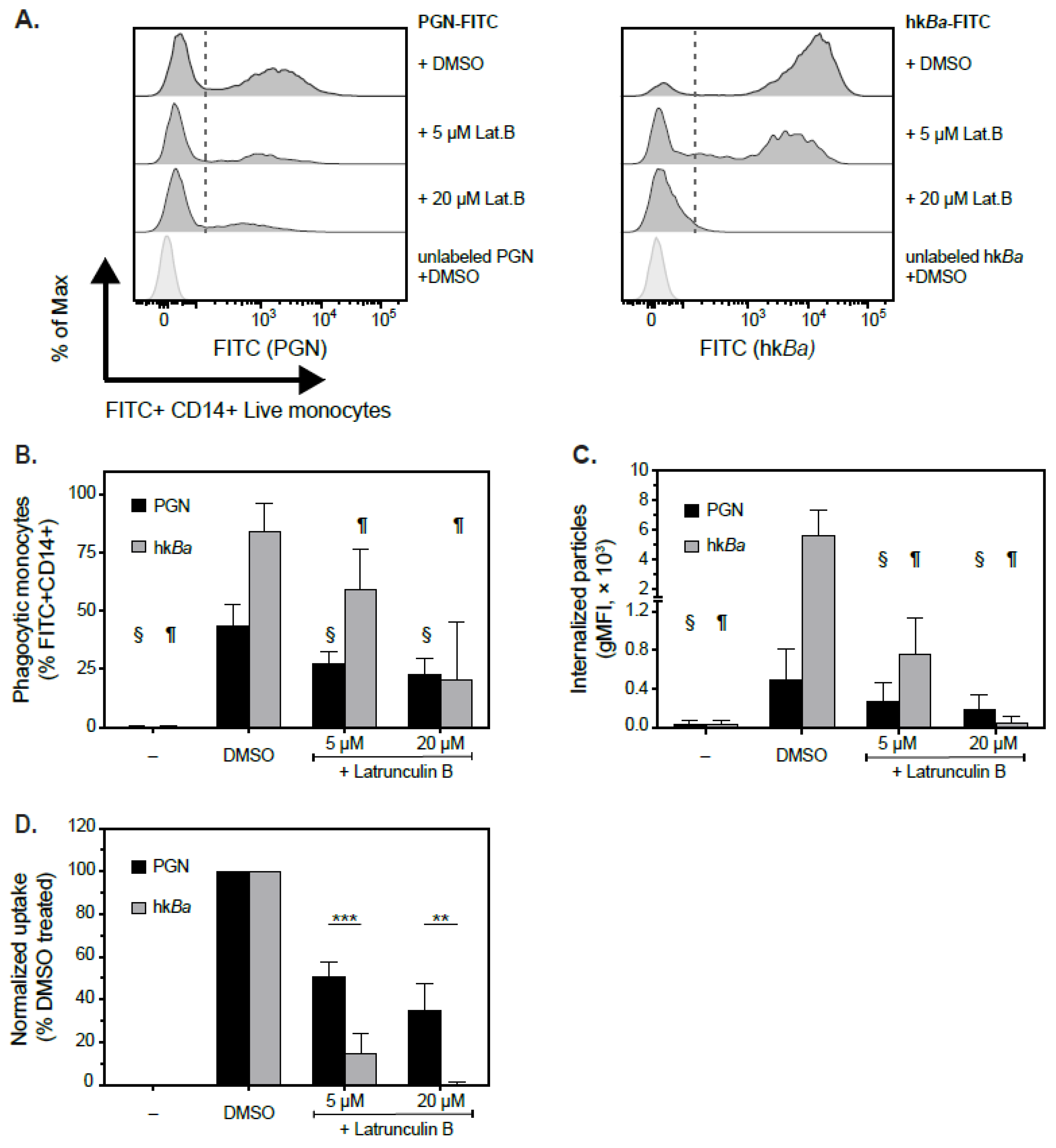
3.2. Pairwise Comparison of PGN and hkBa Internalization by Human Monocytes
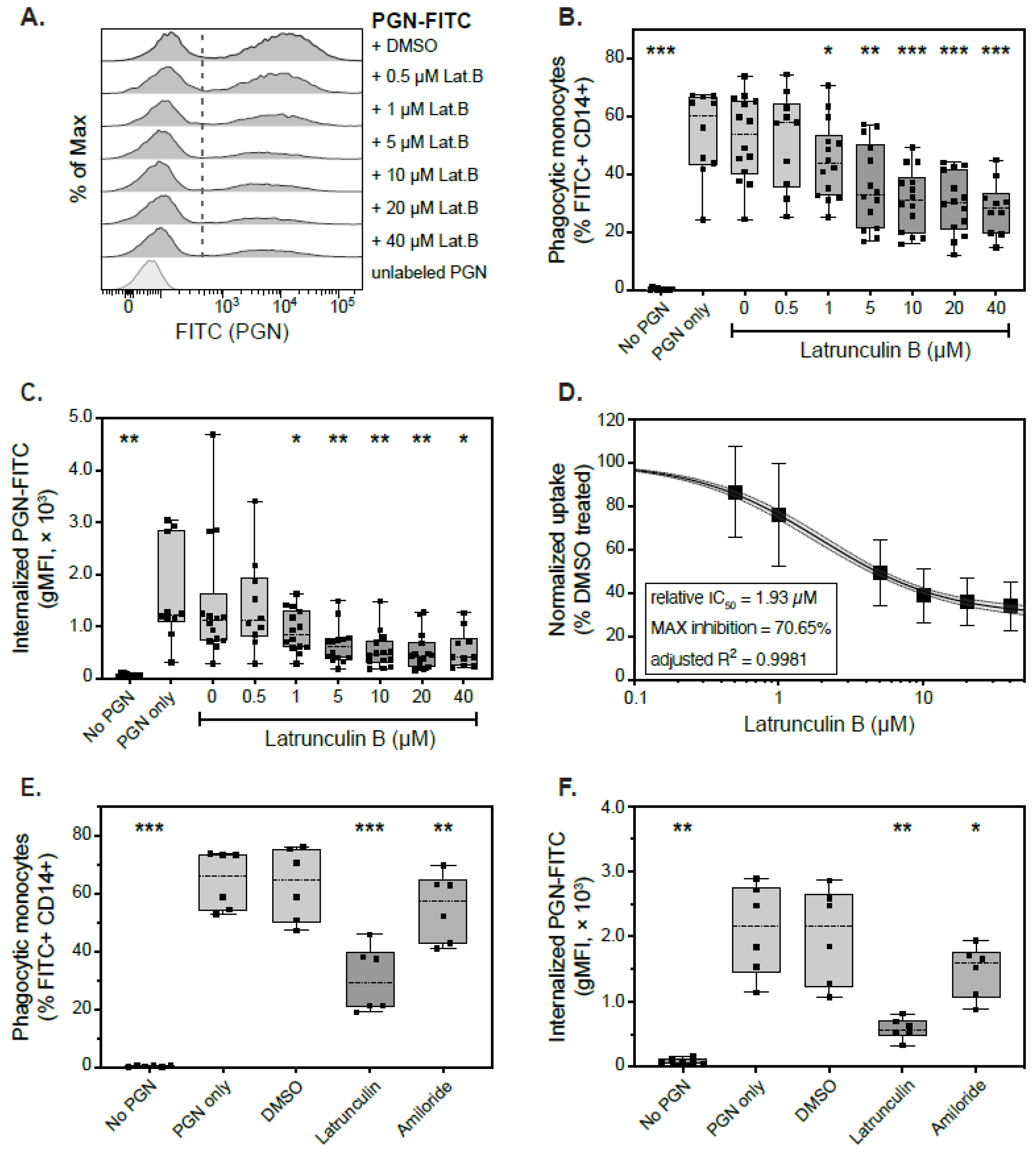
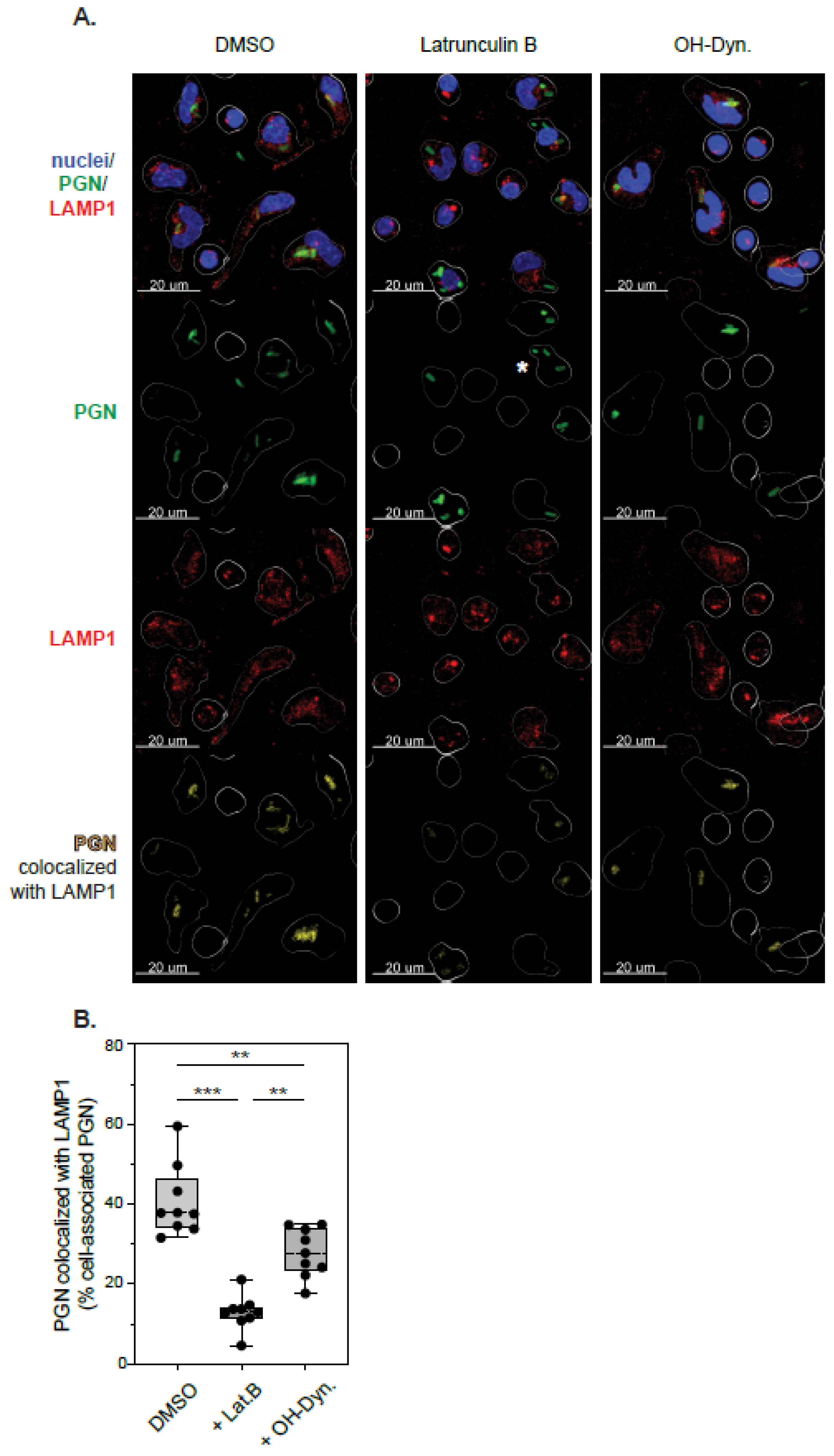
3.3. Actin-Dependent Mechanisms of PGN Uptake
3.4. Actin-Independent Internalization of PGN Uptake
3.5. Phagolysosomal Trafficking of Internalized PGN
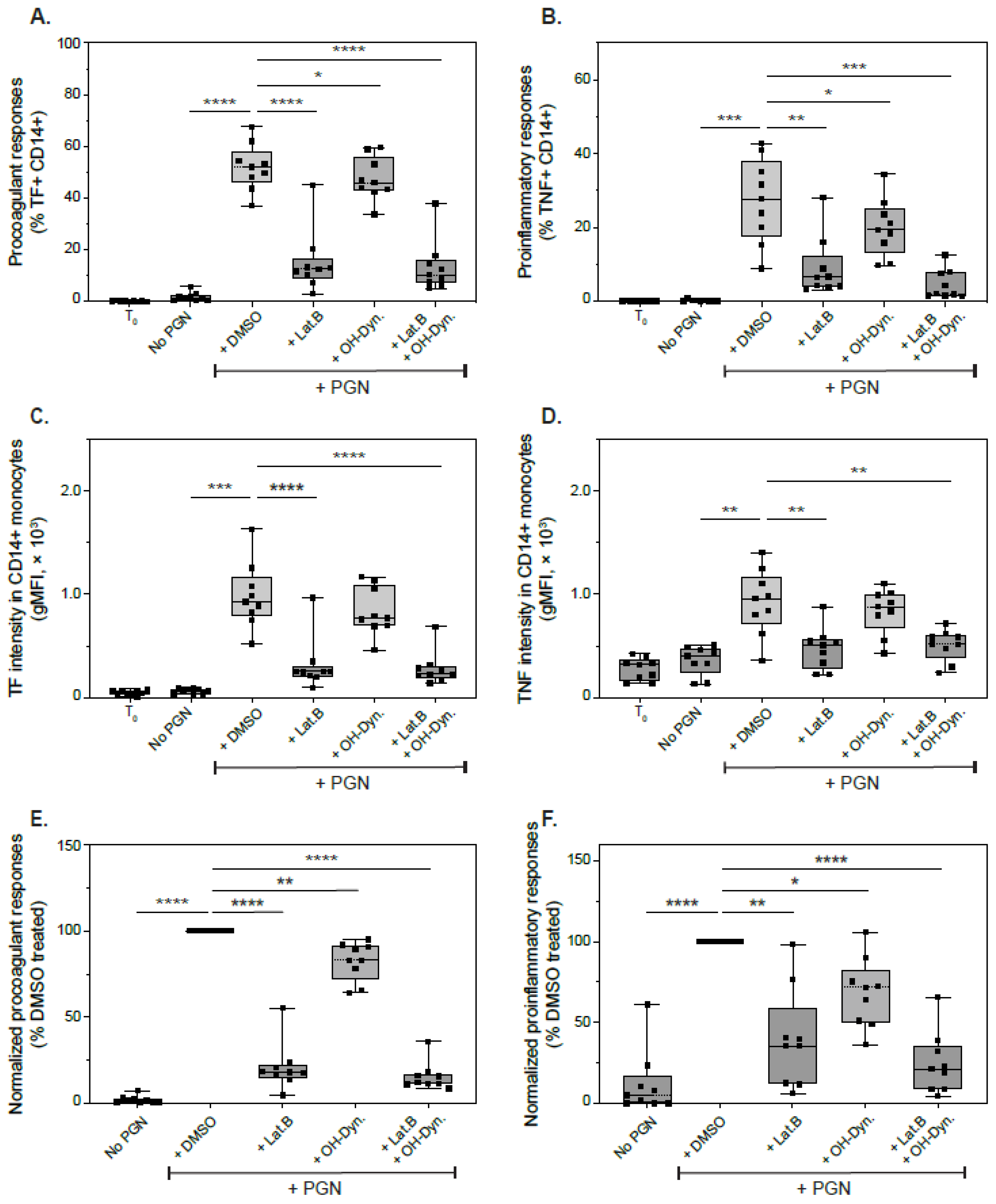
3.6. Effect of Endocytic Inhibitors on Immune Responses to PGN
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lin, A.E.; Guttman, J.A. Hijacking the endocytic machinery by microbial pathogens. Protoplasma 2010, 244, 75–90. [Google Scholar] [CrossRef]
- Flannagan, R.S.; Jaumouille, V.; Grinstein, S. The cell biology of phagocytosis. Annu. Rev. Pathol. 2012, 7, 61–98. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.P.; Mintern, J.D.; Gleeson, P.A. Macropinocytosis in Different Cell Types: Similarities and Differences. Membranes Basel 2020, 10, 177. [Google Scholar] [CrossRef] [PubMed]
- Mayor, S.; Pagano, R.E. Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell Biol. 2007, 8, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Harper, C.B.; Popoff, M.R.; McCluskey, A.; Robinson, P.J.; Meunier, F.A. Targeting membrane trafficking in infection prophylaxis: Dynamin inhibitors. Trends Cell Biol. 2013, 23, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Blanot, D.; De Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, A.J.; Reyes, C.N.; Liang, W.; Becker, C.; Shimada, K.; Wheeler, M.L.; Cho, H.C.; Popescu, N.I.; Coggeshall, K.M.; Arditi, M.; et al. Hexokinase Is an Innate Immune Receptor for the Detection of Bacterial Peptidoglycan. Cell 2016, 166, 624–636. [Google Scholar] [CrossRef] [Green Version]
- Langer, M.; Girton, A.W.; Popescu, N.I.; Burgett, T.; Metcalf, J.P.; Coggeshall, K.M. Neither Lys- and DAP-type peptidoglycans stimulate mouse or human innate immune cells via Toll-like receptor 2. PLoS ONE 2018, 13, e0193207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girardin, S.E.; Travassos, L.H.; Herve, M.; Blanot, D.; Boneca, I.G.; Philpott, D.J.; Sansonetti, P.J.; Mengin-Lecreulx, D. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J. Biol. Chem. 2003, 278, 41702–41708. [Google Scholar] [CrossRef] [Green Version]
