New Antimicrobial Peptide with Two CRAC Motifs: Activity against Escherichia coli and Bacillus subtilis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Peptides and Peptide Stock Solutions
2.2. Bacterial Strains and Cultivation Conditions
2.3. Treatment of Bacterial Cells with Peptides
2.4. Evaluation of the Effects of the Peptides on Cultured Mouse Macrophages IC-21
2.5. Statistical Analysis
3. Results
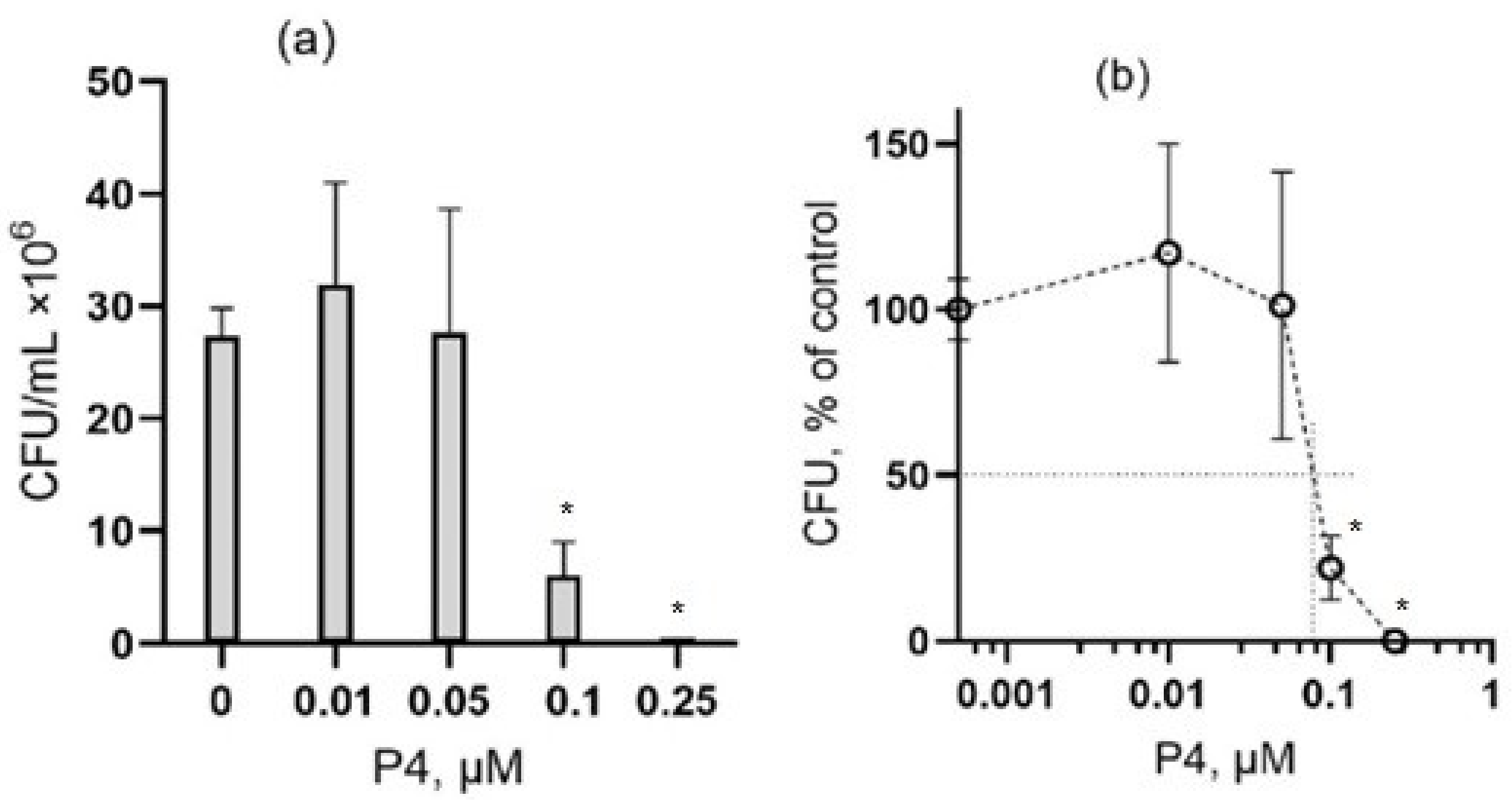
3.1. The Effect of Peptide P4 on E. coli
3.2. The Effect of Peptide P4 on B. subtilis
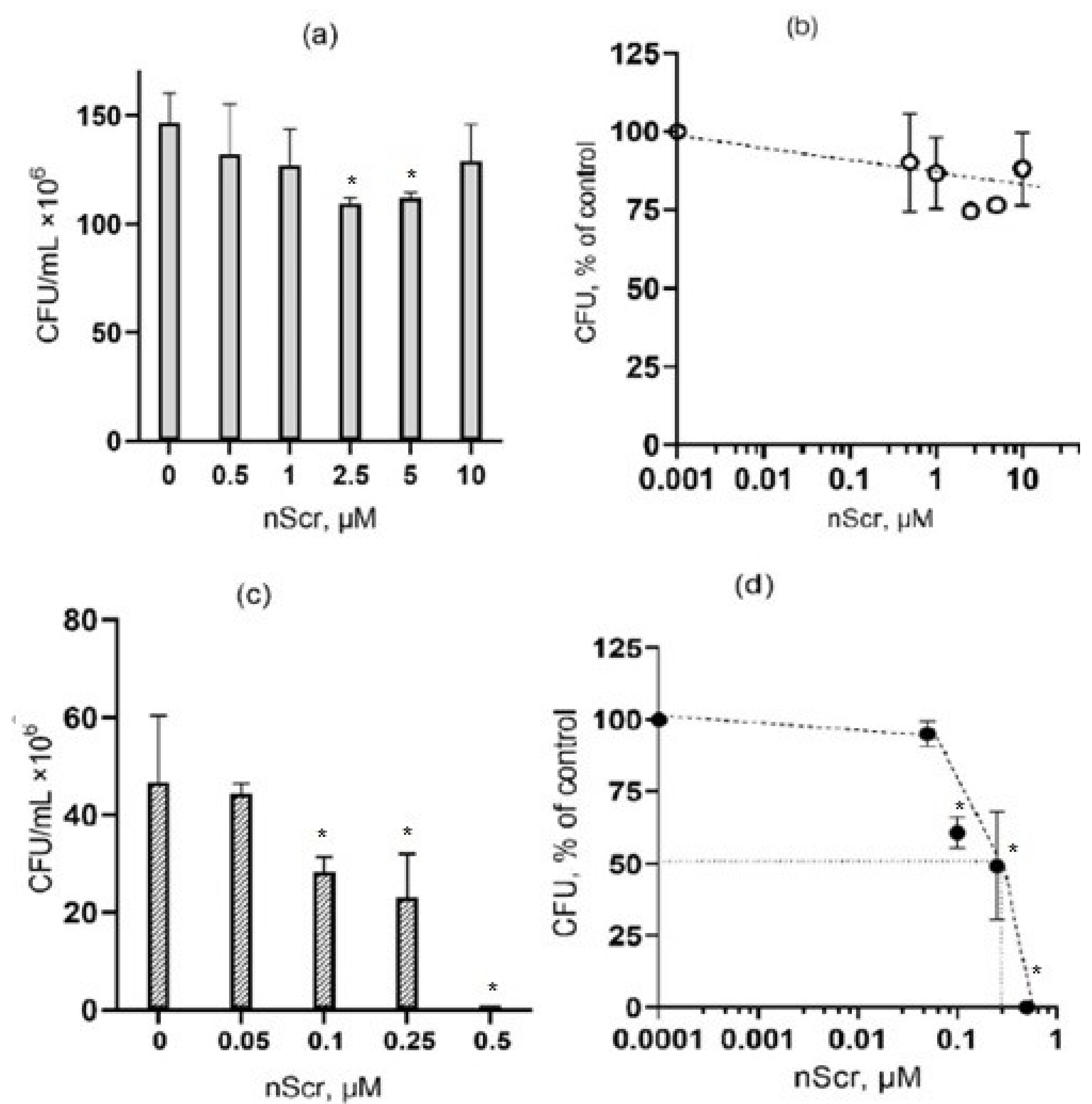
3.3. The Effect of the Scramble Peptide (nScr) on Bacteria
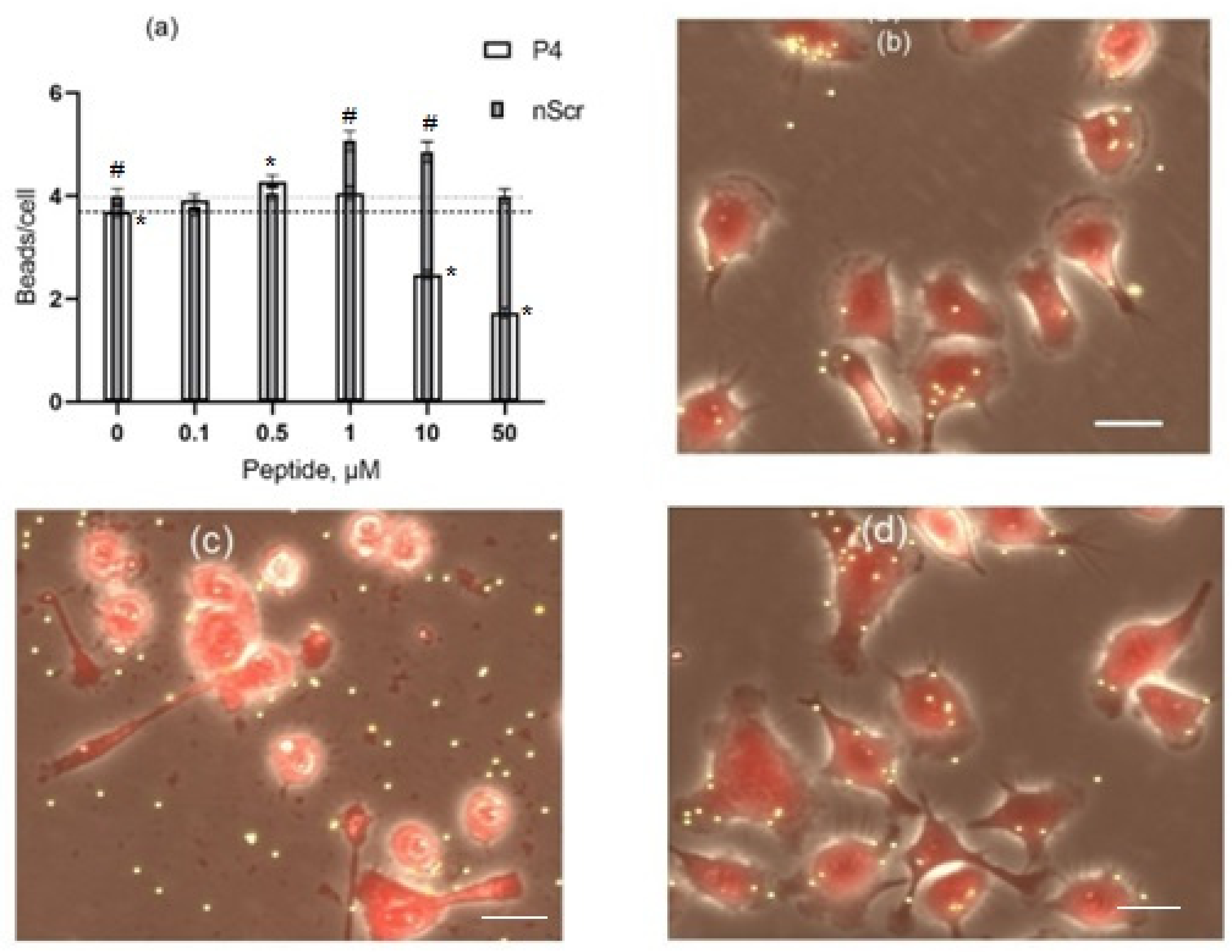
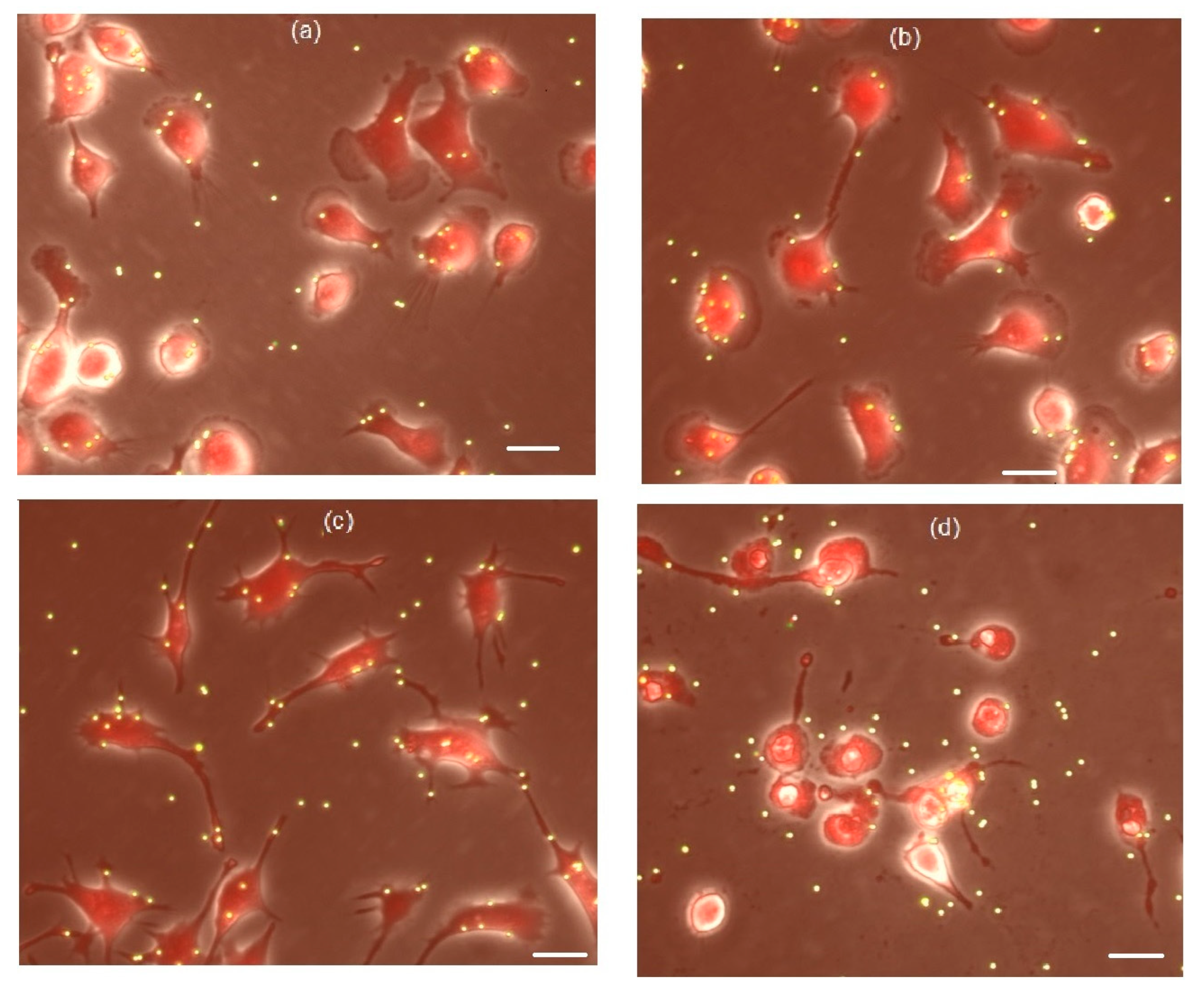
3.4. The Effects of Peptides P4 and nScr on Cultured Macrophages IC-21
3.5. Cholesterol Extractant mβCD Lowers the Cytotoxic Concentration of Peptide P4 on Cultured Macrophages IC-21
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deslouches, B.; Di, Y.P. Antimicrobial peptides: A potential therapeutic option for surgical site infections. Clin. Surg. 2017, 2, 1740. [Google Scholar] [PubMed]
- Steiner, H.; Hultmark, D.; Engstrom, A.; Bennich, H.; Boman, H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 1981, 292, 246–248. [Google Scholar] [CrossRef] [PubMed]
- Pasupuleti, M.; Malmsten, M.; Schmidtchen, A. Antimicrobial peptides: A key component of innate immunity. Crit. Rev. Biotechnol. 2011, 32, 143–171. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Peng, Y. Synergistic effect of clinically used antibiotics and peptide antibiotics against Gram-positive and Gram-negative bacteria. Exp. Ther. Med. 2013, 6, 1000–1004. [Google Scholar] [CrossRef]
- Lee, T.-H.; Hall, K.N.; Aguilar, M.-I. Antimicrobial peptide structure and mechanism of action: A focus on the role of membrane structure. Curr. Topics Medicinal Chem. 2016, 16, 25–39. [Google Scholar] [CrossRef]
- Ganz, T.; Selsted, M.E.; Szklarek, D.; Harwig, S.S.; Daher, K.; Bainton, D.F.; Lehrer, R.I. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Investig. 1985, 76, 1427–1435. [Google Scholar] [CrossRef]
