Degrading Characterization of the Newly Isolated Nocardioides sp. N39 for 3-Amino-5-methyl-isoxazole and the Related Genomic Information
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Culture Medium
2.2. Isolation and Identification of N39 and N40
2.3. Analytic Methods
2.4. Degradation Experiments in Different Conditions
2.5. Genomic DNA Extraction, Sequencing and Comparison
3. Results and Discussion
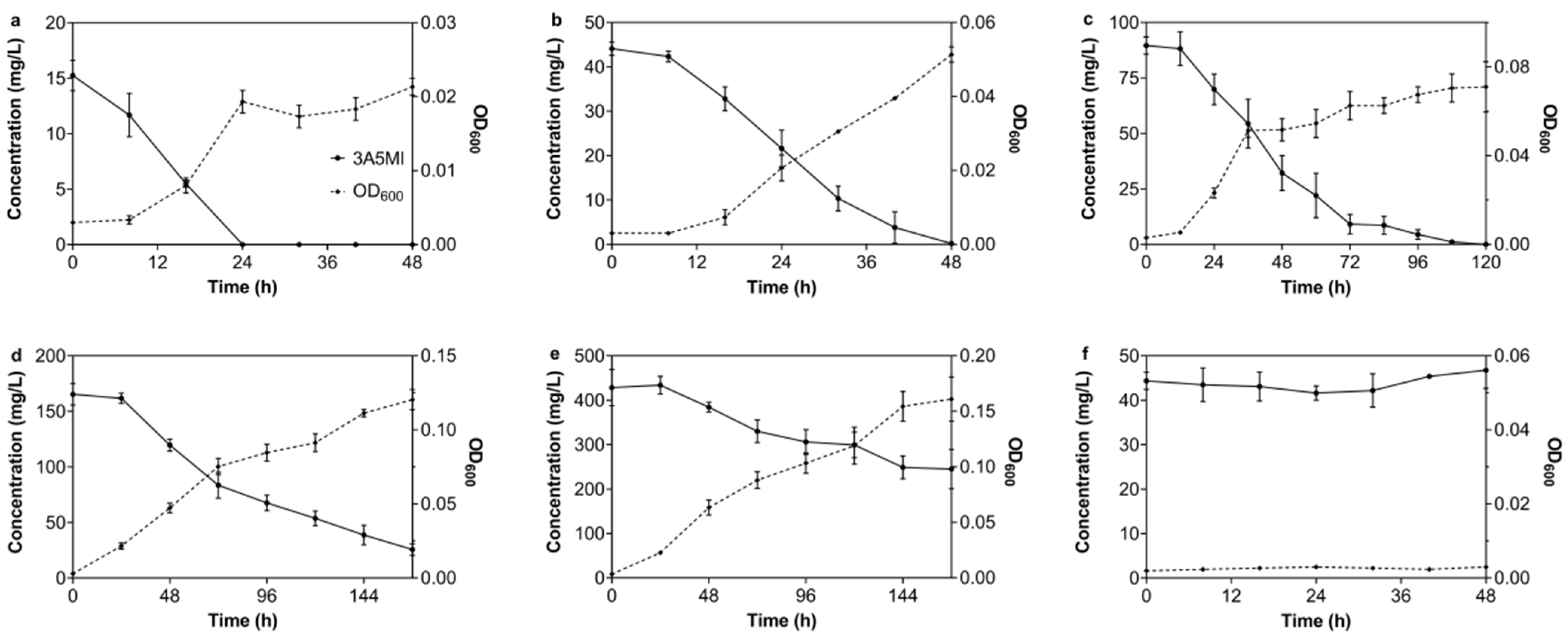
3.1. Isolation of Strains N39 and N40
3.2. Effect of Different Factors on 3A5MI Degradation by N39
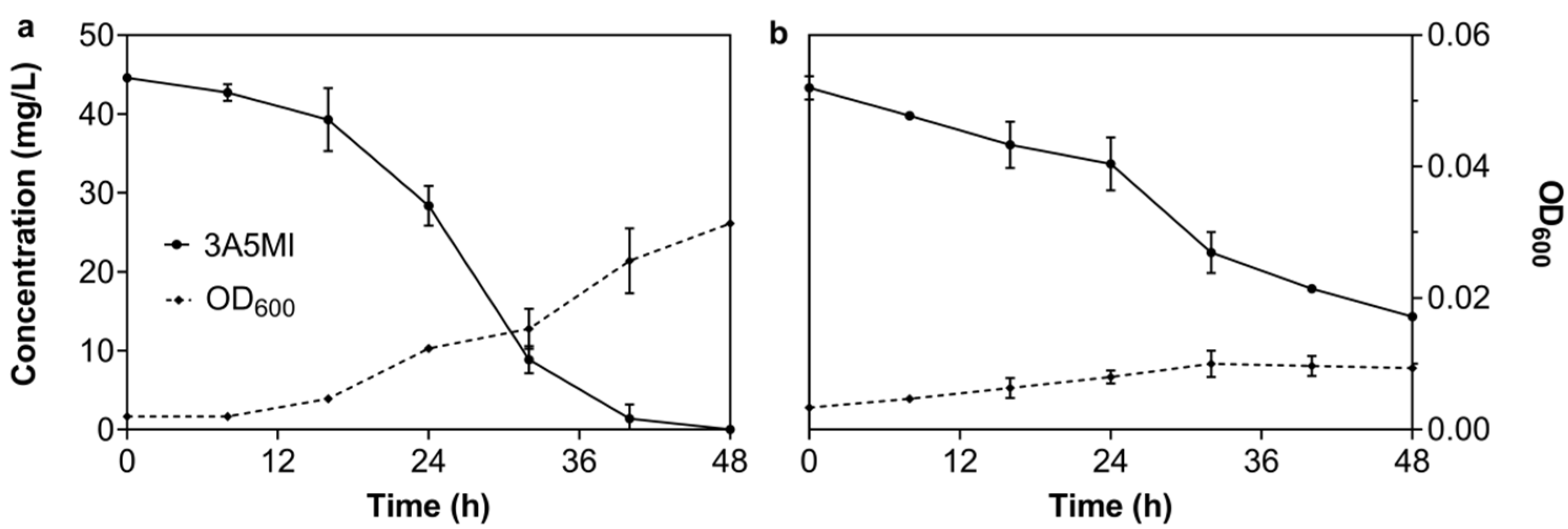
3.2.1. Effect of the Initial 3A5MI Concentration on the Degradation Ability of N39
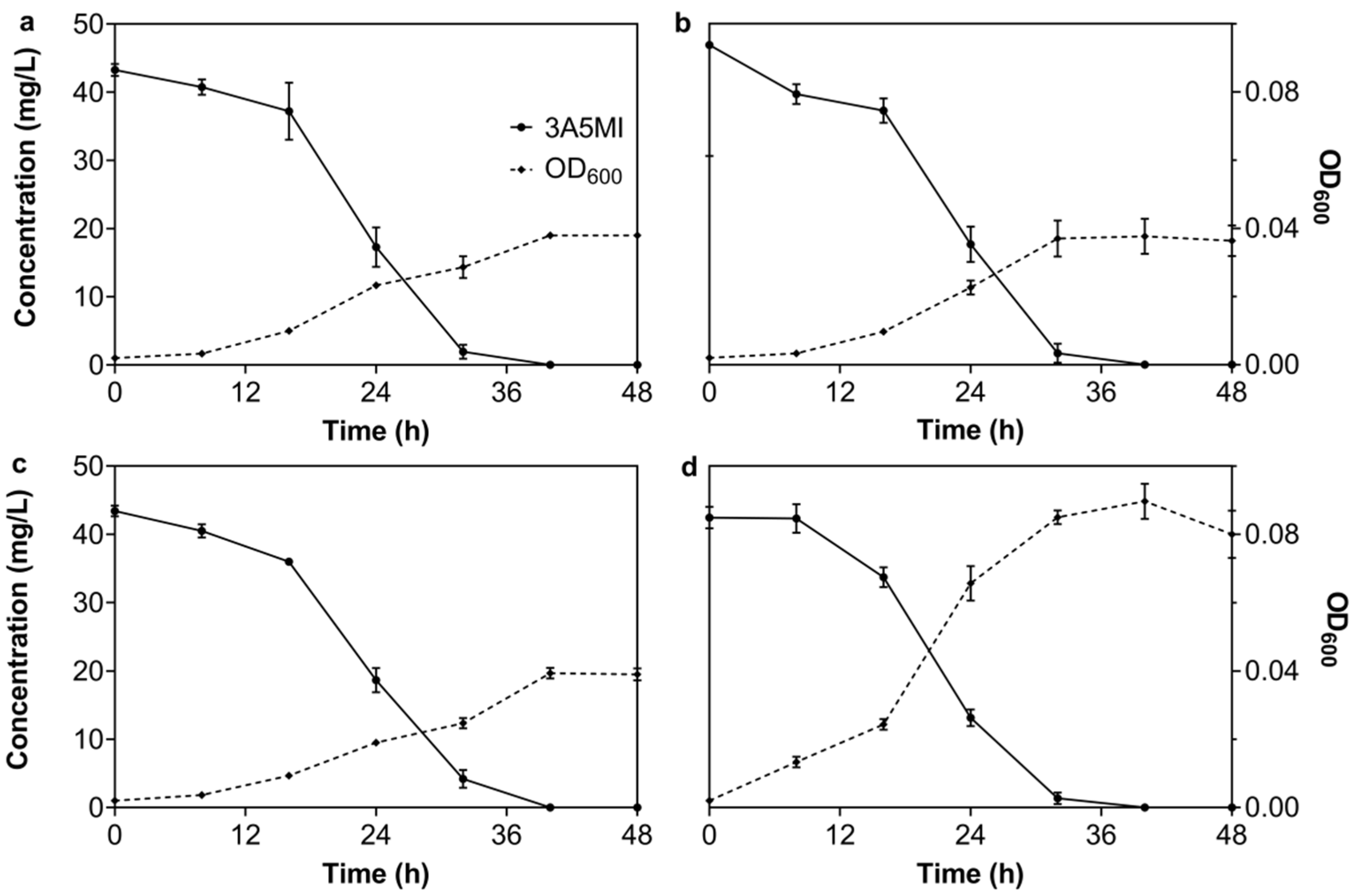
3.2.2. Effect of Temperature on the Degradation of 3A5MI by N39
3.2.3. Effect of the pH Value on the Degradation of 3A5MI by N39
3.2.4. Effect of Dissolved Oxygen on the Degrading Ability of N39
3.2.5. Effect of Additional Carbon or Nitrogen on the Degrading Ability of N39
3.2.6. The Degrading Spectrum of N39 for 3A5MI Analogs
3.3. Genomic Characteristic of the Strains N39 and N40
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Q.-Q.; Ying, G.-G.; Pan, C.-G.; Liu, Y.-S.; Zhao, J.-L. Comprehensive evaluation of antibiotics emission and fate in the river basins of China: Source analysis, multimedia modeling, and linkage to bacterial resistance. Environ. Sci. Technol. 2015, 49, 6772–6782. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-R.; Owens, G.; Kwon, S.-I.; So, K.-H.; Lee, D.-B.; Ok, Y.S. Occurrence and environmental fate of veterinary antibiotics in the terrestrial environment. Water Air Soil Poll. 2011, 214, 163–174. [Google Scholar] [CrossRef]
- Wang, M.; Shen, W.; Yan, L.; Wang, X.-H.; Xu, H. Stepwise impact of urban wastewater treatment on the bacterial community structure, antibiotic contents, and prevalence of antimicrobial resistance. Environ. Pollut. 2017, 231, 1578–1585. [Google Scholar] [CrossRef] [PubMed]