- Sun, D.; Raisley, B.; Langer, M.; Iyer, J.K.; Vedham, V.; Ballard, J.L.; James, J.A.; Metcalf, J.; Coggeshall, K.M. Anti-peptidoglycan antibodies and Fcgamma receptors are the key mediators of inflammation in Gram-positive sepsis. J. Immunol. 2012, 189, 2423–2431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girton, A.W.; Popescu, N.I.; Keshari, R.S.; Burgett, T.; Lupu, F.; Coggeshall, K.M. Serum Amyloid P and IgG Exhibit Differential Capabilities in the Activation of the Innate Immune System in Response to Bacillus anthracis Peptidoglycan. Infect. Immun. 2018, 86, e00076-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keshari, R.S.; Popescu, N.I.; Silasi, R.; Regmi, G.; Lupu, C.; Simmons, J.H.; Ricardo, A.; Coggeshall, K.M.; Lupu, F. Complement C5 inhibition protects against hemolytic anemia and acute kidney injury in anthrax peptidoglycan-induced sepsis in baboons. Proc. Natl. Acad. Sci. USA 2021, 118, e2104347118. [Google Scholar] [CrossRef] [PubMed]
- Royet, J.; Gupta, D.; Dziarski, R. Peptidoglycan recognition proteins: Modulators of the microbiome and inflammation. Nat. Rev. Immunol. 2011, 11, 837–851. [Google Scholar] [CrossRef] [PubMed]
- Chamaillard, M.; Hashimoto, M.; Horie, Y.; Masumoto, J.; Qiu, S.; Saab, L.; Ogura, Y.; Kawasaki, A.; Fukase, K.; Kusumoto, S.; et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 2003, 4, 702–707. [Google Scholar] [CrossRef] [PubMed]
- Faustin, B.; Lartigue, L.; Bruey, J.M.; Luciano, F.; Sergienko, E.; Bailly-Maitre, B.; Volkmann, N.; Hanein, D.; Rouiller, I.; Reed, J.C. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol. Cell 2007, 25, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Popescu, N.I.; Silasi, R.; Keshari, R.S.; Girton, A.; Burgett, T.; Zeerleder, S.S.; Gailani, D.; Gruber, A.; Lupu, F.; Coggeshall, K.M. Peptidoglycan induces disseminated intravascular coagulation in baboons through activation of both coagulation pathways. Blood 2018, 132, 