The Rare Actinobacterium Crossiella sp. Is a Potential Source of New Bioactive Compounds with Activity against Bacteria and Fungi
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling, Isolation, and Identification of Bacteria
2.2. Bacterial and Fungal Inhibition Assays
2.3. Genomic Analysis
3. Results and Discussion
3.1. Caves as a Promising Source of Antimicrobial Compounds (ACs)
3.2. The Case of Altamira Cave
3.3. Isolation of Crossiella Strains
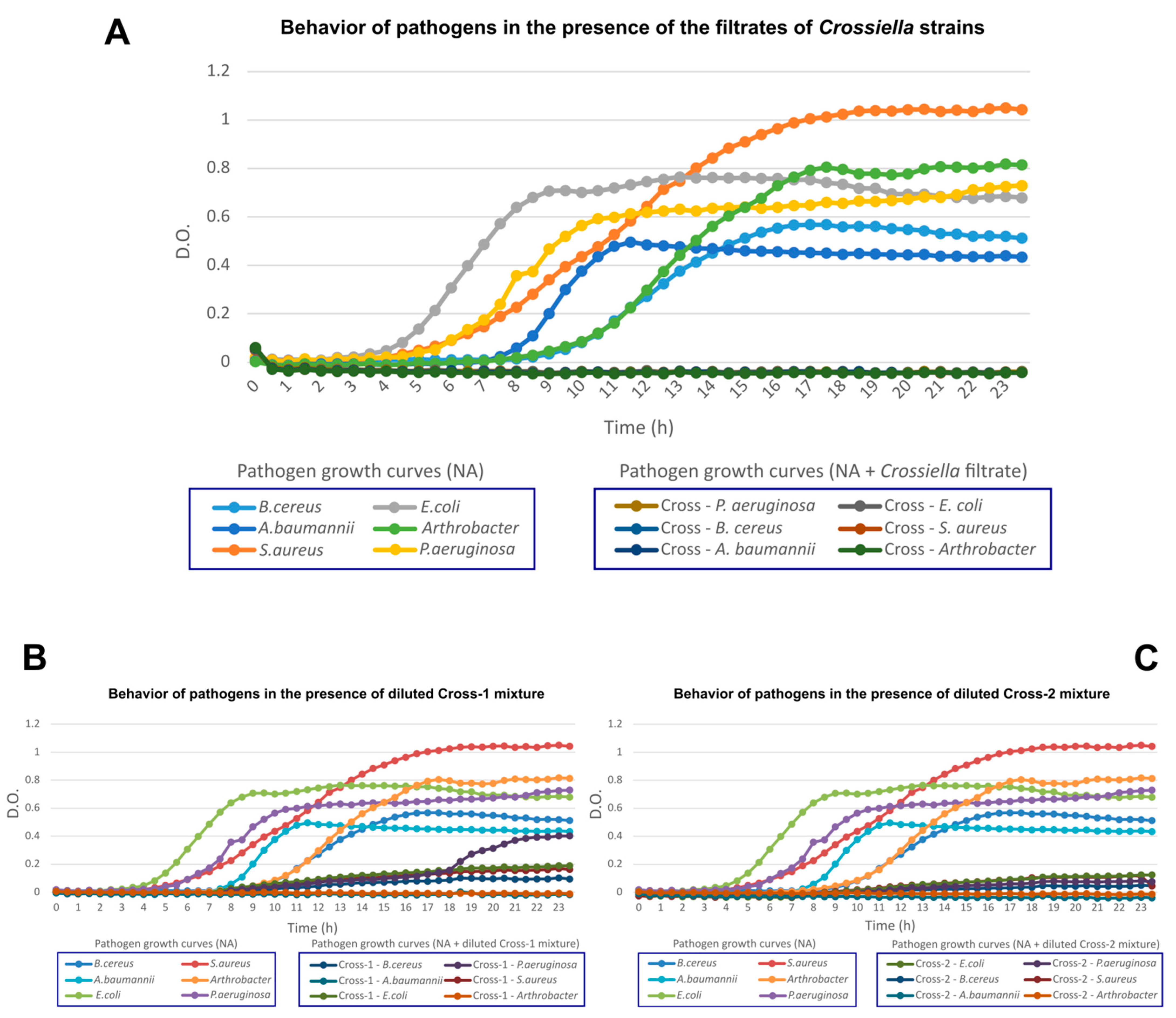
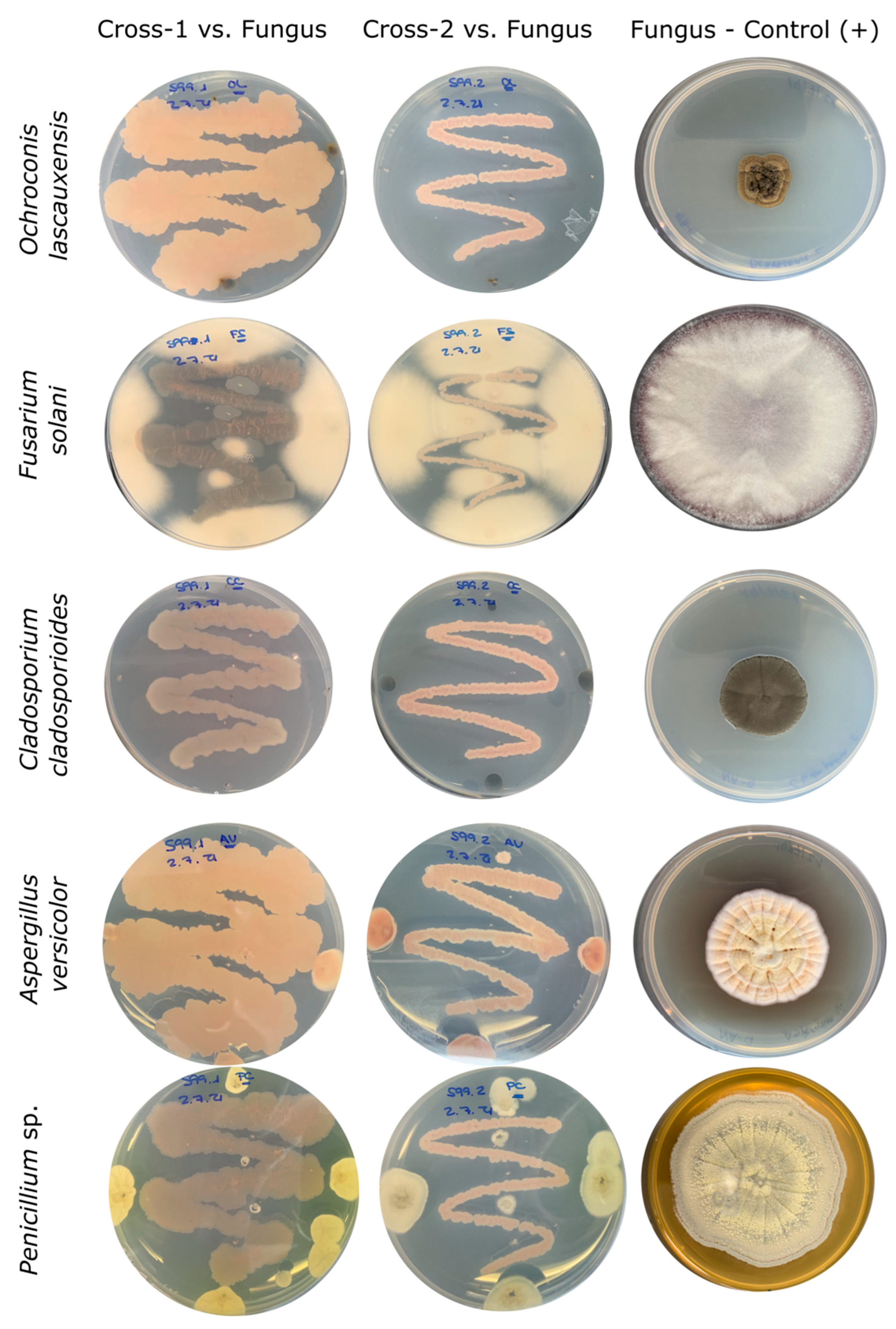
3.4. In Vitro Analyses for Inhibition of Pathogenic Microorganisms
3.5. In Silico Analyses for the Identification of Biosynthetic Gene Clusters
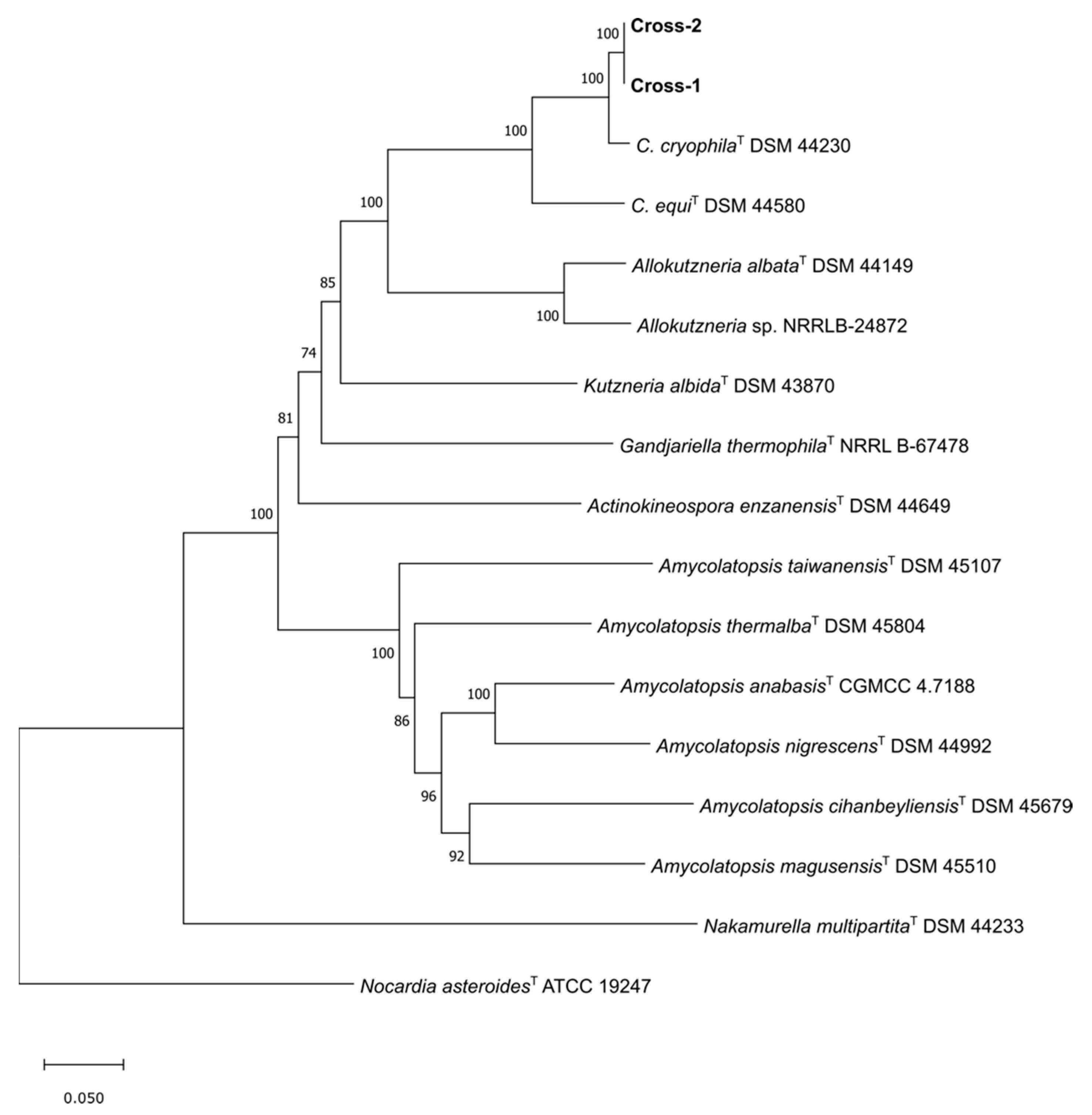
3.5.1. Phylogenomics
3.5.2. Functional Annotations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schatz, A.; Bugle, E.; Waksman, S.A. Streptomycin, a substance exhibiting antibiotic activity against gram-positive and gram-negative bacteria. Exp. Biol. Med. 1944, 55, 66–69. [Google Scholar] [CrossRef]
- Bergmann, W.; Burke, D.C. Marine products. XXXIX. The nucleosides of sponges. III. Spongothymidine and spongouridine. J. Org. Chem. 1955, 20, 1501–1507. [Google Scholar] [CrossRef]
- Rangseekaew, P.; Pathom-Aree, W. Cave Actinobacteria as producers of bioactive metabolites. Front. Microbiol. 2019, 10, 387. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Thakur, D. Antimicrobial biosynthetic potential and diversity of culturable soil actinobacteria from forest ecosystems of Northeast India. Sci. Rep. 2020, 10, 4104. [Google Scholar] [CrossRef]
- McCauley, E.P.; Piña, I.C.; Thompson, A.D.; Bashir, K.; Weinberg, M.; Kurz, S.L.; Crews, P. Highlights of marine natural products having parallel scaffolds found from marine-derived bacteria, sponges, and tunicates. J. Antibiot. 2020, 73, 504–525. [Google Scholar] [CrossRef]
- World Health Organization. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Nakaew, N.; Pathom-Aree, W.; Lumyong, S. Generic diversity of rare actinomycetes from Thai cave soils and their possible use as new bioactive compounds. Actinomycetologica 2009, 23, 21–26. [Google Scholar] [CrossRef]
- Wiseschart, A.; Mhuantong, W.; Tangphatsornruang, S.; Chantasingh, D.; Pootanakit, K. Shotgun metagenomic sequencing from Manao-Pee cave, Thailand, reveals insight into the microbial community structure and its metabolic potential. BMC Microbiol. 2019, 19, 144. [Google Scholar] [CrossRef]
