Acrylamide Elimination by Lactic Acid Bacteria: Screening, Optimization, In Vitro Digestion, and Mechanism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains of Bacteria
2.2. Preparation of Stock and Working Solutions of Acrylamide
2.3. Acrylamide Binding Assay—Preliminary Screening for Media Components and Bacterial Cultures
2.4. Optimization of Acrylamide Removal Using Box–Behnken Design
2.5. In Vitro Digestion by INFOGEST2.0 Model
2.6. Quantification of Acrylamide by LC-MS-MS
2.7. Understanding Mechanism of Acrylamide Binding by LAB
2.7.1. Preparation of Samples and Binding Assay
2.7.2. Transmission Electron Microscopy (TEM) Measurement
2.7.3. Estimation of Zeta Potential
2.7.4. Fourier Transform Infrared Spectroscopy (FTIR) Analysis
2.7.5. Scanning Electron Microscopy Coupled with Energy-Dispersive X-ray Spectroscopy (SEM-EDS)
2.8. Statistical Analysis
3. Results
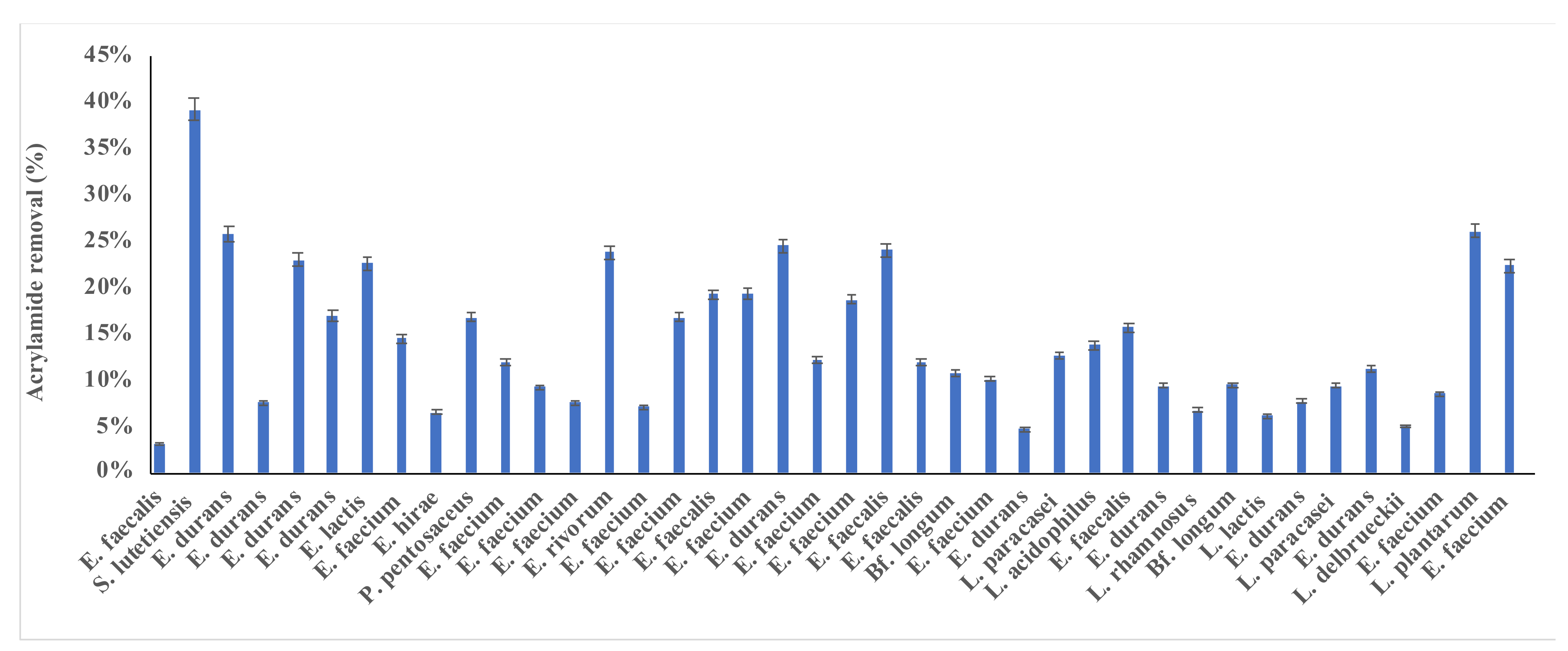
3.1. Screening of Acrylamide Removal by LAB
3.2. Optimization of Acrylamide Removal
- (1)
- Regression equation in uncoded units of L. plantarum
3.3. Acrylamide Removal Under In Vitro Digestion
3.4. Mechanisms of Acrylamide Removal
3.4.1. Estimation of Zeta Potential of Bacterial Cells
3.4.2. Fourier Transform Infrared Spectroscopy (FTIR) Analysis
3.4.3. Scanning Electron Microscopy Coupled with Energy-Dispersive X-ray Spectroscopy (SEM-EDS)
3.4.4. Transmission Electron Microscopy (TEM) Measurement
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- International Agency for Research on Cancer. IARC monographs on the evaluation of carcinogenic risks to humans. In Some Industrial Chemicals; International Agency for Research on Cancer: Lyon, France, 1994; pp. 389–433. [Google Scholar]
- Wu, B.; Chai, X.; He, A.; Huang, Z.; Chen, S.; Rao, P.; Ke, L.; Xiang, L. Inhibition of acrylamide toxicity in vivo by arginine-glucose maillard reaction products. Food Chem. Toxicol. 2021, 154, 112315. [Google Scholar] [CrossRef] [PubMed]
- Mottram, S.D.; Wedzicha, B.L.; Dodson, A.T. Food chemistry: Acrylamide is formed in the Maillard reaction. Nature 2002, 419, 448. [Google Scholar] [CrossRef] [PubMed]
- HEATOX Project. Guidelines to Authorities and Consumer Organisations on Home Cooking and Consumption; Food Quality and Safety: Oxford, UK, 2006. [Google Scholar]
- Codex Alimentarius. Codex Alimentarius. Code of Practice for the Reduction of Acrylamide in Foods. In Prevention and Reduction of Food and Feed Contamination; CAC/RCP 67–2009; FAO: Rome, Italy, 2009. [Google Scholar]