- Eisenhauer, P.B.; Harwig, S.S.; Szklarek, D.; Ganz, T.; Selsted, M.E.; Lehrer, R.I. Purification and antimicrobial properties of three defensins from rat neutrophils. Infect. Immun. 1989, 57, 2021–2027. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389. [Google Scholar] [CrossRef]
- Zhou, C.; Qi, X.; Li, P.; Chen, W.; Mouad, L.; Chang, M.W.; Leong, S.S.; Chang-Park, M.B. High potency and broad-spectrum antimicrobial peptides synthesized via ring-opening polymerization of a-amino acid-N-carboxyanhydrides. Biomacromolecules 2010, 11, 60–67. [Google Scholar] [CrossRef]
- Abd-El-Aziz, A.S.; Agatemor, C.; Etkin, N.; Overy, D.P.; Lanteigne, M.; McQuillan, K. Antimicrobial organometallic dendrimers with tunable activity against multidrug-resistant bacteria. Biomacromolecules 2015, 16, 3694–3703. [Google Scholar] [CrossRef]
- Shang, D.; Liu, Y.; Jiang, F.; Ji, F.; Wang, H.; Han, X. Synergistic antibacterial activity of designed Trp-containing antibacterial peptides in combination with antibiotics against multidrug-resistant Staphylococcus epidermidis. Front. Microbiol. 2019, 10, 2719. [Google Scholar] [CrossRef]
- Jiang, S.; Deslouches, B.; Chen, C.; Di, M.E.; Di, Y.P. Antibacterial properties and efficacy of a novel SPLUNC1-derived antimicrobial peptide, α4-Short, in a murine model of respiratory infection. mBio 2019, 10, e00226-e19. [Google Scholar] [CrossRef]
- Antimicrobial Peptide Database. Available online: https://aps.unmc.edu/ (accessed on 26 July 2022).
- The Database of Antimicrobial Activity and Structure of Peptides (DBAASP). Available online: https://dbaasp.org/ (accessed on 26 July 2022).
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucl. Acids Res. 2021, 49, D288–D297. [Google Scholar] [CrossRef]
- Wimley, W.C. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 2010, 5, 905–917. [Google Scholar] [CrossRef]
- Jean-Francois, F.; Castano, S.; Desbat, B.; Odaert, B.; Roux, M.; Metz-Boutigue, M.-H.; Dufourc, E.J. Aggregation of cateslytin beta-sheets on negatively charged lipids promotes rigid membrane domains. A new mode of action for antimicrobial peptides? Biochemistry 2008, 47, 6394–6402. [Google Scholar] [CrossRef]