849–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jutras, B.L.; Lochhead, R.B.; Kloos, Z.A.; Biboy, J.; Strle, K.; Booth, C.J.; Govers, S.K.; Gray, J.; Schumann, P.; Vollmer, W.; et al. Borrelia burgdorferi peptidoglycan is a persistent antigen in patients with Lyme arthritis. Proc. Natl. Acad. Sci. USA 2019, 116, 13498–13507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Heel, D.A.; Ghosh, S.; Butler, M.; Hunt, K.A.; Lundberg, A.M.; Ahmad, T.; McGovern, D.P.; Onnie, C.; Negoro, K.; Goldthorpe, S.; et al. Muramyl dipeptide and toll-like receptor sensitivity in NOD2-associated Crohn’s disease. Lancet 2005, 365, 1794–1796. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, J.; Xu, X.; Wang, H.; Qiao, Y.; Chu, W.C.; Xu, S.; Chai, L.; Cottier, F.; Pavelka, N.; et al. Antibody neutralization of microbiota-derived circulating peptidoglycan dampens inflammation and ameliorates autoimmunity. Nat. Microbiol. 2019, 4, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Iyer, J.K.; Khurana, T.; Langer, M.; West, C.M.; Ballard, J.D.; Metcalf, J.P.; Merkel, T.J.; Coggeshall, K.M. Inflammatory cytokine response to Bacillus anthracis peptidoglycan requires phagocytosis and lysosomal trafficking. Infect. Immun. 2010, 78, 2418–2428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paradis-Bleau, C.; Markovski, M.; Uehara, T.; Lupoli, T.J.; Walker, S.; Kahne, D.E.; Bernhardt, T.G. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 2010, 143, 1110–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Typas, A.; Banzhaf, M.; Van den Berg van Saparoea, B.; Verheul, J.; Biboy, J.; Nichols, R.J.; Zietek, M.; Beilharz, K.; Kannenberg, K.; Von Rechenberg, M.; et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 2010, 143, 1097–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irazoki, O.; Hernandez, S.B.; Cava, F. Peptidoglycan Muropeptides: Release, Perception, and Functions as Signaling Molecules. Front. Microbiol. 2019, 10, 500. [Google Scholar] [CrossRef] [PubMed]