- Yasir, M. Analysis of bacterial communities and characterization of antimicrobial strains from cave microbiota. Braz. J. Microbiol. 2018, 49, 248–257. [Google Scholar] [CrossRef]
- Paun, V.I.; Lavin, P.; Chifiriuc, M.C.; Purcarea, C. First report on antibiotic resistance and antimicrobial activity of bacteria isolates from 13,000-year old cave ice core. Sci. Rep. 2021, 11, 514. [Google Scholar] [CrossRef]
- Martin-Pozas, T.; Gonzalez-Pimentel, J.L.; Jurado, V.; Sanchez-Moral, S.; Cuezva, S.; Dominguez-Moñino, I.; Fernandez-Cortes, A.; Cañaveras, J.; Saiz-Jimenez, C. Microbial activity in subterranean ecosystems: Recent advances. Appl. Sci. 2020, 10, 8130. [Google Scholar] [CrossRef]
- Gohain, A.; Manpoong, C.; Saikia, R.; De Mandal, S. Actinobacteria: Diversity and biotechnological applications. In Recent Advancements in Microbial Diversity; De Mandal, S., Bhatt, P., Eds.; Academic Press: London, UK, 2020; pp. 217–231. [Google Scholar] [CrossRef]
- Zada, S.; Sajjad, W.; Rafiq, M.; Ali, S.; Hu, Z.; Wang, H.; Cai, R. Cave microbes as a potential source of drugs development in the modern era. Microb. Ecol. 2021, 1–14. [Google Scholar] [CrossRef]
- Groth, I.; Vettermann, R.; Schuetze, B.; Schumann, P.; Saiz-Jimenez, C. Actinomycetes in Karstic caves of northern Spain (Altamira and Tito Bustillo). J. Microbiol. Methods 1999, 36, 115–122. [Google Scholar] [CrossRef]
- Groth, I.; Schumann, P.; Laiz, L.; Sanchez-Moral, S.; Cañaveras, J.C.; Saiz-Jimenez, C. Geomicrobiological Study of the Grotta dei Cervi, Porto Badisco, Italy. Geomicrobiol. J. 2001, 241–258. [Google Scholar] [CrossRef]
- Herold, K.; Gollmick, F.A.; Groth, I.; Roth, M.; Menzel, K.D.; Möllmann, U.; Gräfe, U.; Hertweck, C. Cervimycin A-D: A polyketide glycoside complex from a cave bacterium can defeat vancomycin resistance. Chem. Eur. J. 2005, 11, 5523–5530. [Google Scholar] [CrossRef] [PubMed]
- Calaforra Chordi, J.M.; Berrocal Pérez, J.A. El Karst de Andalucía. Geoespeleología, Bioespeleología y Presencia Humana; Consejería de Medio Ambiente, Junta de Andalucía: Sevilla, Spain, 2008. [Google Scholar]
- Cuezva, S.; Jurado, V.; Fernandez-Cortes, A.; Garcia-Anton, E.; Rogerio-Candelera, M.A.; Ariño, X.; Benavente, D.; Cañaveras, J.C.; Saiz-Jimenez, C.; Sanchez-Moral, S. Scientific data suggest Altamira Cave should remain closed. In Microbial Life of Cave Systems; Engel, A.S., Ed.; DeGruiter: Berlin, Germany, 2017; pp. 303–320. [Google Scholar] [CrossRef]
- Jurado, V.; Groth, I.; Gonzalez, J.M.; Laiz, L.; Saiz-Jimenez, C. Agromyces subbeticus sp. nov., isolated from a cave in southern Spain. Int. J. Syst. Evol. Microbiol. 2005, 55, 1897–1901. [Google Scholar] [CrossRef]