- Valcárcel, Y.; Alonso, S.G.; Rodríguez-Gil, J.; Gil, A.; Catalá, M. Detection of pharmaceutically active compounds in the rivers and tap water of the Madrid Region (Spain) and potential ecotoxicological risk. Chemosphere 2011, 84, 1336–1348. [Google Scholar] [CrossRef] [PubMed]
- Cabeza, Y.; Candela, L.; Ronen, D.; Teijon, G. Monitoring the occurrence of emerging contaminants in treated wastewater and groundwater between 2008 and 2010. The Baix Llobregat (Barcelona, Spain). J. Hazard. Mater. 2012, 239, 32–39. [Google Scholar] [CrossRef]
- Bartelt-Hunt, S.; Snow, D.D.; Damon-Powell, T.; Miesbach, D. Occurrence of steroid hormones and antibiotics in shallow groundwater impacted by livestock waste control facilities. J. Contam. Hydrol. 2011, 123, 94–103. [Google Scholar] [CrossRef]
- Lapworth, D.J.; Baran, N.; Stuart, M.E.; Ward, R.S. Emerging organic contaminants in groundwater: A review of sources, fate and occurrence. Environ. Pollut. 2012, 163, 287–303. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Xie, Y.; Wang, J.; Christakos, G.; Si, J.; Zhao, H.; Ding, Y.; Li, J. Influence of planting patterns on fluoroquinolone residues in the soil of an intensive vegetable cultivation area in northern China. Sci. Total Environ. 2013, 458, 63–69. [Google Scholar] [CrossRef]
- Hu, X.; Zhou, Q.; Luo, Y. Occurrence and source analysis of typical veterinary antibiotics in manure, soil, vegetables and groundwater from organic vegetable bases, northern China. Environ. Pollut. 2010, 158, 2992–2998. [Google Scholar] [CrossRef]
- Martínez-Carballo, E.; González-Barreiro, C.; Scharf, S.; Gans, O. Environmental monitoring study of selected veterinary antibiotics in animal manure and soils in Austria. Environ. Pollut. 2007, 148, 570–579. [Google Scholar] [CrossRef]
- Li, W.; Shi, Y.; Gao, L.; Liu, J.; Cai, Y. Occurrence of antibiotics in water, sediments, aquatic plants, and animals from Baiyangdian Lake in North China. Chemosphere 2012, 89, 1307–1315. [Google Scholar] [CrossRef]