- Bechinger, B.; Gorr, S.U. Antimicrobial peptides: Mechanisms of action and resistance. J. Dent. Res. 2017, 96, 254–260. [Google Scholar] [CrossRef]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by [alpha]-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta. 1999, 1462, 55–70. [Google Scholar] [CrossRef]
- Huang, Z.; London, E. Cholesterol lipids and cholesterol-containing lipid rafts in bacteria. Chem. Phys. Lipids. 2016, 199, 11–16. [Google Scholar] [CrossRef]
- Guzmán-Flores, J.E.; Steinemann-Hernández, L.; González de la Vara, L.E.; Gavilanes-Ruiz, M.; Romeo, T.; Alvarez, A.F.; Georgellis, D. Proteomic analysis of Escherichia coli detergent-resistant membranes (DRM). PLoS ONE 2019, 14, e0223794. [Google Scholar] [CrossRef]
- Rohmer, M.; Bouvier-Nave, P.; Ourisson, G. Distribution of hopanoid triterpenes in prokaryotes. Microbiology 1984, 130, 1137–1150. [Google Scholar] [CrossRef]
- Sáenz, J.P.; Grosser, D.; Bradley, A.S.; Lagny, T.J.; Lavrynenko, O.; Broda, M.; Simons, K. Hopanoids as functional analogues of cholesterol in bacterial membranes. Proc. Natl. Acad. Sci. USA 2015, 112, 11971–11976. Available online: www.pnas.org/cg (accessed on 26 July 2022). [CrossRef]
- Sang, P.; Shi, Y.; Teng, P.; Cao, A.; Xu, H.; Li, Q.; Cai, J. Antimicrobial AApeptides. Curr. Top. Med. Chem. 2017, 17, 1266–1279. [Google Scholar] [CrossRef]
- Gomes, B.; Augusto, M.T.; Felício, M.R.; Hollmann, A.; Franco, O.L.; Gonçalves, S.; Santos, N.C. Designing improved active peptides for therapeutic approaches against infectious diseases. Biotechnol. Adv. 2018, 36, 415–429. [Google Scholar] [CrossRef]
- Sandhu, A.K.; Yang, Y.; Li, W.-W. In vivo antibacterial efficacy of antimicrobial peptides modified metallic implants---Systematic review and meta-analysis. ACS Biomater. Sci. Eng. 2022, 8, 1749–1762. [Google Scholar] [CrossRef]
