Recombinant PilS: Cloning, Expression and Biochemical Characterization of a Pil-Fimbriae Subunit
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Plasmids
2.2. PilS Amplification
2.3. pilS Gene Cloning
2.4. Expression of PilS
2.5. Refolding at High Hydrostatic Pressure (HHP) of PilS-IBs
2.6. Purification of PilS
2.7. PilS Biochemical Characterization
2.7.1. In Gel Digestion
2.7.2. Mass Spectrometry and In Silico Analysis
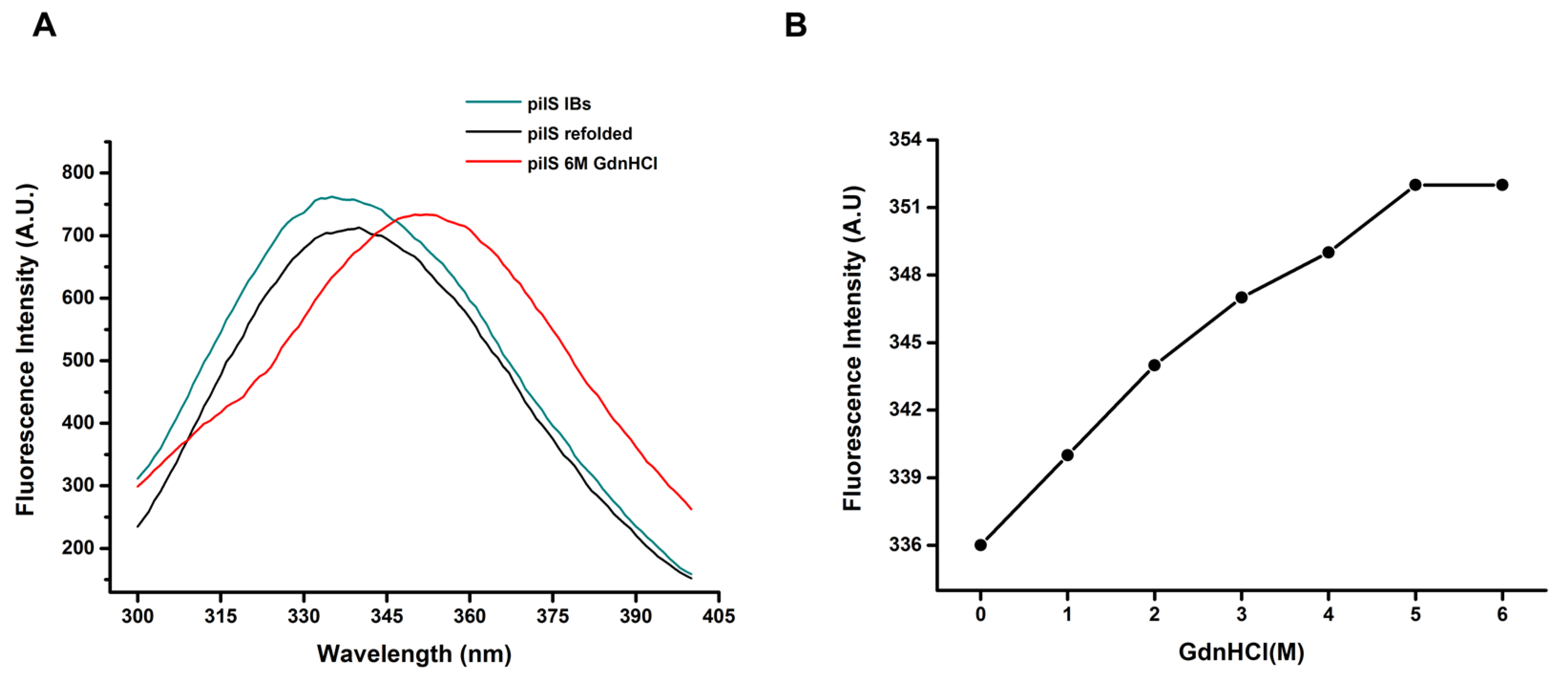
2.7.3. Fluorescence Spectroscopy
2.7.4. Circular Dichroism
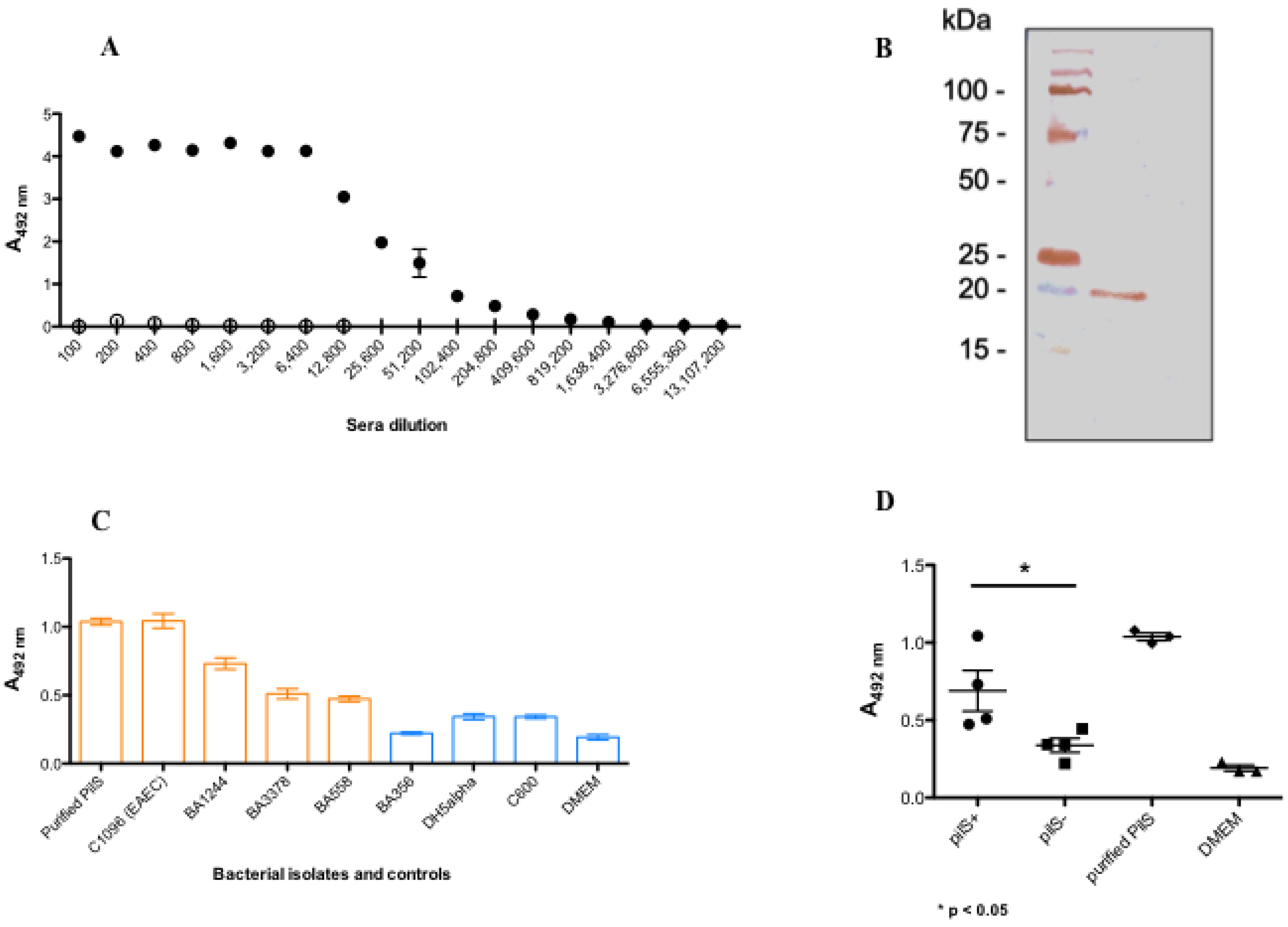
2.8. Generation of PilS Polyclonal Antibody
2.9. Detection of PilS in Bacterial Cells by Using Polyclonal Antibody
3. Results
3.1. Cloning and Sequencing of PilS
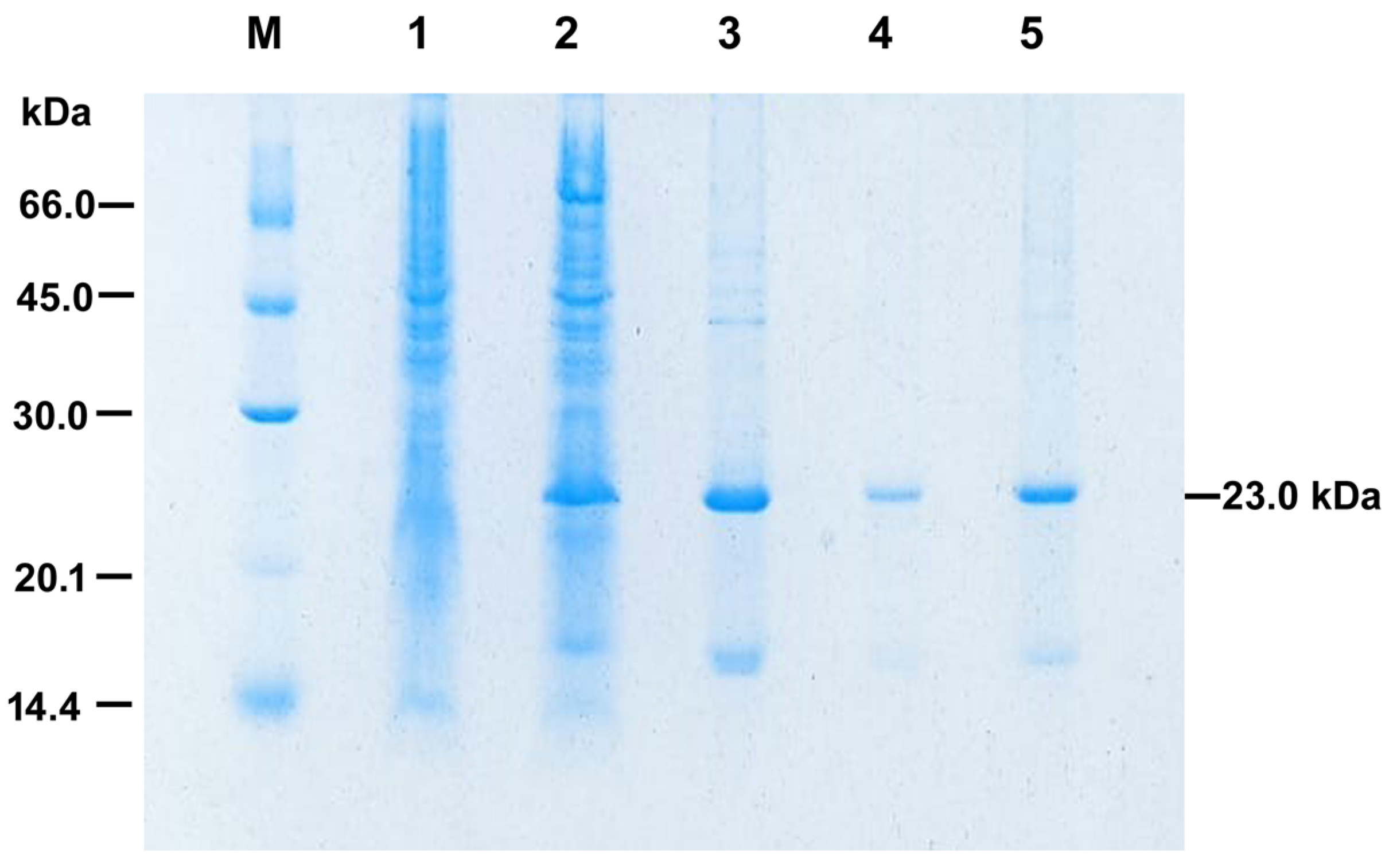
3.2. Solubilization and Refolding at HHP of PilS
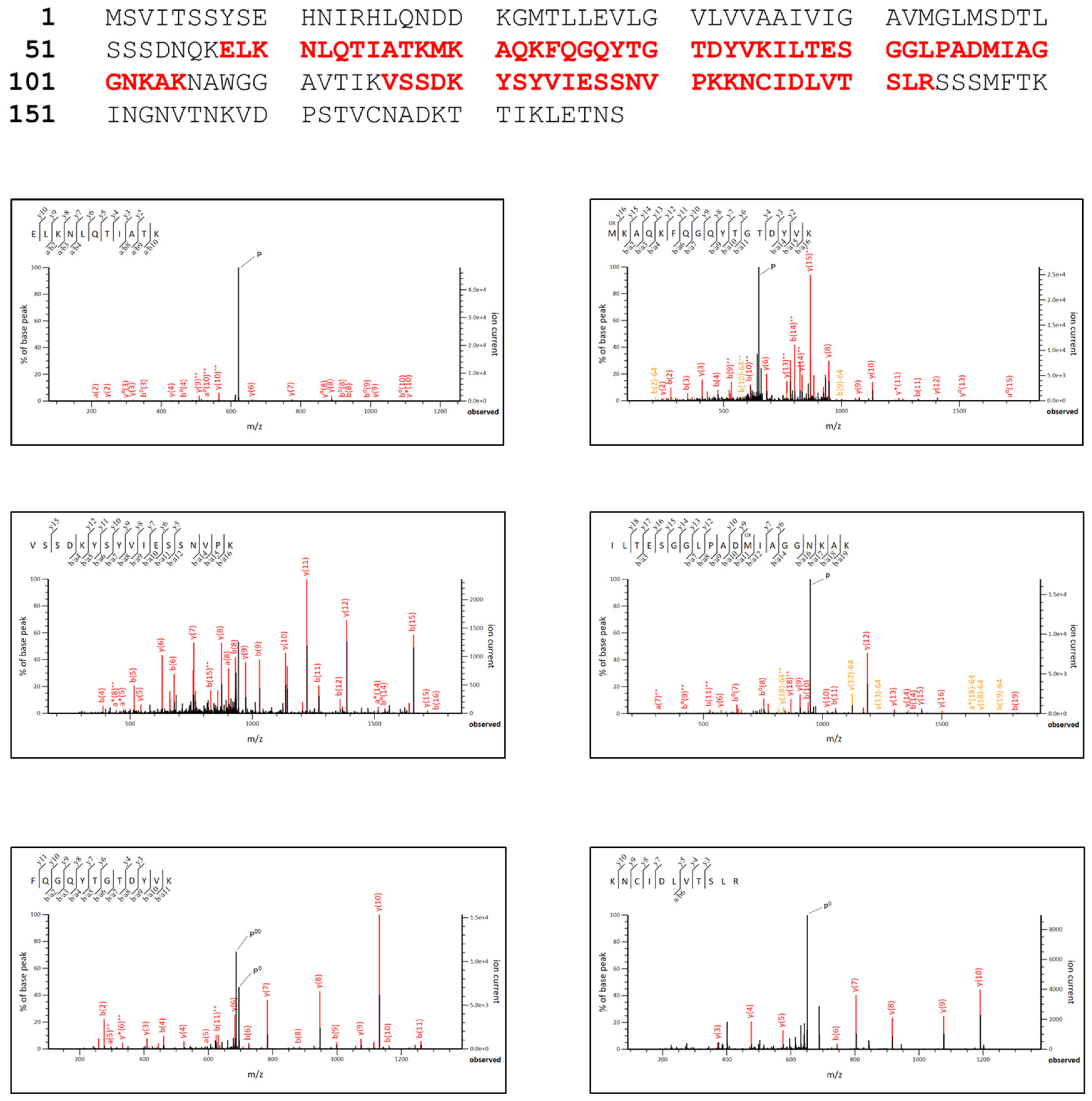
3.3. rPilS Identification
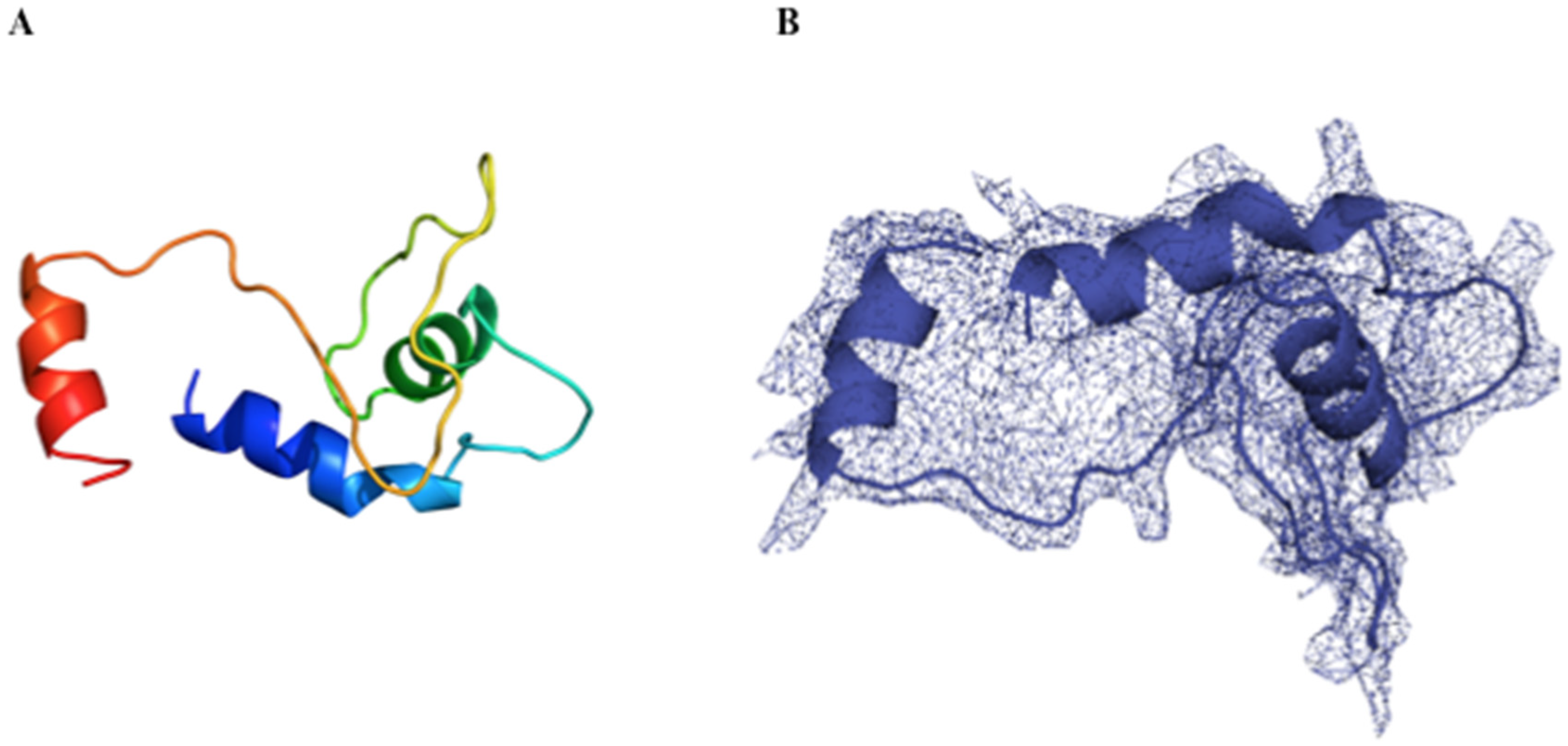
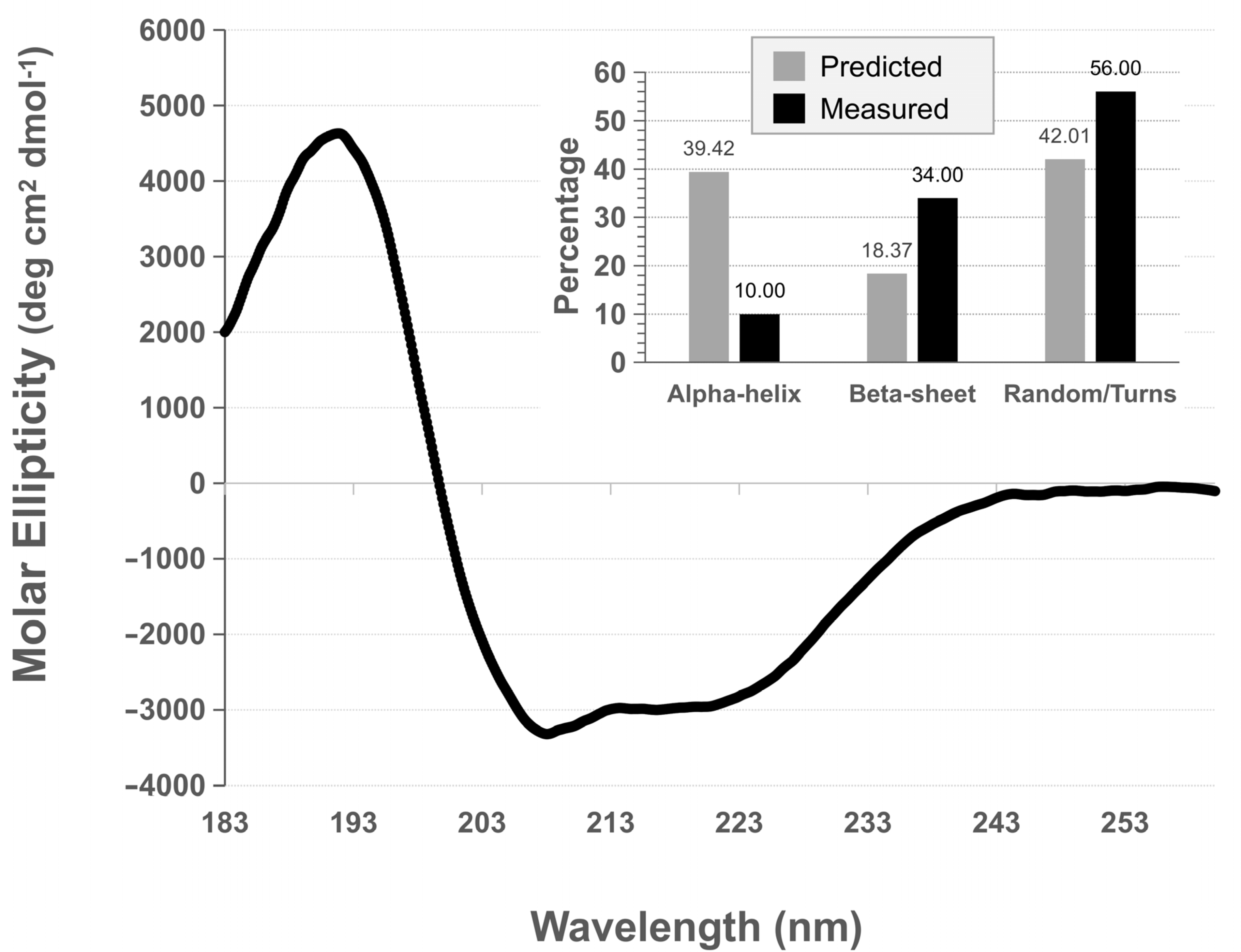
3.4. rPilS Structure
3.5. rPilS Recovered by HHP Generates a Polyclonal Antibody Able to Recognize Strains Harboring PilS
4. Discussion
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kline, K.A.; Fälker, S.; Dahlberg, S.; Normark, S.; Henriques-Normark, B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe 2009, 5, 580–592. [Google Scholar] [CrossRef] [Green Version]
- DuPont, H.L. Persistent Diarrhea: A Clinical Review. JAMA 2016, 315, 2712–2723. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proft, T.; Baker, E.N. Pili in Gram-negative and Gram-positive bacteria—Structure, assembly and their role in disease. Cell. Mol. Life Sci. 2009, 66, 613–635. [Google Scholar] [CrossRef] [PubMed]