- Marina-Garcia, N.; Franchi, L.; Kim, Y.G.; Hu, Y.; Smith, D.E.; Boons, G.J.; Nunez, G. Clathrin- and dynamin-dependent endocytic pathway regulates muramyl dipeptide internalization and NOD2 activation. J. Immunol. 2009, 182, 4321–4327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, N.; Lill, J.R.; Phung, Q.; Jiang, Z.; Bakalarski, C.; De Maziere, A.; Klumperman, J.; Schlatter, M.; Delamarre, L.; Mellman, I. Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature 2014, 509, 240–244. [Google Scholar] [CrossRef]
- Pathirana, R.D.; Kaparakis-Liaskos, M. Bacterial membrane vesicles: Biogenesis, immune regulation and pathogenesis. Cell Microbiol. 2016, 18, 1518–1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaparakis, M.; Turnbull, L.; Carneiro, L.; Firth, S.; Coleman, H.A.; Parkington, H.C.; Le Bourhis, L.; Karrar, A.; Viala, J.; Mak, J.; et al. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell Microbiol. 2010, 12, 372–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thay, B.; Damm, A.; Kufer, T.A.; Wai, S.N.; Oscarsson, J. Aggregatibacter actinomycetemcomitans outer membrane vesicles are internalized in human host cells and trigger NOD1- and NOD2-dependent NF-kappaB activation. Infect. Immun. 2014, 82, 4034–4046. [Google Scholar] [CrossRef] [Green Version]
- Mayer-Scholl, A.; Hurwitz, R.; Brinkmann, V.; Schmid, M.; Jungblut, P.; Weinrauch, Y.; Zychlinsky, A. Human neutrophils kill Bacillus anthracis. PLoS Pathog. 2005, 1, e23. [Google Scholar] [CrossRef] [Green Version]
- Keshari, R.S.; Silasi, R.; Popescu, N.I.; Patel, M.M.; Chaaban, H.; Lupu, C.; Coggeshall, K.M.; Mollnes, T.E.; DeMarco, S.J.; Lupu, F. Inhibition of complement C5 protects against organ failure and reduces mortality in a baboon model of Escherichia coli sepsis. Proc. Natl. Acad. Sci. USA 2017, 114, E6390–E6399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, J.K.; Coggeshall, K.M. Cutting edge: Primary innate immune cells respond efficiently to polymeric peptidoglycan, but not to peptidoglycan monomers. J. Immunol. 2011, 186, 3841–3845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popescu, N.I.; Girton, A.; Burgett, T.; Lovelady, K.; Coggeshall, K.M. Monocyte procoagulant responses to anthrax peptidoglycan are reinforced by proinflammatory cytokine signaling. Blood Adv. 2019, 3, 2436–2447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popescu, N.I.; Keshari, R.S.; Cochran, J.; Coggeshall, K.M.; Lupu, F. C3 Opsonization of Anthrax Bacterium and Peptidoglycan Supports Recognition and Activation of Neutrophils. Microorganisms 2020, 8, 1039. [Google Scholar] [CrossRef]