- Jurado, V.; Gonzalez, J.M.; Laiz, L.; Saiz-Jimenez, C. Aurantimonas altamirensis sp. nov., a member of the order Rhizobiales isolated from Altamira Cave. Int. J. Syst. Evol. Microbiol. 2006, 56, 2583–2585. [Google Scholar] [CrossRef]
- Jurado, V.; Boiron, P.; Kroppenstedt, R.M.; Laurent, F.; Couble, A.; Laiz, L.; Klenk, H.-P.; Gonzalez, J.M.; Saiz-Jimenez, C.; Mouniée, D.; et al. Nocardia altamirensis sp. nov., isolated from Altamira cave, Cantabria, Spain. Int. J. Syst. Evol. Microbiol. 2008, 58, 2210–2214. [Google Scholar] [CrossRef]
- Jurado, V.; Kroppenstedt, R.M.; Saiz-Jimenez, C.; Klenk, H.-P.; Mouniée, D.; Laiz, L.; Couble, A.; Pötter, G.; Boiron, P.; Rodríguez-Nava, V. Hoyosella altamirensis gen. nov., sp. nov., a new member of the order Actinomycetales isolated from a cave biofilm. Int. J. Syst. Evol. Microbiol. 2009, 59, 3105–3110. [Google Scholar] [CrossRef]
- Nováková, A.; Hubka, V.; Saiz-Jimenez, C.; Kolarik, M. Aspergillus baeticus sp. nov. and Aspergillus thesauricus sp. nov., two species in section Usti from Spanish caves. Int. J. Syst. Evol. Microbiol. 2012, 62, 2778–2785. [Google Scholar] [CrossRef]
- Dominguez-Moñino, I.; Jurado, V.; Hermosin, B.; Saiz-Jimenez, C. Paracoccus cavernae sp. nov., isolated from a show cave. Int. J. Syst. Evol. Microbiol. 2016, 66, 2265–2270. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Moñino, I.; Jurado, V.; Gonzalez-Pimentel, J.L.; Miller, A.Z.; Hermosin, B.; Saiz-Jimenez, C. Bacillus onubensis sp. nov., isolated from the air of two Andalusian caves. Syst. Appl. Microbiol. 2018, 41, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Patricio, S.; Gonzalez-Pimentel, J.L.; Miller, A.Z.; Hermosin, B.; Saiz-Jimenez, C. Paracoccus onubensis sp. nov., a novel alphaproteobacterium isolated from the wall of a show cave. Int. J. Syst. Evol. Microbiol. 2021, 71, 004942. [Google Scholar] [CrossRef] [PubMed]
- Dorrestein, P.C.; Kelleher, N.L. Dissecting non-ribosomal and polyketide biosynthetic machineries using electrospray ionization Fourier-Transform mass spectrometry. Nat. Prod. Rep. 2006, 23, 893–918. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta 2013, 1830, 3670–3695. [Google Scholar] [CrossRef]
- Gonzalez-Pimentel, J.L.; Miller, A.Z.; Jurado, V.; Laiz, L.; Pereira, M.F.C.; Saiz-Jimenez, C. Yellow colored mats from lava tube of La Palma (Canary Islands, Spain) are dominated by metabolically active Actinobacteria. Sci. Rep. 2018, 8, 1944. [Google Scholar] [CrossRef]