- Flaherty, C.M.; Dodson, S.I. Effects of pharmaceuticals on Daphnia survival, growth, and reproduction. Chemosphere 2005, 61, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Che, B.; Duan, A.; Mao, J.; Dahlgren, R.A.; Zhang, M.; Zhang, H.; Zeng, A.; Wang, X. Toxicity evaluation of beta-diketone antibiotics on the development of embryo-larval zebrafish (Danio rerio). Environ. Toxicol. 2014, 29, 1134–1146. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Asselman, J.; Jeong, T.-Y.; Yu, S.; De Schamphelaere, K.A.C.; Kim, S.D. Multigenerational effects of the antibiotic tetracycline on transcriptional responses of daphnia magna and its relationship to higher levels of biological organizations. Environ. Sci. Technol. 2017, 51, 12898–12907. [Google Scholar] [CrossRef] [PubMed]
- Eggen, T.; Asp, T.N.; Grave, K.; Hormazabal, V. Uptake and translocation of metformin, ciprofloxacin and narasin in forage- and crop plants. Chemosphere 2011, 85, 26–33. [Google Scholar] [CrossRef]
- Drury, B.; Scott, J.; Rosi-Marshall, E.J.; Kelly, J.J. Triclosan Exposure Increases Triclosan Resistance and Influences Taxonomic Composition of Benthic Bacterial Communities. Environ. Sci. Technol. 2013, 47, 8923–8930. [Google Scholar] [CrossRef] [Green Version]
- Cycoń, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the Soil Environment-Degradation and Their Impact on Microbial Activity and Diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef]
- Thiele-Bruhn, S.; Beck, I.C. Effects of sulfonamide and tetracycline antibiotics on soil microbial activity and microbial biomass. Chemosphere 2005, 59, 457–465. [Google Scholar] [CrossRef]
- Kummerer, K. Antibiotics in the aquatic environment—A review—Part I. Chemosphere 2009, 75, 417–434. [Google Scholar] [CrossRef]
- Pérez, S.; Eichhorn, P.; Aga, D.S. Evaluating the biodegradability of sulfamethazine, sulfamethoxazole, sulfathiazole, and trimethoprim at different stages of sewage treatment. Environ. Toxicol. Chem. 2005, 24, 1361–1367. [Google Scholar] [CrossRef]
- Reis, A.C.; Kolvenbach, B.A.; Nunes, O.C.; Corvini, P.F. Biodegradation of antibiotics: The new resistance determinants—Part I. New Biotechnol. 2020, 54, 34–51. [Google Scholar] [CrossRef]