- Langer, M.; Malykhin, A.; Maeda, K.; Chakrabarty, K.; Williamson, K.S.; Feasley, C.L.; West, C.M.; Metcalf, J.P.; Coggeshall, K.M. Bacillus anthracis peptidoglycan stimulates an inflammatory response in monocytes through the p38 mitogen-activated protein kinase pathway. PLoS ONE 2008, 3, e3706. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, J.A.; Bothun, A.M.; Annis, S.N.; Sheehan, H.; Ray, S.; Gao, Y.; Ivanov, A.R.; Khrapko, K.; Tilly, J.L.; Woods, D.C. A nanoscale, multi-parametric flow cytometry-based platform to study mitochondrial heterogeneity and mitochondrial DNA dynamics. Commun. Biol. 2019, 2, 258. [Google Scholar] [CrossRef]
- Peng, G.E.; Wilson, S.R.; Weiner, O.D. A pharmacological cocktail for arresting actin dynamics in living cells. Mol. Biol. Cell 2011, 22, 3986–3994. [Google Scholar] [CrossRef] [PubMed]
- Dejonghe, W.; Sharma, I.; Denoo, B.; De Munck, S.; Lu, Q.; Mishev, K.; Bulut, H.; Mylle, E.; De Rycke, R.; Vasileva, M.; et al. Disruption of endocytosis through chemical inhibition of clathrin heavy chain function. Nat. Chem. Biol. 2019, 15, 641–649. [Google Scholar] [CrossRef]
- Harper, C.B.; Martin, S.; Nguyen, T.H.; Daniels, S.J.; Lavidis, N.A.; Popoff, M.R.; Hadzic, G.; Mariana, A.; Chau, N.; McCluskey, A.; et al. Dynamin inhibition blocks botulinum neurotoxin type A endocytosis in neurons and delays botulism. J. Biol. Chem. 2011, 286, 35966–35976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, X.; Marchioni, F.; Evelyn, C.R.; Sipes, N.; Zhou, X.; Seibel, W.; Wortman, M.; Zheng, Y. Small-molecule inhibitors targeting G-protein-coupled Rho guanine nucleotide exchange factors. Proc. Natl. Acad. Sci. USA 2013, 110, 3155–3160. [Google Scholar] [CrossRef] [Green Version]