- Giltner, C.L.; Nguyen, Y.; Burrows, L.L. Type IV pilin proteins: Versatile molecular modules. Microbiol. Mol. Biol. Rev. 2012, 76, 740–772. [Google Scholar] [CrossRef] [Green Version]
- Albers, S.V.; Pohlschröder, M. Diversity of archaeal type IV pilin-like structures. Extremophiles 2009, 13, 403–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alphonse, S.; Durand, E.; Douzi, B.; Waegele, B.; Darbon, H.; Filloux, A.; Voulhoux, R.; Bernard, C. Structure of the Pseudomonas aeruginosa XcpT pseudopilin, a major component of the type II secretion system. J. Struct. 2010, 169, 75–80. [Google Scholar] [CrossRef]
- Aroeti, B.; Friedman, G.; Zlotkin-Rivkin, E.; Donnenberg, M.S. Retraction of enteropathogenic E. coli type IV pili promotes efficient host cell colonization, effector translocation and tight junction disruption. Gut Microbes 2012, 3, 267–271. [Google Scholar] [CrossRef] [Green Version]
- Strom, M.S.; Lory, S. Structure-function and biogenesis of the type IV pili. Annu. Rev Microbiol. 1993, 47, 565–596. [Google Scholar] [CrossRef]
- Collyn, F.; Léty, M.A.; Nair, S.; Escuyer, V.; Ben Younes, A.; Simonet, M.; Marceau, M. Yersinia pseudotuberculosis harbors a type IV pilus gene cluster that contributes to pathogenicity. Infect. Immun. 2002, 70, 6196–6205. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.R.; Komano, T. The plasmid R64 thin pilus identified as a type IV pilus. J. Bacteriol. 1997, 179, 3594–3603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishiwa, A.; Komano, T. PilV adhesins of plasmid R64 thin pili specifically bind to the lipopolysaccharides of recipient cells. J. Mol. Biol. 2004, 343, 615–625. [Google Scholar] [CrossRef]