- Deng, Y.; Mao, Y.; Li, B.; Yang, C.; Zhang, T. Aerobic Degradation of Sulfadiazine by Arthrobacter spp.: Kinetics, Pathways, and Genomic Characterization. Environ. Sci. Technol. 2016, 50, 9566–9575. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Li, A.; Cui, D.; Cai, R.; Ma, F.; Wang, Y. Biodegradation and metabolic pathway of sulfamethoxazole by Pseudomonas psychrophila HA-4, a newly isolated cold-adapted sulfamethoxazole-degrading bacterium. Appl. Microbiol. Biot. 2014, 98, 4671–4681. [Google Scholar] [CrossRef] [PubMed]
- Mulla, S.I.; Hu, A.; Sun, Q.; Li, J.; Suanon, F.; Ashfaq, M.; Yu, C.-P. Biodegradation of sulfamethoxazole in bacteria from three different origins. J. Environ. Manag. 2018, 206, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Li, F.; Zhou, Q. Degradation mechanisms of sulfamethoxazole and its induction of bacterial community changes and antibiotic resistance genes in a microbial fuel cell. Bioresour. Technol. 2019, 289, 121632. [Google Scholar] [CrossRef]
- Bouju, H.; Ricken, B.; Beffa, T.; Corvini, P.F.-X.; Kolvenbach, B.A. Isolation of bacterial strains capable of sulfamethoxazole mineralization from an acclimated membrane bioreactor. Appl. Environ. Microbiol. 2012, 78, 277–279. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, N.; Mishra, P. The synthetic and therapeutic expedition of isoxazole and its analogs. Med. Chem. Res. 2018, 27, 1309–1344. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.K.; Shin, K.J.; Yoo, K.H.; Seo, K.J.; Hong, C.Y.; Lee, C.-S.; Park, S.Y.; Kim, D.J.; Park, S.W. Synthesis and antibacterial activity of new carbapenems containing isoxazole moiety. Bioorg. Med. Chem. Lett. 2000, 10, 95–99. [Google Scholar] [CrossRef]
- Zimecki, M.; Bąchor, U.; Mączyński, M. Isoxazole derivatives as regulators of immune functions. Molecules 2018, 23, 2724. [Google Scholar] [CrossRef] [Green Version]