- Sanchez-Moral, S. Estudio Integral del Estado de Conservación de la Cueva de Altamira y su Arte Paleolítico (2007–2009). Perspectivas Futuras de Conservación; Monografías No. 24, Museo Nacional y Centro de Investigación de Altamira; Ministerio de Educación, Cultura y Deporte: Madrid, Spain, 2014. [Google Scholar]
- Porca, E.; Jurado, V.; Martin-Sanchez, P.M.; Hermosin, B.; Bastian, F.; Alabouvette, C.; Saiz-Jimenez, C. Aerobiology: An ecological indicator for early detection and control of fungal outbreaks in caves. Ecol. Indic. 2011, 11, 1594–1598. [Google Scholar] [CrossRef]
- Dominguez-Moñino, I.; Jurado, V.; Rogerio-Candelera, M.A.; Hermosin, B.; Saiz-Jimenez, C. Airborne bacteria in show caves from Southern Spain. Microb. Cell 2021, 8, 247–255. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y. Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Jonsbu, E.; McIntyre, M.; Nielsen, J. The influence of carbon sources and morphology on nystatin production by Streptomyces noursei. J. Biotechnol. 2002, 95, 133–144. [Google Scholar] [CrossRef]
- Saudagar, P.S.; Singhal, R.S. Optimization of nutritional requirements and feeding strategies for clavulanic acid production by Streptomyces clavuligerus. Bioresour. Technol. 2007, 98, 2010–2017. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.R. Bacterial inhibition of fungal growth and pathogenicity. Microb. Ecol. Health Dis. 1999, 11, 129–142. [Google Scholar] [CrossRef]
- Dominguez-Moñino, I.; Jurado, V.; Rogerio-Candelera, M.A.; Hermosin, B.; Saiz-Jimenez, C. Airborne fungi in show caves from Southern Spain. Appl. Sci. 2021, 11, 5027. [Google Scholar] [CrossRef]
- Martin-Sanchez, P.M.; Nováková, A.; Bastian, F.; Alabouvette, C.; Saiz-Jimenez, C. Two new species of the genus Ochroconis, O. lascauxensis and O. anomala isolated from black stains in Lascaux Cave, France. Fungal Biol. 2012, 116, 574–589. [Google Scholar] [CrossRef] [PubMed]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Glöckner, F.O.; Peplies, J. JSpeciesWS: A Web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Eren, A.M.; Kiefl, E.; Shaiber, A.; Veseli, I.; Miller, S.E.; Schechter, M.S.; Fink, I.; Pan, J.N.; Yousef, M.; Fogarty, E.C.; et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 2021, 6, 3–6. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Martin-Sanchez, P.M.; Jurado, V.; Porca, E.; Bastian, F.; Lacanette, D.; Alabouvette, C.; Saiz-Jimenez, C. Aerobiology of Lascaux Cave (France). Int. J. Speleol. 2014, 43, 295–303. [Google Scholar] [CrossRef]