- Martínez-Villaluenga, C.; Hernández-Ledesma, B. Peptides for health benefits 2019. Int. J. Mol. Sci. 2020, 21, 2543. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Kenworthy, A.K.; Sanders, C.R. Cholesterol as a co-solvent and a ligand for membrane proteins. Protein Sci. 2014, 23, 1–22. [Google Scholar] [CrossRef]
- Sezgin, E.; Levental, I.; Mayor, S.; Eggeling, C. The mystery of membrane organization: Composition, regulation, and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.P.; Jiang, T.; Sun, C.; Lihan, M.; Pant, S.; Mahinthichaichan, P.; Trifan, A.; Tajkhorshid, E. Characterization of lipid-protein interactions and lipid-mediated modulation of membrane protein function through molecular simulation. Chem. Rev. 2019, 19, 6086–6161. [Google Scholar] [CrossRef] [PubMed]
- Enkavi, G.; Javanainen, M.; Kulig, W.; Róg, T.; Vattulainen, I. Multiscale simulations of biological membranes: The challenge to understand biological phenomena in a living substance. Chem. Rev. 2019, 119, 5607–5774. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Papadopoulos, V. Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology 1998, 139, 4991–4997. [Google Scholar] [CrossRef]
- Papadopoulos, V.; Baraldi, M.; Guilarte, T.R.; Knudsen, T.B.; Lacapere, J.J.; Lindemann, P.L.; Norenberg, M.D.; Nutt, D.; Weizman, A.; Zhang, M.R.; et al. Translocator protein (18kDa): New nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol. Sci. 2006, 27, 402–409. [Google Scholar] [CrossRef]
- Li, H.; Yao, Z.; Degenhardt, B.; Teper, G.; Papadopoulos, V. Cholesterol binding at the cholesterol recognition/interaction amino acid consensus (CRAC) of the peripheral-type benzodiazepine receptor and inhibition of steroidogenesis by an HIV TAT-CRAC peptide. Proc. Natl. Acad. Sci. USA. 2001, 98, 1267–1272. [Google Scholar] [CrossRef]
- Fantini, J.; Epand, R.M.; Barrantes, F.J. Cholesterol-Recognition Motifs in Membrane Proteins. In Direct Mechanisms in Cholesterol Modulation of Protein Function; Rosenhouse-Dantsker, A., Bukiya, A.N., Eds.; Series Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2019; Volume 1135, pp. 3–25. [Google Scholar] [CrossRef]