- European Food Safety Authority (EFSA). Scientific opinion on acrylamide in food: EFSA Panel on Contaminants in the Food Chain. EFSA J. 2015, 13, 4104. [Google Scholar]
- Albedwawi, A.S.; Turner, M.S.; Olaimat, A.N.; Osaili, T.M.; Al-Nabulsi, A.A.; Liu, S.-Q.; Shah, N.P.; Ayyash, M.M. An overview of microbial mitigation strategies for acrylamide: Lactic acid bacteria, yeast, and cell-free extracts. LWT 2021, 143, 111159. [Google Scholar] [CrossRef]
- Van Der Fels-Klerx, H.; Capuano, E.; Nguyen, H.; Mogol, B.A.; Kocadağlı, T.; Tas, N.; Hamzalıoğlu, A.; Van Boekel, M.; Gökmen, V. Acrylamide and 5-hydroxymethylfurfural formation during baking of biscuits: NaCl and temperature–time profile effects and kinetics. Food Res. Int. 2014, 57, 210–217. [Google Scholar] [CrossRef]
- Xu, C.; Yagiz, Y.; Marshall, S.; Li, Z.; Simonne, A.; Lu, J.; Marshall, M.R. Application of muscadine grape (Vitis rotundifolia Michx.) pomace extract to reduce carcinogenic acrylamide. Food Chem. 2015, 182, 200–208. [Google Scholar] [CrossRef]
- Xu, F.; Oruna-Concha, M.-J.; Elmore, J.S. The use of asparaginase to reduce acrylamide levels in cooked food. Food Chem. 2016, 210, 163–171. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, M.; Adhikari, B. Recent developments in frying technologies applied to fresh foods. Trends Food Sci. Technol. 2020, 98, 68–81. [Google Scholar] [CrossRef]
- Khorshidian, N.; Yousefi, M.; Shadnoush, M.; Siadat, S.D.; Mohammadi, M.; Mortazavian, A.M. Using probiotics for mitigation of acrylamide in food products: A mini review. Curr. Opin. Food Sci. 2020, 32, 67–75. [Google Scholar] [CrossRef]
- Bartkiene, E.; Jakobsone, I.; Juodeikiene, G.; Vidmantiene, D.; Pugajeva, I.; Bartkevics, V. Study on the reduction of acrylamide in mixed rye bread by fermentation with bacteriocin-like inhibitory substances producing lactic acid bacteria in combination with Aspergillus niger glucoamylase. Food Control 2013, 30, 35–40. [Google Scholar] [CrossRef]
- Bartkiene, E.; Jakobsone, I.; Pugajeva, I.; Bartkevics, V.; Zadeike, D.; Juodeikiene, G. Reducing of acrylamide formation in wheat biscuits supplemented with flaxseed and lupine. LWT 2016, 65, 275–282. [Google Scholar] [CrossRef]
- Mousavinejad, G.; Rezaei, K.; Khodaiyan, F. Reducing acrylamide in fried potato pancake using baker’s yeast, lactobacilli and microalgae. Qual. Assur. Saf. Crop. Foods 2015, 7, 779–787. [Google Scholar] [CrossRef]