- Montano, E.T.; Henderson, L.O. Studies of antibiotic production by cave bacteria. In Cave Microbiomes: A Novel Resource for Drug Discovery; Cheeptham, N., Ed.; Springer: New York, NY, USA, 2013; pp. 109–130. [Google Scholar] [CrossRef]
- Jurado, V.; Fernandez-Cortes, A.; Cuezva, S.; Laiz, L.; Cañaveras, J.C.; Sanchez-Moral, S.; Saiz-Jimenez, C. The fungal colonisation of rock-art caves: Experimental evidence. Naturwissenschaften 2009, 96, 1027–1034. [Google Scholar] [CrossRef]
- Garcia-Anton, E.; Cuezva, S.; Jurado, V.; Porca, E.; Miller, A.Z.; Fernandez-Cortes, A.; Saiz-Jimenez, C.; Sanchez-Moral, S. Combining stable isotope (δ13C) of trace gases and aerobiological data to monitor the entry and dispersion of microorganisms in caves. Environ. Sci. Pollut. Res. 2014, 21, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Bérdy, J. Bioactive microbial metabolites. A personal view. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Moral, S.; Jurado, V.; Fernandez-Cortes, A.; Cuezva, S.; Martin-Pozas, T.; Gonzalez-Pimentel, J.L.; Ontañon, R.; Saiz-Jimenez, C. Environment-driven control of fungi in subterranean ecosystems: The case of La Garma Cave (northern Spain). Int. Microbiol. 2021, 24, 573–591. [Google Scholar] [CrossRef] [PubMed]
- Donahue, J.M.; Williams, N.M.; Sells, S.F.; Labeda, D.P. Crossiella equi sp. nov., isolated from equine placentas. Int. J. Syst. Evol. Microbiol. 2002, 52, 2169–2173. [Google Scholar] [CrossRef]
- Card, C.; Lopate, C. Infectious diseases of the puerperal period. In Current Therapy in Large Animal Theriogenology, 2nd ed.; Youngquist, R.S., Threlfall, W.R., Eds.; Saunders: Philadelphia, PA, USA, 2007; pp. 138–144. [Google Scholar] [CrossRef]
- Labeda, D.P.; Lechevalier, M.P. Amendment of the genus Saccharothrix Labeda et al. 1984 and descriptions of Saccharothrix espanaensis sp. nov., Saccharothrix cryophilis sp. nov., and Saccharothrix mutabilis comb. nov. Int. J. Syst. Bacteriol. 1989, 39, 420–423. [Google Scholar] [CrossRef]
- Labeda, D.P. Crossiella gen. nov., a new genus related to Streptoalloteichus. Int. J. Syst. Evol. Microbiol. 2001, 51, 1575–1579. [Google Scholar] [CrossRef]
- Takahashi, A.; Hotta, K.; Saito, N.; Morioka, M.; Okami, Y.; Umezawa, H. Production of novel antibiotic, dopsisamine, by a new subspecies of Nocardiopsis mutabilis with multiple antibiotic resistance. J. Antibiot. 1986, 39, 175–183. [Google Scholar] [CrossRef]