- Miller, C.M.; Brown, A.C.; Mittal, J. Disorder in cholesterol-binding functionality of CRAC peptides: A molecular dynamics study. J. Phys. Chem. B 2014, 118, 13169–13174. [Google Scholar] [CrossRef]
- Brown, A.C.; Balashova, N.V.; Epand, R.M.; Epand, R.F.; Bragin, A.; Kachlany, S.C.; Walters, M.J.; Du, Y.; Boesze-Battaglia, K.; Lally, E.T. Aggregatibacter actinomycetemcomitans leukotoxin utilizes a cholesterol recognition/amino acid consensus site for membrane association. J. Biol. Chem. 2013, 288, 23607–23621. [Google Scholar] [CrossRef]
- Vazquez, R.F.; Mat´e, S.M.; Bak´as, L.S.; Fern´andez, M.M.; Malchoidi, E.L.; Herlax, V.S. Novel evidence for the specific interaction between cholesterol and α-haemolysin of Escherichia coli. Biochem. J. 2014, 458, 481–489. [Google Scholar] [CrossRef]
- Koufos, E.; Chang, E.H.; Rasti, E.S.; Krueger, E.; Brown, A.C. Use of a cholesterol recognition amino acid consensus peptide to inhibit binding of a bacterial toxin to cholesterol. Biochemistry 2016, 55, 4787–4797. [Google Scholar] [CrossRef]
- Bi, X.; Wang, C.; Dong, W.; Zhu, W.; Shang, D. Antimicrobial properties and interaction of two Trp-substituted cationic antimicrobial peptides with a lipid bilayer. J. Antibiot. 2014, 67, 361–368. [Google Scholar] [CrossRef]
- Bellm, L.; Lehrer, R.I.; Ganz, T. Protegrins: New antibiotics of mammalian origin. Expert Opin. Investig. Drugs 2000, 9, 1731–1742. [Google Scholar] [CrossRef]
- Zasloff, M. Magainins, a class of antimicrobial peptides from Xenopus skin: Isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA 1987, 84, 5449–5453. [Google Scholar] [CrossRef]
- Mangoni, M.L.; Papo, N.; Barra, D.; Simmaco, M.; Bozzi, A.; Di Giulio, A.; Rinaldi, A.C. Effects of the antimicrobial peptide temporin L on cell morphology, membrane permeability and viability of Escherichia coli. Biochem. J. 2004, 380, 859–865. [Google Scholar] [CrossRef]
- Rinaldi, A.C.; Mangoni, M.L.; Rufo, A.; Luzi, C.; Barra, D.; Zhao, H.; Kinnunen, P.K.; Bozzi, A.; Di Giulio, A.; Simmaco, M. Temporin L: Antimicrobial, haemolytic and cytotoxic activities, and effects on membrane permeabilization in lipid vesicles. Biochem. J. 2002, 368, 91–100. [Google Scholar] [CrossRef]