- Munhoz, D.D.; Nara, J.M.; Freitas, N.C.; Moraes, C.T.P.; Nunes, K.O.; Yamamoto, B.B.; Vasconcellos, F.M.; Martínez-Laguna, Y.; Girón, J.A.; Martins, F.H.; et al. Distribution of Major Pilin Subunit Genes Among Atypical Enteropathogenic Escherichia coli and Influence of Growth Media on Expression of the ecp Operon. Front. Microbiol. 2018, 9, 942. [Google Scholar] [CrossRef]
- Garcia, B.G.; Castro, F.S.; Vieira, M.A.M.; Girão, D.M.; Uenishi, L.T.; Cergole-Novella, M.C.; Dos Santos, L.F.; Piazza, R.M.F.; Hernandes, R.T.; Gomes, T.A.T. Distribution of the pilS gene in Escherichia coli pathovars, its transfer ability and influence in the typical enteropathogenic E. coli adherence phenotype. Int. J. Med. Microbiol. 2019, 309, 66–72. [Google Scholar] [CrossRef]
- Zhang, X.L.; Tsui, I.S.; Yip, C.M.; Fung, A.W.; Wong, D.K.; Dai, X.; Yang, Y.; Hackett, J.; Morris, C. Salmonella enterica serovar typhi uses type IVB pili to enter human intestinal epithelial cells. Infect. Immun. 2000, 68, 3067–3073. [Google Scholar] [CrossRef] [Green Version]
- Srimanote, P.; Paton, A.W.; Paton, J.C. Characterization of a novel type IV pilus locus encoded on the large plasmid of locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 2002, 70, 3094–3100. [Google Scholar] [CrossRef] [Green Version]
- Komano, T. Shufflons: Multiple inversion systems and integrons. Annu. Rev. Genet. 1999, 33, 171–191. [Google Scholar] [CrossRef] [PubMed]
- Sakai, D.; Komano, T. Genes required for plasmid R64 thin-pilus biogenesis: Identification and localization of products of the pilK, pilM, pilO, pilP, pilR, and pilT genes. J. Bacteriol. 2002, 184, 444–451. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Furuya, N.; Ishikura, M.; Isobe, T.; Haino-Fukushima, K.; Ogawa, T.; Komano, T. Purification and characterization of thin pili of IncI1 plasmids ColIb-P9 and R64: Formation of PilV-specific cell aggregates by type IV pili. J. Bacteriol. 1998, 180, 2842–2848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudley, E.G.; Abe, C.; Ghigo, J.M.; Latour-Lambert, P.; Hormazabal, J.C.; Nataro, J.P. An IncI1 plasmid contributes to the adherence of the atypical enteroaggregative Escherichia coli strain C1096 to cultured cells and abiotic surfaces. Infect. Immun. 2006, 74, 2102–2114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rathore, A.S.; Bade, P.; Joshi, V.; Pathak, M.; Pattanayek, S.K. Refolding of biotech therapeutic proteins expressed in bacteria: Review. J. Chem. Technol. Biotechnol. 2013, 10, 1794–1806. [Google Scholar] [CrossRef]
- Baeshen, M.N.; Al-Hejin, A.M.; Bora, R.S.; Ahmed, M.M.M.; Ramadan, H.A.I.; Saini, K.S.; Baeshen, N.A.; Redwan, E.M. Production of Biopharmaceuticals in E. coli: Current Scenario and Future Perspectives. J. Microbiol. Biotechnol. 2015, 25, 953–962. [Google Scholar] [CrossRef]
- Bueris, V.; Sircili, M.P.; Taddei, C.R.; dos Santos, M.F.; Franzolin, M.R.; Martinez, M.B.; Ferrer, S.R.; Barreto, M.L.; Trabulsi, L.R. Detection of diarrheagenic Escherichia coli from children with and without diarrhea in Salvador, Bahia, Brazil. Mem. Inst. Oswaldo Cruz 2007, 102, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Abe, C.M.; Trabulsi, L.R.; Blanco, J.; Blanco, M.; Dahbi, G.; Blanco, J.E.; Mora, A.; Franzolin, M.R.; Taddei, C.R.; Martinez, M.B.; et al. Virulence features of atypical enteropathogenic Escherichia coli identified by the eae(+) EAF-negative stx(-) genetic profile. Diagn. Microbiol. Infect. Dis. 2009, 64, 357–365. [Google Scholar] [CrossRef]