- Thottacherry, J.J.; Kosmalska, A.J.; Kumar, A.; Vishen, A.S.; Elosegui-Artola, A.; Pradhan, S.; Sharma, S.; Singh, P.P.; Guadamillas, M.C.; Chaudhary, N.; et al. Mechanochemical feedback control of dynamin independent endocytosis modulates membrane tension in adherent cells. Nat. Commun. 2018, 9, 4217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karnik, R.; Ludlow, M.J.; Abuarab, N.; Smith, A.J.; Hardy, M.E.; Elliott, D.J.; Sivaprasadarao, A. Endocytosis of HERG is clathrin-independent and involves arf6. PLoS ONE 2013, 8, e85630. [Google Scholar] [CrossRef] [PubMed]
- Benabdi, S.; Peurois, F.; Nawrotek, A.; Chikireddy, J.; Caneque, T.; Yamori, T.; Shiina, I.; Ohashi, Y.; Dan, S.; Rodriguez, R.; et al. Family-wide Analysis of the Inhibition of Arf Guanine Nucleotide Exchange Factors with Small Molecules: Evidence of Unique Inhibitory Profiles. Biochemistry 2017, 56, 5125–5133. [Google Scholar] [CrossRef]
- Dziarski, R.; Tapping, R.I.; Tobias, P.S. Binding of bacterial peptidoglycan to CD14. J. Biol. Chem. 1998, 273, 8680–8690. [Google Scholar] [CrossRef] [Green Version]
- Dixon, T.C.; Meselson, M.; Guillemin, J.; Hanna, P.C. Anthrax. N. Engl. J. Med. 1999, 341, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Popov, S.G. New Candidate Anthrax Pathogenic Factors; Springer: Totowa, NJ, USA, 2008; pp. 25–36. [Google Scholar] [CrossRef]
- Coggeshall, K.M.; Lupu, F.; Ballard, J.; Metcalf, J.P.; James, J.A.; Farris, D.; Kurosawa, S. The sepsis model: An emerging hypothesis for the lethality of inhalation anthrax. J. Cell Mol. Med. 2013, 17, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, D.A.; Hicks, C.W.; Cui, X.; Li, Y.; Eichacker, P.Q. Anthrax infection. Am. J. Respir. Crit. Care Med. 2011, 184, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Qiu, P.; Li, Y.; Shiloach, J.; Cui, X.; Sun, J.; Trinh, L.; Kubler-Kielb, J.; Vinogradov, E.; Mani, H.; Al-Hamad, M.; et al. Bacillus anthracis cell wall peptidoglycan but not lethal or edema toxins produces changes consistent with disseminated intravascular coagulation in a rat model. J. Infect. Dis. 2013, 208, 978–989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, D.; Popescu, N.I.; Raisley, B.; Keshari, R.S.; Dale, G.L.; Lupu, F.; Coggeshall, K.M. Bacillus anthracis peptidoglycan activates human platelets through FcgammaRII and complement. Blood 2013, 122, 571–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canton, J.; Schlam, D.; Breuer, C.; Gutschow, M.; Glogauer, M.; Grinstein, S. Calcium-sensing receptors signal constitutive macropinocytosis and facilitate the uptake of NOD2 ligands in macrophages. Nat. Commun. 