- Stadler, L.B.; Ernstoff, A.S.; Aga, D.S.; Love, N.G. Micropollutant fate in wastewater treatment: Redefining “removal”. Environ. Sci. Technol. 2012, 46, 10485–10486. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Y.; Ma, J.; Zhao, F. Rapid degradation of sulphamethoxazole and the further transformation of 3-amino-5-methylisoxazole in a microbial fuel cell. Water Res. 2016, 88, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Mao, D.; Luo, Y.; Xu, L. Sulfamethoxazole biodegradation and biotransformation in the water-sediment system of a natural river. Bioresour. Technol. 2011, 102, 7069–7076. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Williams, M.; Smith, L.; Chen, D.; Kookana, R. Dissipation of sulfamethoxazole and trimethoprim antibiotics from manure-amended soils. J. Environ. Sci. Health B 2012, 47, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-W.; Tsai, L.-L.; Chang, B.-V. Anaerobic degradation of sulfamethoxazole in mangrove sediments. Sci. Total Environ. 2018, 643, 1446–1455. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ye, Q.; Gan, J. Influence of organic amendment on fate of acetaminophen and sulfamethoxazole in soil. Environ. Pollut. 2015, 206, 543–550. [Google Scholar] [CrossRef]
- Qi, M.; Liang, B.; Zhang, L.; Ma, X.; Yan, L.; Dong, W.; Kong, D.; Zhang, L.; Zhu, H.; Gao, S.-H.; et al. Microbial interactions drive the complete catabolism of the antibiotic sulfamethoxazole in activated sludge microbiomes. Environ. Sci. Technol. 2021, 55, 3270–3282. [Google Scholar] [CrossRef]
- Qi, M.; Ma, X.; Liang, B.; Zhang, L.; Kong, D.; Li, Z.; Wang, A. Complete genome sequences of the antibiotic sulfamethoxazole-mineralizing bacteria Paenarthrobacter sP27 and Norcardiodes sN27. Environ. Res. 2022, 204 Pt B, 112013. [Google Scholar] [CrossRef]
- Yan, L.; Yan, N.; Gao, X.Y.; Liu, Y.; Liu, Z.P. Degradation of amoxicillin by newly isolated Bosea sAds-6. Sci. Total Environ. 2022, 828, 154411. [Google Scholar] [CrossRef]
- Oxford Nanopore Technologies, Genomic DNA by Ligation (SQK-LSK109), Oxford, UK, 2020. Available online: https://store.nanoporetech.com/us/media/wysiwyg/pdfs/SQK-LSK109/Genomic_DNA_by_Ligation_SQK-LSK109_-minion.pdf (accessed on 8 January 2021).
- Marçais, G.; Delcher, A.L.; Phillippy, A.; Coston, R.; Salzberg, S.; Zimin, A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef] [Green Version]