- Dunina-Barkovskaya, A.Y..; Vishnyakova, K.S.; Baratova, L.A.; Radyukhin, V.A. Modulation of cholesterol-dependent activity of macrophages IC-21 by a peptide containing two CRAC motifs from protein M1 of influenza virus. Biochem. Suppl. Ser. A Membr. Cell Biol. 2019, 13, 268–276. [Google Scholar] [CrossRef]
- Dunina-Barkovskaya, A.Y..; Vishnyakova, K.S. Modulation of the cholesterol-dependent activity of macrophages IC-21 by CRAC peptides with substituted motif-forming amino acids. Biochem. Suppl. Series A Membr. Cell Biol. 2020, 14, 331–343. [Google Scholar] [CrossRef]
- Dunina-Barkovskaya, A.Y..; Vishnyakova, K.S.; Golovko, A.O.; Arutyunyan, A.M.; Baratova, L.A.; Bathishchev, O.V.; Radyukhin, V.A. Amphipathic CRAC-containing peptides derived from the influenza virus A M1 protein modulate cholesterol-dependent activity of cultured IC-21 macrophages. Biochemistry 2018, 83, 982–991. [Google Scholar]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1972; 466p, ISBN 100879691069. [Google Scholar]
- Safronova, N.A.; Koksharova, O.A. Bacteria Rhodococcus sp. as potential destructors of detonation nanodiamonds. Nanotechnol Russia. 2018, 13, 439–442. [Google Scholar] [CrossRef]
- Golovkina, M.S.; Skachkov, I.V.; Metelev, M.V.; Kuzevanov, A.V.; Vishniakova, K.S.; Kireev, I.I.; Dunina-Barkovskaya, A.Y.. Serum-induced inhibition of the phagocytic activity of cultured macrophages IC-21. Biochem. Suppl. Ser. A Membr. Cell Biol. 2009, 4, 412–419. [Google Scholar] [CrossRef]
- Zidovetzki, R.; Levitan, I. Use of cyclodextrins to manipulate plasma membrane cholesterol content: Evidence, misconceptions and control strategies. Biochim. Biophys. Acta Biomembranes. 2007, 1768, 1311–1324. [Google Scholar] [CrossRef] [PubMed]
- Kurkov, S.V.; Loftsson, T. Cyclodextrins. Int. J. Pharm. 2013, 453, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Subczynski, W.K.; Pasenkiewicz-Gierula, M.; Widomska, J.; Mainali, L.; Raguz, M. High cholesterol/low cholesterol: Effects in biological membranes: A review. Cell Biochem. Biophys. 2017, 75, 369–385. [Google Scholar] [CrossRef]