2016, 7, 11284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loh, L.N.; Gao, G.; Tuomanen, E.I. Dissecting Bacterial Cell Wall Entry and Signaling in Eukaryotic Cells: An Actin-Dependent Pathway Parallels Platelet-Activating Factor Receptor-Mediated Endocytosis. mBio 2017, 8, e02030-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boucrot, E.; Ferreira, A.P.; Almeida-Souza, L.; Debard, S.; Vallis, Y.; Howard, G.; Bertot, L.; Sauvonnet, N.; McMahon, H.T. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature 2015, 517, 460–465. [Google Scholar] [CrossRef]
- Crowley, M.T.; Costello, P.S.; Fitzer-Attas, C.J.; Turner, M.; Meng, F.; Lowell, C.; Tybulewicz, V.L.; DeFranco, A.L. A critical role for Syk in signal transduction and phagocytosis mediated by Fcgamma receptors on macrophages. J. Exp. Med. 1997, 186, 1027–1039. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tohyama, Y.; Kadono, T.; He, J.; Miah, S.M.; Hazama, R.; Tanaka, C.; Tohyama, K.; Yamamura, H. Protein-tyrosine kinase Syk is required for pathogen engulfment in complement-mediated phagocytosis. Blood 2006, 107, 4554–4562. [Google Scholar] [CrossRef] [PubMed]
- Caron, E.; Hall, A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science 1998, 282, 1717–1721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.; Kono, H.; Hirose, N.; Okada, M.; Yamamoto, T.; Yamamoto, K.; Honda, Z. Differential involvement of Src family kinases in Fc gamma receptor-mediated phagocytosis. J. Immunol. 2000, 165, 473–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abrami, L.; Bischofberger, M.; Kunz, B.; Groux, R.; Van der Goot, F.G. Endocytosis of the anthrax toxin is mediated by clathrin, actin and unconventional adaptors. PLoS Pathog. 2010, 6, e1000792. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.; Werling, D.; Hope, J.C.; Taylor, G.; Howard, C.J. Caveolae and caveolin in immune cells: Distribution and functions. Trends Immunol. 2002, 23, 158–164. [Google Scholar] [CrossRef]