- Qiu, J.; Zhang, Y.; Zhao, L.; He, Q.; Jiang, J.; Hong, Q.; He, J. Isolation and characterization of the cotinine-degrading bacterium Nocardioides sStrain JQ2195. J. Hazard. Mater. 2018, 353, 158–165. [Google Scholar] [CrossRef]
- Omotayo, A.E.; Ilori, M.O.; Obayori, O.S.; Amund, O.O. Influence of pH, temperature and nutrient addition on the degradation of atrazine by Nocardioides spisolated from agricultural soil in Nigeria. Malays. J. Microbiol. 2016, 12, 270–278. [Google Scholar]
- Roh, S.G.; Lee, C.; Kim, M.K.; Kang, H.J.; Kim, Y.S.; Kim, M.J.; Malik, A.; Kim, S.B. Nocardioides euryhalodurans snov., Nocardioides seonyuensis snov. and Nocardioides eburneiflavus snov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2020, 70, 2682–2689. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.-H.; Yang, C.-W.; Chang, B.-V. Anaerobic degradation of sulfamethoxazole by mixed cultures from swine and sewage sludge. Environ. Technol. 2019, 40, 210–218. [Google Scholar] [CrossRef]
- Herzog, B.; Lemmer, H.; Horn, H.; Müller, E. Characterization of pure cultures isolated from sulfamethoxazole-acclimated activated sludge with respect to taxonomic identification and sulfamethoxazole biodegradation potential. BMC Microbiol. 2013, 13, 276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.; Liu, Q.; Maeng, S.; Choi, W.J.; Chang, Y.; Im, W.-T. Nocardioides convexus snov. and Nocardioides anomalus snov., isolated from soil and mineral water. Int. J. Syst. Evol. Microbiol. 2020, 70, 6402–6407. [Google Scholar] [CrossRef]
- Yoon, J.H.; Cho, Y.G.; Lee, S.T.; Suzuki, K.; Nakase, T.; Park, Y.H. Nocardioides nitrophenolicus snov., a ρ-nitrophenol-degrading bacterium. Int. J. Syst. Bacteriol. 1999, 49 Pt 2, 675–680. [Google Scholar] [CrossRef] [Green Version]
- Britten, R.J. Cases of ancient mobile element DNA insertions that now affect gene regulation. Mol. Phylogenet. Evol. 1996, 5, 13–17. [Google Scholar] [CrossRef]


| Strain | Genes | CDS | Gaps | Sequence | Contig Length (bp) | GC Content (%) | Reference |
|---|---|---|---|---|---|---|---|