- Cobeljić, M.; Miljković-Selimović, B.; Paunović-Todosijević, D.; Velicković, Z.; Lepsanović, Z.; Zec, N.; Savić, D.; Ilić, R.; Konstantinović, S.; Jovanović, B.; et al. Enteroaggregative Escherichia coli associated with an outbreak of diarrhoea in a neonatal nursery ward. Epidemiol. Infect. 1996, 117, 11–16. [Google Scholar] [CrossRef] [Green Version]
- Sambrook, J.; Fritsch, E.R.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Hanahan, D. Techniques for transformation of E. coli. In DNA Cloning: A Practical Approach; Glover, D.M., Ed.; IRL Press: Oxford, UK, 1985. [Google Scholar]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Jensen, E.B.; Carlsen, S. Production of recombinant human growth hormone in Escherichia coli: Expression of different precursors and physiological effects of glucose, acetate, and salts. Biotechnol. Bioeng. 1990, 36, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chura-Chambi, R.M.; Cordeiro, Y.; Malavasi, N.V.; Lemke, L.S.; Rodrigues, D.; Morganti, L. An analysis of the factors that affect the dissociation of inclusion bodies and the refolding of endostatin under high pressure. Process. Biochem. 2013, 48, 250–259. [Google Scholar] [CrossRef] [Green Version]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef]
- Whitmore, L.; Wallace, B.A. Dichroweb: An online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004, 32, W668–W673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lobley, A.; Whitmore, L.; Wallace, B.A. DICHROWEB: An interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 2002, 18, 211–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Studier, F.W. Analysis of bacteriophage T7 early RNAs and proteins on slab gels. J. Mol. Biol. 1973, 79, 237–248. [Google Scholar] [CrossRef]