- Caetano, T.; van der Donk, W.; Mendo, S. Bacteroidetes can be a rich source of novel lanthipeptides: The case study of Pedobacter lusitanus. Microbiol. Res. 2020, 235, 126441. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Alam, K.; Zhao, Y.; Hao, J.; Yang, Q.; Zhang, Y.; Li, R.; Li, A. Mining and biosynthesis of bioactive lanthipeptides from microorganisms. Front. Bioeng. Biotechnol. 2021, 9, 692466. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, Y.; Zhai, G.; Fu, S.; Xia, Y.; Hu, B.; Cai, X.; Zhang, Y.; Li, Y.; Deng, Z.; et al. A cell-free platform based on nisin biosynthesis for discovering novel lanthipeptides and guiding their overproduction in vivo. Adv. Sci. 2020, 7, 2001616. [Google Scholar] [CrossRef] [PubMed]
- Kisil, O.V.; Efimenko, T.A.; Efremenkova, O.V. Looking back to Amycolatopsis: History of the antibiotic discovery and future prospects. Antibiotics 2021, 10, 1254. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, J.; Li, G.; Yang, Y.; Ding, W. Current advancements in sactipeptide natural products. Front. Chem. 2021, 9, 595991. [Google Scholar] [CrossRef] [PubMed]
- Engelberg-Kulka, H.; Hazan, R. Cannibals defy starvation and avoid sporulation. Science 2003, 301, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Salomón, R.A.; Farías, R.N. Microcin 25, a novel antimicrobial peptide produced by Escherichia coli. J. Bacteriol. 1992, 174, 7428–7435. [Google Scholar] [CrossRef] [PubMed]
- Knappe, T.A.; Linne, U.; Zirah, S.; Rebuffat, S.; Xie, X.; Marahiel, M.A. Isolation and structural characterization of capistruin, a lasso peptide predicted from the genome sequence of Burkholderia thailandensis E264. J. Am. Chem. Soc. 2008, 130, 11446–11454. [Google Scholar] [CrossRef]
- Zimmermann, M.; Hegemann, J.D.; Xie, X.; Marahiel, M.A. The astexin-1 lasso peptides: Biosynthesis, stability, and structural studies. Chem. Biol. 2013, 20, 558–569. [Google Scholar] [CrossRef]
- Jomon, K.; Kuroda, Y.; Ajisaka, M.; Sakai, H. A new antibiotic, ikarugamycin. J. Antibiot. 1972, 25, 271–280. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, W.; Zhang, Q.; Shi, T.; Ma, L.; Zhu, Y.; Li, S.; Zhang, H.; Zhao, Y.-L.; Shi, R.; et al. Mechanistic insights into polycycle formation by reductive cyclization in ikarugamycin biosynthesis. Angew. Chem. 2014, 53, 4840–4844. [Google Scholar] [CrossRef] [PubMed]
- Tietz, J.; Schwalen, C.; Patel, P.; Maxson, T.; Blair, P.M.; Tai, H.-C.; Zakai, U.I.; Mitchell, D.A. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 2017, 13, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Qiao, C.; Wilson, D.J.; Bennett, E.M.; Aldrich, C.C. A mechanism-based aryl carrier protein/thiolation domain affinity probe. J. Am. Chem. Soc. 2007, 129, 6350–6351. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, R.; Ebert-Khosla, S.; Hopwood, D.A.; Khosla, C. Engineered biosynthesis of novel polyketides. Science 1993, 262, 1546–1550. [Google Scholar] [CrossRef]