| N39 | 6157 | 6076 | 0 | Chromosome | 6,092,424 | 72.28 | This study |
| Plasmid I | 110,939 | 69.60 | |||||
| Plasmid II | 61,745 | 69.89 | |||||
| Plasmid III | 3820 | 78.27 | |||||
| N40 | 6152 | 6071 | 0 | Chromosome | 6,090,985 | 72.29 | This study |
| Plasmid 1 | 110,947 | 69.60 | |||||
| Plasmid 2 | 61,745 | 69.89 | |||||
| N27 | 6658 | 6603 | Chromosome | 6,104,465 | 72.29 | [37] | |
| Plasmid A | 294,133 | 67.26 | |||||
| Plasmid B | 109,667 | 69.95 | |||||
| Plasmid C | 80,343 | 69.90 |
| Growth Yield (×10−4 AU/mg) | 20 °C | 25 °C | 30 °C | 35 °C | 40 °C |
|---|---|---|---|---|---|
| 0 h | 0.00 a ± 0.00 b | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| 8 h | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| 16 h | 2.08 ± 3.84 | 0.56 ± 0.96 | 3.67 ± 0.68 | 4.56 ± 2.06 | 0.00 ± 0.00 |
| 24 h | 1.23 ± 1.30 | 2.55 ± 1.49 | 7.10 ± 0.52 | 6.95 ± 1.29 | 0.00 ± 0.00 |
| 32 h | 1.51 ± 1.58 | 2.84 ± 0.35 | 8.60 ± 1.00 | 7.33 ± 0.39 | 0.00 ± 0.00 |
| 40 h | 2.21 ± 4.08 | 2.38 ± 0.40 | 9.77 ± 0.91 | 7.77 ± 0.32 | 0.00 ± 0.00 |
| 48 h | 3.43 ± 8.39 | 4.28 ± 0.06 | 11.03 ± 0.90 | 8.86 ± 0.71 | 0.00 ± 0.00 |
| Growth rate (×10−4 AU/h) | 20 °C | 25 °C | 30 °C | 35 °C | 40 °C |
| 0 h | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| 8 h | 0.00 ± 1.25 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| 16 h | 0.42 ± 0.36 | 0.21 ± 0.36 | 2.71 ± 1.30 | 2.50 ± 0.63 | 0.00 ± 0.00 |
| 24 h | 0.42 ± 0.00 | 0.83 ± 0.42 | 7.36 ± 1.46 | 6.39 ± 0.96 | 0.00 ± 0.00 |
| 32 h | 0.21 ± 0.18 | 1.88 ± 0.00 | 10.00 ± 2.44 | 7.40 ± 0.18 | 0.00 ± 0.00 |
| 40 h | 1.83 ± 0.29 | 1.83 ± 0.38 | 9.83 ± 1.23 | 7.58 ± 0.14 | 0.00 ± 0.00 |
| 48 h | 2.43 ± 0.48 | 3.75 ± 0.55 | 10.07 ± 0.43 | 7.92 ± 0.42 | 0.00 ± 0.00 |
| Growth Yield (×10−4 AU/mg) | pH = 5.0 | pH = 6.0 | pH = 7.0 | pH = 8.0 | pH = 9.0 |
|---|---|---|---|---|---|
| 0 h | 0.00 a ± 0.00 b | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| 8 h | 0.00 ± 0.00 | 4.56 ± 5.66 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| 16 h | 0.00 ± 0.00 | 0.35 ± 2.32 | 3.67 ± 0.68 | 1.91 ± 1.51 | 3.22 ± 0.88 |