- Raffy, S.; Teissié, J. Control of lipid membrane stability by cholesterol content. Biophys. J. 1999, 76, 2072–2080. [Google Scholar] [CrossRef]
- Zhang, X.; Barraza, K.M.; Beauchamp, J.L. Cholesterol provides nonsacrificial protection of membrane lipids from chemical damage at air–water interface. Proc. Natl. Acad. Sci. USA 2018, 115, 3255–3260. [Google Scholar] [CrossRef]
- Yeliseev, A.; Iyer, M.R.; Joseph, T.T.; Coffey, N.J.; Cinar, R.; Zoubak, L.; Kunos, G.; Gawrisch, K. Cholesterol as a modulator of cannabinoid receptor CB2 signaling. Sci. Rep. 2021, 11, 3706. [Google Scholar] [CrossRef]
- Rout, A.K.; Wu, X.; Starich, M.R.; Strub, M.-P.; Hammer, J.A.; Tjandra, N. The structure of melanoregulin reveals a role for cholesterol recognition in the protein’s ability to promote dynein function. Structure 2018, 26, 1373–1383. [Google Scholar] [CrossRef]
- Kiriakidi, S.; Kolocouris, A.; Liapakis, G.; Ikram, S.; Durdagi, S.; Mavromoustakos, T. Effects of Cholesterol on GPCR Function: Insights from Computational and Experimental Studies. In Direct Mechanisms in Cholesterol Modulation of Protein Function; Rosenhouse-Dantsker, A., Bukiya, A.N., Eds.; Series Advances in Experimental Medicine and Biology; Springer Nature: Cham, Switzerland, 2019; Volume 1135, pp. 89–103. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Sugishita, K.; Fujii, N.; Miyajima, K. Molecular basis for membrane selectivity of an antimicrobial peptide, magainin 2. Biochemistry 1995, 34, 3423–3429. [Google Scholar] [CrossRef]
- Shahmiri, M.; Enciso, M.; Mechler, A. Controls and constrains of the membrane disrupting action of Aurein 1.2. Sci. Rep. 2015, 5, 16378. [Google Scholar] [CrossRef]
- Omardien, S.; Drijfhout, J.W.; Vaz, F.M.; Wenzel, M.; Hamoen, L.W.; Zaat, S.A.J.; Brul, S. Bactericidal activity of amphipathic cationic antimicrobial peptides involves altering the membrane fluidity when interacting with the phospholipid bilayer. Biochim. Biophys. Acta Biomembr. 2018, 1860, 2404–2415. [Google Scholar] [CrossRef]