- Staunton, J.; Weissman, K.J. Polyketide biosynthesis: A millennium review. Nat. Prod. Rep. 2001, 18, 380–416. [Google Scholar] [CrossRef]
- Yu, D.; Xu, F.; Zeng, J.; Zhan, J. Type III polyketide synthases in natural product biosynthesis. IUBMB Life 2012, 64, 285–295. [Google Scholar] [CrossRef]
- Umezawa, H.; Takeuchi, T. Nitrosporin, antibiotic from Streptomyces nitrosporeus active to Gram positive bacteria. Jpn. J. Med. 1951, 4, 173–179. [Google Scholar] [CrossRef]
- Yang, X.; Van Der Donk, W.A. Ribosomally synthesized and post-translationally modified peptide natural products: New insights into the role of leader and core peptides during biosynthesis. Chemistry 2013, 19, 7662–7677. [Google Scholar] [CrossRef]
- Kudo, F.; Kasama, Y.; Hirayama, T.; Eguchi, T. Cloning of the pactamycin biosynthetic gene cluster and characterization of a crucial glycosyltransferase prior to a unique cyclopentane ring formation. J. Antibiot. 2007, 60, 492–503. [Google Scholar] [CrossRef]
- Xiao, J.; Zhang, Q.; Zhu, Y.; Li, S.; Zhang, G.; Zhang, H.; Saurav, K.; Zhang, C. Characterization of the sugar-O-methyltransferase LobS1 in lobophorin biosynthesis. Appl. Microbiol. Biotechnol. 2013, 97, 9043–9053. [Google Scholar] [CrossRef]
- Rutkowski, J.; Brzezinski, B. Structures and properties of naturally occurring polyether antibiotics. BioMed Res. Int. 2013, 2013, 162513. [Google Scholar] [CrossRef] [PubMed]
- Kevin II, D.A.; Meujo, D.A.; Hamann, M.T. Polyether ionophores: Broad-spectrum and promising biologically active molecules for the control of drug-resistant bacteria and parasites. Expert. Opin. Drug. Discov. 2009, 4, 109–146. [Google Scholar] [CrossRef] [PubMed]
- Migita, A.; Watanabe, M.; Hirose, Y.; Watanabe, K.; Tokiwano, T.; Kinashi, H.; Oikawa, H. Identification of a gene cluster of polyether antibiotic lasalocid from Streptomyces lasaliensis. Biosci. Biotechnol. Biochem. 2009, 73, 169–176. [Google Scholar] [CrossRef]
- Park, H.R.; Furihata, K.; Hayakawa, Y.; Shin-Ya, K. Versipelostatin, a novel GRP78/Bip molecular chaperone down-regulator of microbial origin. Tetrahedron Lett. 2002, 43, 6941–6945. [Google Scholar] [CrossRef]

| Source | Tested Strains | AC Strains | % |
|---|---|---|---|
| Altamira Cave (rock) | 289 | 77 | 26.6 |