- Chura-Chambi, R.M.; Prieto-da-Silva, A.R.B.; Di Lela, M.M.; Oliveira, J.E.; Abreu, P.E.A.; Meireles, L.R.; de Andrade, H.F., Jr.; Morganti, L. High level SARS-CoV-2 nucleocapsid refolding using mild condition for inclusion bodies solubilization: Application of high pressure at pH 9.0. PLoS ONE 2022, 17, e0262591. [Google Scholar] [CrossRef] [PubMed]
- Chura-Chambi, R.M.; da Silva, C.M.R.; Pereira, L.R.; Bartolini, P.; Ferreira, L.C.S.; Morganti, L. Protein refolding based on high hydrostatic pressure and alkaline pH: Application on a recombinant dengue virus NS1 protein. PLoS ONE 2019, 14, e0211162. [Google Scholar] [CrossRef]
- Singh, S.M.; Sharma, A.; Upadhyay, A.K.; Singh, A.; Garg, L.C.; Panda, A.K. Solubilization of inclusion body proteins using n-propanol and its refolding into bioactive form. Protein Expr. Purif. 2012, 81, 75–82. [Google Scholar] [CrossRef]
- Crisman, R.L.; Randolph, T.W. Refolding of proteins from inclusion bodies is favored by a diminished hydrophobic effect at elevated pressures. Biotechnol. Bioeng. 2009, 1022, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.L.; Oliveira, A.C.; Vieira, T.C.; de Oliveira, G.A.; Suarez, M.C.; Foguel, D. High-pressure chemical biology and biotechnology. Chem. Rev. 2014, 114, 7239–7267. [Google Scholar] [CrossRef] [PubMed]
- Yongsawatdigul, J.; Park, J.W. Effects of alkali and acid solubilization on gelation characteristics of rockfish muscle proteins. J. Food Sci. 2004, 69, C499–C505. [Google Scholar] [CrossRef]