| 24 h | 0.00 ± 0.00 | 2.82 ± 1.11 | 7.10 ± 0.52 | 5.07 ± 1.90 | 6.03 ± 1.10 |
| 32 h | 0.00 ± 0.00 | 3.76 ± 0.90 | 8.60 ± 1.00 | 7.10 ± 1.52 | 8.41 ± 1.35 |
| 40 h | 0.68 ± 2.21 | 5.42 ± 1.34 | 9.77 ± 0.91 | 8.73 ± 1.05 | 9.07 ± 0.41 |
| 48 h | 6.35 ± 4.35 | 7.32 ± 1.13 | 11.03 ± 0.90 | 9.55 ± 1.52 | 9.52 ± 0.44 |
| Growth rate (×10−4 AU/h) | pH = 5.0 | pH = 6.0 | pH = 7.0 | pH = 8.0 | pH = 9.0 |
| 0 h | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| 8 h | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| 16 h | 0.00 ± 0.00 | 0.00 ± 0.63 | 2.71 ± 1.30 | 0.83 ± 0.36 | 1.88 ± 0.00 |
| 24 h | 0.00 ± 0.00 | 1.39 ± 0.64 | 7.36 ± 1.46 | 4.44 ± 0.87 | 7.50 ± 1.25 |
| 32 h | 0.00 ± 0.00 | 2.81 ± 0.94 | 10.00 ± 2.44 | 8.65 ± 0.18 | 11.67 ± 1.57 |
| 40 h | 0.17 ± 0.38 | 5.00 ± 1.50 | 9.83 ± 1.23 | 10.17 ± 0.52 | 10.08 ± 0.14 |
| 48 h | 1.04 ± 0.42 | 6.46 ± 0.63 | 10.07 ± 0.43 | 9.24 ± 0.64 | 8.82 ± 0.32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, L.; Liang, B.; Qi, M.-Y.; Wang, A.-J.; Liu, Z.-P. Degrading Characterization of the Newly Isolated Nocardioides sp. N39 for 3-Amino-5-methyl-isoxazole and the Related Genomic Information. Microorganisms 2022, 10, 1496. https://doi.org/10.3390/microorganisms10081496
Yan L, Liang B, Qi M-Y, Wang A-J, Liu Z-P. Degrading Characterization of the Newly Isolated Nocardioides sp. N39 for 3-Amino-5-methyl-isoxazole and the Related Genomic Information. Microorganisms. 2022; 10(8):1496. https://doi.org/10.3390/microorganisms10081496
Chicago/Turabian StyleYan, Lei, Bin Liang, Meng-Yuan Qi, Ai-Jie Wang, and Zhi-Pei Liu. 2022. "Degrading Characterization of the Newly Isolated Nocardioides sp. N39 for 3-Amino-5-methyl-isoxazole and the Related Genomic Information" Microorganisms 10, no. 8: 1496. https://doi.org/10.3390/microorganisms10081496
APA StyleYan, L., Liang, B., Qi, M.-Y., Wang, A.-J., & Liu, Z.-P. (2022). Degrading Characterization of the Newly Isolated Nocardioides sp. N39 for 3-Amino-5-methyl-isoxazole and the Related Genomic Information. Microorganisms, 10(8), 1496. https://doi.org/10.3390/microorganisms10081496