| Tito Bustillo Cave (rock) | 181 | 42 | 23.2 |
| Ardales Cave (air) | 89 | 9 | 10.1 |
| Maravillas Grotta (air) | 200 | 14 | 7.0 |
| Tesoro Cave (air) | 89 | 4 | 4.5 |
| Tesoro Cave (rock) | 8 | 2 | 25.0 |
| La Palma Island lava tubes (rock) | 160 | 30 | 23.8 |
| Viento Cave (Tenerife Island) (rock) | 31 | 7 | 22.8 |
| Total | 1047 | 178 | 17.0 |
| Total (rocks) | 669 | 151 | 22.6 |
| Total (air) | 378 | 27 | 7.1 |
| Cluster Type | Closest Species | Similarity (%) |
|---|---|---|
| Lanthipeptide-class-I | Amycolatopsis marina | 91–92–93.8 |
| Sactipeptide | Micromonospora ureilytica | 49.2 |
| Lasso peptide (1) | Nocardiopsis alba-Actinokineospora iranica | 52.3–50.4 |
| Furan | Thermobifida halotolerans | 41.3 |
| Lasso peptide (2) | Streptomyces sp. | 55–50 |
| Lanthipeptide-class-V | Crossiella cryophila-Rubrobacter xylanophilus-Blastococcus sp. | 88.4–29.9–31.1 |
| Lasso peptide (3) | Saccharothrix espanaensis | 63.4–51.1 |
| NRPS (1) | Actinoplanes sp.-Crossiella cryophila | 56.3–91.2 |
| T1PKSs (1) | Streptomyces nitrosporeus | 53.9 |
| Lanthipeptide-class-III | Streptomyces curacoi | 50.2 |
| NRPS (2) | Streptomyces sp. | 53.1 |
| T1PKSs (2) (Region on contig edge) | Streptomyces sp. | 49.1–48.9 |
| T1PKSs (3) (Region on contig edge) | Umezawaea tangerina-Kutzneria buriramensis-Lentzea waywayandensis | 65–49.7–52.7 |
| Lasso peptide (4) (Region on contig edge) | Amycolatopsis rhizosphaerae | 52.4–53.4–51.4 |
| T1PKSs (4) (Region on contig edge) | Streptomyces versipellis | 55.3–50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez-Pimentel, J.L.; Dominguez-Moñino, I.; Jurado, V.; Laiz, L.; Caldeira, A.T.; Saiz-Jimenez, C. The Rare Actinobacterium Crossiella sp. Is a Potential Source of New Bioactive Compounds with Activity against Bacteria and Fungi. Microorganisms 2022, 10, 1575. https://doi.org/10.3390/microorganisms10081575
Gonzalez-Pimentel JL, Dominguez-Moñino I, Jurado V, Laiz L, Caldeira AT, Saiz-Jimenez C. The Rare Actinobacterium Crossiella sp. Is a Potential Source of New Bioactive Compounds with Activity against Bacteria and Fungi. Microorganisms. 2022; 10(8):1575. https://doi.org/10.3390/microorganisms10081575
Chicago/Turabian StyleGonzalez-Pimentel, Jose Luis, Irene Dominguez-Moñino, Valme Jurado, Leonila Laiz, Ana Teresa Caldeira, and Cesareo Saiz-Jimenez. 2022. "The Rare Actinobacterium Crossiella sp. Is a Potential Source of New Bioactive Compounds with Activity against Bacteria and Fungi" Microorganisms 10, no. 8: 1575. https://doi.org/10.3390/microorganisms10081575