- Rivas-Jimenez, L.; Ramírez-Ortiz, K.; González-Córdova, A.; Vallejo-Cordoba, B.; Garcia, H.; Hernandez-Mendoza, A. Evaluation of acrylamide-removing properties of two Lactobacillus strains under simulated gastrointestinal conditions using a dynamic system. Microbiol. Res. 2016, 190, 19–26. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Nematollahi, A.; Meybodi, N.M.; Khaneghah, A.M. An overview of the combination of emerging technologies with conventional methods to reduce acrylamide in different food products: Perspectives and future challenges. Food Control 2021, 127, 108144. [Google Scholar] [CrossRef]
- Alkalbani, N.; Turner, M.; Ayyash, M.M. Isolation, identification, and potential probiotic characterization of isolated lactic acid bacteria and in vitro investigation of the cytotoxicity, antioxidant, and antidiabetic activities in fermented sausage. Microb. Cell Factories 2019, 18, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Ayyash, M.; Abushelaibi, A.; Al-Mahadin, S.; Enan, M.; El-Tarabily, K.; Shah, N. In-vitro investigation into probiotic characterisation of Streptococcus and Enterococcus isolated from camel milk. LWT 2017, 87, 478–487. [Google Scholar] [CrossRef]
- Abushelaibi, A.; Al-Mahadin, S.; El-Tarabily, K.; Shah, N.P.; Ayyash, M. Characterization of potential probiotic lactic acid bacteria isolated from camel milk. LWT 2017, 79, 316–325. [Google Scholar] [CrossRef]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef]
- Shen, Y.; Zhao, S.; Zhao, X.; Sun, H.; Shao, M.; Xu, H. In vitro adsorption mechanism of acrylamide by lactic acid bacteria. LWT 2018, 100, 119–125. [Google Scholar] [CrossRef]
- Ge, N.; Xu, J.; Peng, B.; Pan, S. Adsorption mechanism of tenuazonic acid using inactivated lactic acid bacteria. Food Control 2017, 82, 274–282. [Google Scholar] [CrossRef]
- Lin, Z.; Ye, Y.; Li, Q.; Xu, Z.; Wang, M. A further insight into the biosorption mechanism of Au(III) by infrared spectrometry. BMC Biotechnol. 2011, 11, 98. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Chen, G.; Li, Y. Effect of added sugars and amino acids on acrylamide formation in white pan bread. Cereal Chem. 2019, 96, 545–553. [Google Scholar] [CrossRef]
- Alexandraki, V.; Tsakalidou, E.; Papadimitriou, K.; Holzapfel, W. Status and Trend of the Conservation and Sustainable Use of Micro-organism in Food Processes; FAO: Rome, Italy, 2013; p. 65. [Google Scholar]