- da Silva, C.M.R.; Chura-Chambi, R.M.; Ramos Pereira, L.; Cordeiro, Y.; de Souza Ferreira, L.C.; Morganti, L. Association of high pressure and alkaline condition for solubilization of inclusion bodies and refolding of the NS1 protein from zika virus. BMC Biotechnol. 2018, 18, 78. [Google Scholar] [CrossRef] [Green Version]
- Arakawa, T.; Ejima, D.; Tsumoto, K.; Obeyama, N.; Tanaka, Y.; Kita, Y.; Timasheff, S.N. Suppression of protein interactions by arginine: A proposed mechanism of the arginine effects. Biophys. Chem. 2007, 127, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, M.; Mattick, J.S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: A general system for the formation of surface-associated protein complexes. Mol. Microbiol. 1993, 2, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-J.; Wang, A.H.-J.; Jennings, M.P. Discovery of virulence factors of pathogenic bacteria. Curr. Opin. Chem. Biol. 2008, 12, 93–101. [Google Scholar] [CrossRef]
- Hernandes, R.T.; Velsko, I.; Sampaio, S.C.; Elias, W.P.; Robins-Browne, R.M.; Gomes, T.A.; Girón, J.A. Fimbrial adhesins produced by atypical enteropathogenic Escherichia coli strains. Appl. Environ. Microbiol. 2011, 77, 8391–8399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, M.S.; Wigneshweraraj, S. Regulation of virulence gene expression. Virulence 2014, 5, 832–834. [Google Scholar] [CrossRef] [PubMed]
- Bakour, S.; Sankar, S.A.; Rathored, J.; Biagini, P.; Raoult, D.; Fournier, P.E. Identification of virulence factors and antibiotic resistance markers using bacterial genomics. Future Microbiol. 2016, 11, 455–466. [Google Scholar] [CrossRef] [PubMed]


| Strains | Characteristic | Reference |
| BA558 | aEPEC pilS+ | [13] |
| BA1244 | aEPEC pilS+ | [13] |
| BA3378 | aEPEC pilS+ | [13] |