- Anselmo, S.; Sancataldo, G.; Mørck Nielsen, H.; Foderà, V.; Vetri, V. Peptide–membrane interactions monitored by fluorescence lifetime imaging: A study case of transportan 10. Langmuir 2021, 37, 13148–13159. [Google Scholar] [CrossRef]
- Armas, F.; Di Stasi, A.; Mardirossian, M.; Romani, A.A.; Benincasa, M.; Scocchi, M. Effects of lipidation on a proline-rich antibacterial peptide. Int. J. Mol. Sci. 2021, 22, 7959. [Google Scholar] [CrossRef]
- Tereshchenkov, A.G.; Shishkina, A.V.; Karpenko, V.V.; Chertkov, V.A.; Konevega, A.L.; Kasatsky, P.S.; Bogdanov, A.A.; Sumbatyan, N.V. New fluorescent macrolide derivatives for studying interactions of antibiotics and their analogs with the ribosomal exit tunnel. Biochemistry 2016, 81, 1163–1172. [Google Scholar] [CrossRef]

| Strain | P4, IC50 1 (μM) | nScr, IC50 1 (μM) |
|---|---|---|
| B. subtilis strain168 (Gram-positive) | 0.07 ± 0.01 | 0.4 ± 0.2 |
| E. coli strain MC4100 (Gram-negative) | 1.9 ± 0.4 | no inhibition |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koksharova, O.; Safronova, N.; Dunina-Barkovskaya, A. New Antimicrobial Peptide with Two CRAC Motifs: Activity against Escherichia coli and Bacillus subtilis. Microorganisms 2022, 10, 1538. https://doi.org/10.3390/microorganisms10081538
Koksharova O, Safronova N, Dunina-Barkovskaya A. New Antimicrobial Peptide with Two CRAC Motifs: Activity against Escherichia coli and Bacillus subtilis. Microorganisms. 2022; 10(8):1538. https://doi.org/10.3390/microorganisms10081538
Chicago/Turabian StyleKoksharova, Olga, Nina Safronova, and Antonina Dunina-Barkovskaya. 2022. "New Antimicrobial Peptide with Two CRAC Motifs: Activity against Escherichia coli and Bacillus subtilis" Microorganisms 10, no. 8: 1538. https://doi.org/10.3390/microorganisms10081538
APA StyleKoksharova, O., Safronova, N., & Dunina-Barkovskaya, A. (2022). New Antimicrobial Peptide with Two CRAC Motifs: Activity against Escherichia coli and Bacillus subtilis. Microorganisms, 10(8), 1538. https://doi.org/10.3390/microorganisms10081538