- Serrano-Niño, J.; Cavazos-Garduño, A.; González-Córdova, A.; Vallejo-Cordoba, B.; Hernández-Mendoza, A.; Garcia, H.S. In vitro Study of the Potential Protective Role of L actobacillus Strains by Acrylamide Binding. J. Food Saf. 2014, 34, 62–68. [Google Scholar] [CrossRef]
- Luz, C.; Ferrer, J.; Mañes, J.; Meca, G. Toxicity reduction of ochratoxin A by lactic acid bacteria. Food Chem. Toxicol. 2018, 112, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Jin, H.; Lan, J.; Zhang, R.; Ren, H.; Zhang, X.; Yu, G. Detoxification of zearalenone by three strains of lactobacillus plantarum from fermented food in vitro. Food Control 2015, 54, 158–164. [Google Scholar] [CrossRef]
- Hernández-Mendoza, A.; Garcia, H.S.; Steele, J.L. Screening of Lactobacillus casei strains for their ability to bind aflatoxin B. Food Chem. Toxicol. 2009, 47, 1064–1068. [Google Scholar] [CrossRef]
- Zhai, Q.; Yin, R.; Yu, L.; Wang, G.; Tian, F.; Yu, R.; Zhao, J.; Liu, X.; Chen, Y.Q.; Zhang, H.; et al. Screening of lactic acid bacteria with potential protective effects against cadmium toxicity. Food Control 2015, 54, 23–30. [Google Scholar] [CrossRef]
- Wang, L.; Yue, T.; Yuan, Y.; Wang, Z.; Ye, M.; Cai, R. A new insight into the adsorption mechanism of patulin by the heat-inactive lactic acid bacteria cells. Food Control 2015, 50, 104–110. [Google Scholar] [CrossRef]






| Runs | Temperature (°C) (X1) | pH (X2) | Time (h) (X3) | NaCl (g/100 mL) (X4) | (%) Growth | |
|---|---|---|---|---|---|---|
| S. lutetiensis | L. plantarum | |||||
| 1 | 42 | 6.5 | 18 | 0.0 | 17.6 | 52.9 |
| 2 | 42 | 5.5 | 18 | 1.5 | 35.1 | 53.7 |
| 3 | 42 | 5.5 | 14 | 1.5 | 24.8 | 56.2 |
| 4 | 37 | 6.5 | 18 | 1.5 | 21.4 | 8.0 |
| 5 | 37 | 5.5 | 22 | 0.0 | 12.9 | 15.5 |
| 6 | 32 | 5.5 | 22 | 1.5 | 25.7 | 16.8 |
| 7 | 37 | 4.5 | 14 | 3.0 | 24.4 | 20.0 |
| 8 | 37 | 4.5 | 18 | 1.5 | 35.8 | 24.7 |
| 9 | 42 | 5.5 | 18 | 1.5 | 28.8 | 54.0 |
| 10 | 37 | 5.5 | 18 | 0.0 | 27.6 | 34.6 |
| 11 | 37 | 5.5 | 14 | 1.5 | 33.6 | 45.0 |
| 12 | 32 | 6.5 | 18 | 3.0 | 21.8 | 8.3 |
| 13 | 37 | 6.5 | 22 | 1.5 | 24.9 | 42.5 |
| 14 | 37 | 6.5 | 14 | 1.5 | 35.0 | 39.7 |
| 15 | 37 | 5.5 | 18 | 0.0 | 33.6 | 43.0 |
| 16 | 32 | 5.5 | 18 | 0.0 | 46.5 | 7.9 |
| 17 | 37 | 6.5 | 18 | 0.0 | 27.5 | 41.0 |
| 18 | 37 | 5.5 | 14 | 3.0 | 25.4 | 35.5 |
| 19 | 42 | 4.5 | 18 | 3.0 | 27.3 | 55.5 |
| 20 | 32 | 5.5 | 14 | 1.5 | 33.6 | 9.3 |
| 21 | 37 | 4.5 | 18 | 1.5 | 30.0 | 38.5 |
| 22 | 37 | 5.5 | 22 | 1.5 | 24.6 | 47.0 |
| 23 | 32 | 4.5 | 18 | 3.0 | 15.8 | 14.7 |
| 24 | 37 | 5.5 | 18 | 3.0 | 27.6 | 50.6 |
| 25 | 37 | 4.5 | 22 | 1.5 | 32.7 | 51.4 |