| BA356 | aEPEC pilS- | [13] |
| C600 | Non-pathogenic E. coli–Negative control | [27] |
| DH5alpha | Non-pathogenic E. coli–Negative control | [27] |
| C1096 | aEAEC | [20,26] |
| JM109 | E. coli strain used for cloning and plasmid maintenance | [28] |
| BL21 (DE3) | E. coli strain used as expression host | [29] |
| Plasmids | Characteristic | Reference |
| pGEM-T | Replication vector | Promega, Madison, WI, USA |
| pET28a | Expression vector | Novagen, Darmstadt, Hesse, Germany |
| Primer | Annealing Temperature | Fragment |
|---|---|---|
| (F) GGATCCATGAGCGTCATAACCTGTTC | 50 °C | 537 pb |
| (R) AAGCTTTTAACTGTTGGTTTCCAGTTT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munhoz, D.D.; Silva, J.C.A.; Freitas, N.C.; Iwai, L.K.; Aires, K.A.; Ozaki, C.Y.; Souza, C.S.; Rocha, L.B.; Silva, M.A.; Henrique, I.M.; et al. Recombinant PilS: Cloning, Expression and Biochemical Characterization of a Pil-Fimbriae Subunit. Microorganisms 2022, 10, 1174. https://doi.org/10.3390/microorganisms10061174
Munhoz DD, Silva JCA, Freitas NC, Iwai LK, Aires KA, Ozaki CY, Souza CS, Rocha LB, Silva MA, Henrique IM, et al. Recombinant PilS: Cloning, Expression and Biochemical Characterization of a Pil-Fimbriae Subunit. Microorganisms. 2022; 10(6):1174. https://doi.org/10.3390/microorganisms10061174
Chicago/Turabian StyleMunhoz, Danielle D., Jessika C. A. Silva, Natalia C. Freitas, Leo K. Iwai, Karina A. Aires, Christiane Y. Ozaki, Cristiane S. Souza, Letícia B. Rocha, Miriam A. Silva, Izabella M. Henrique, and et al. 2022. "Recombinant PilS: Cloning, Expression and Biochemical Characterization of a Pil-Fimbriae Subunit" Microorganisms 10, no. 6: 1174. https://doi.org/10.3390/microorganisms10061174