- Baorto, D.M.; Gao, Z.; Malaviya, R.; Dustin, M.L.; Van der Merwe, A.; Lublin, D.M.; Abraham, S.N. Survival of FimH-expressing enterobacteria in macrophages relies on glycolipid traffic. Nature 1997, 389, 636–639. [Google Scholar] [CrossRef] [PubMed]
- Peyron, P.; Bordier, C.; N’Diaye, E.N.; Maridonneau-Parini, I. Nonopsonic phagocytosis of Mycobacterium kansasii by human neutrophils depends on cholesterol and is mediated by CR3 associated with glycosylphosphatidylinositol-anchored proteins. J. Immunol. 2000, 165, 5186–5191. [Google Scholar] [CrossRef] [Green Version]
- Zimnicka, A.M.; Husain, Y.S.; Shajahan, A.N.; Sverdlov, M.; Chaga, O.; Chen, Z.; Toth, P.T.; Klomp, J.; Karginov, A.V.; Tiruppathi, C.; et al. Src-dependent phosphorylation of caveolin-1 Tyr-14 promotes swelling and release of caveolae. Mol. Biol. Cell 2016, 27, 2090–2106. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popescu, N.I.; Cochran, J.; Duggan, E.; Kluza, J.; Silasi, R.; Coggeshall, K.M. Internalization of Polymeric Bacterial Peptidoglycan Occurs through Either Actin or Dynamin Dependent Pathways. Microorganisms 2022, 10, 552. https://doi.org/10.3390/microorganisms10030552
Popescu NI, Cochran J, Duggan E, Kluza J, Silasi R, Coggeshall KM. Internalization of Polymeric Bacterial Peptidoglycan Occurs through Either Actin or Dynamin Dependent Pathways. Microorganisms. 2022; 10(3):552. https://doi.org/10.3390/microorganisms10030552
Chicago/Turabian StylePopescu, Narcis I., Jackie Cochran, Elizabeth Duggan, Jędrzej Kluza, Robert Silasi, and Kenneth Mark Coggeshall. 2022. "Internalization of Polymeric Bacterial Peptidoglycan Occurs through Either Actin or Dynamin Dependent Pathways" Microorganisms 10, no. 3: 552. https://doi.org/10.3390/microorganisms10030552
APA StylePopescu, N. I., Cochran, J., Duggan, E., Kluza, J., Silasi, R., & Coggeshall, K. M. (2022). Internalization of Polymeric Bacterial Peptidoglycan Occurs through Either Actin or Dynamin Dependent Pathways. Microorganisms, 10(3), 552. https://doi.org/10.3390/microorganisms10030552