| 26 | 32 | 5.5 | 18 | 1.5 | 20.1 | 12.9 |
| 27 | 42 | 5.5 | 22 | 1.5 | 23.6 | 56.8 |
| Source | S. lutetiensis | L. plantarum | |||
|---|---|---|---|---|---|
| DF | F-Value | p-Value | F-Value | p-Value | |
| Model | 14 | 2.31 | 0.077 | 7.00 | 0.001 |
| Linear | 4 | 1.89 | 0.177 | 15.96 | 0.000 |
| Temp | 1 | 0.01 | 0.911 | 62.51 | 0.000 |
| pH | 1 | 1.59 | 0.232 | 0.34 | 0.569 |
| Time | 1 | 6.97 | 0.022 | 0.39 | 0.546 |
| Salt | 1 | 0.65 | 0.437 | 1.84 | 0.200 |
| Square | 4 | 0.62 | 0.658 | 3.30 | 0.048 |
| Temp × Temperature | 1 | 0.01 | 0.922 | 4.86 | 0.048 |
| pH × pH | 1 | 1.51 | 0.243 | 2.63 | 0.131 |
| Time × Time | 1 | 0.45 | 0.513 | 4.42 | 0.057 |
| Salt × Salt | 1 | 1.03 | 0.330 | 2.55 | 0.136 |
| 2-Way Interaction | 6 | 3.15 | 0.043 | 3.07 | 0.047 |
| Temperature × pH | 1 | 1.45 | 0.251 | 0.00 | 0.988 |
| Temperature × Time | 1 | 0.38 | 0.550 | 0.16 | 0.695 |
| Temperature × Salt | 1 | 9.07 | 0.011 | 0.18 | 0.678 |
| pH × Time | 1 | 0.00 | 0.980 | 0.59 | 0.457 |
| pH × Salt | 1 | 4.11 | 0.065 | 0.25 | 0.627 |
| Time × Salt | 1 | 4.17 | 0.064 | 13.49 | 0.003 |
| Error | 12 | ||||
| Lack-of-Fit | 9 | 1.84 | 0.335 | 1.92 | 0.321 |
| Pure Error | 3 | ||||
| Total | 26 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albedwawi, A.S.; Al Sakkaf, R.; Yusuf, A.; Osaili, T.M.; Al-Nabulsi, A.; Liu, S.-Q.; Palmisano, G.; Ayyash, M.M. Acrylamide Elimination by Lactic Acid Bacteria: Screening, Optimization, In Vitro Digestion, and Mechanism. Microorganisms 2022, 10, 557. https://doi.org/10.3390/microorganisms10030557
Albedwawi AS, Al Sakkaf R, Yusuf A, Osaili TM, Al-Nabulsi A, Liu S-Q, Palmisano G, Ayyash MM. Acrylamide Elimination by Lactic Acid Bacteria: Screening, Optimization, In Vitro Digestion, and Mechanism. Microorganisms. 2022; 10(3):557. https://doi.org/10.3390/microorganisms10030557
Chicago/Turabian StyleAlbedwawi, Amal S., Reem Al Sakkaf, Ahmed Yusuf, Tareq M. Osaili, Anas Al-Nabulsi, Shao-Quan Liu, Giovanni Palmisano, and Mutamed M. Ayyash. 2022. "Acrylamide Elimination by Lactic Acid Bacteria: Screening, Optimization, In Vitro Digestion, and Mechanism" Microorganisms 10, no. 3: 557. https://doi.org/10.3390/microorganisms10030557
APA StyleAlbedwawi, A. S., Al Sakkaf, R., Yusuf, A., Osaili, T. M., Al-Nabulsi, A., Liu, S.-Q., Palmisano, G., & Ayyash, M. M. (2022). Acrylamide Elimination by Lactic Acid Bacteria: Screening, Optimization, In Vitro Digestion, and Mechanism. Microorganisms, 10(3), 557. https://doi.org/10.3390/microorganisms10030557








