Differed Growth Stage Dynamics of Root-Associated Bacterial and Fungal Community Structure Associated with Halophytic Plant Lycium ruthenicum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Areas and Sample Collection
2.2. Soil Chemical Properties Determination
2.3. DNA Extraction, PCR Amplification, and Sequencing
2.4. Sequence Analysis
2.5. Statistical Analysis
3. Results
3.1. Soil Chemical Properties
3.2. Statistic of Sequencing Data
3.3. Microbial Community Structure of Bacteria and Fungi
3.3.1. Bacterial Community Structure
3.3.2. Fungal Community Structure
3.4. Microbial Community Structure Exhibiting Distinct and Overlapping in Three Spatial Root Compartments
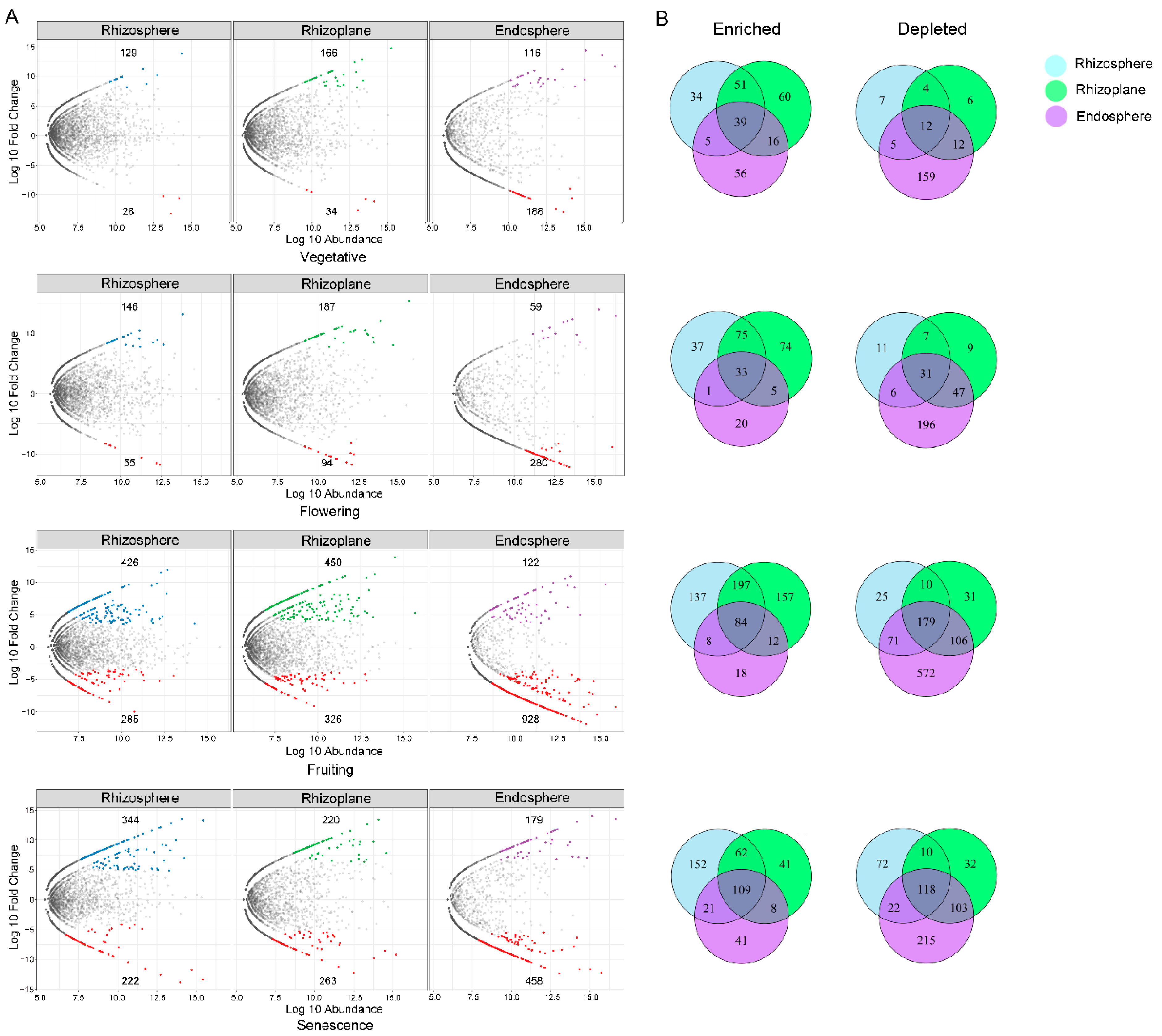
3.4.1. Association of Significantly Enriched Bacterial OTUs with Different Rhizocompartments
3.4.2. Association of Significantly Enriched Fungal OTUs with Different Rhizocompartments
3.5. Compartment-Specific Biomarkers Identification in Each Root Compartment
3.6. Growth Stage Dynamics of Microbial Communities in Each Rhizocompartment
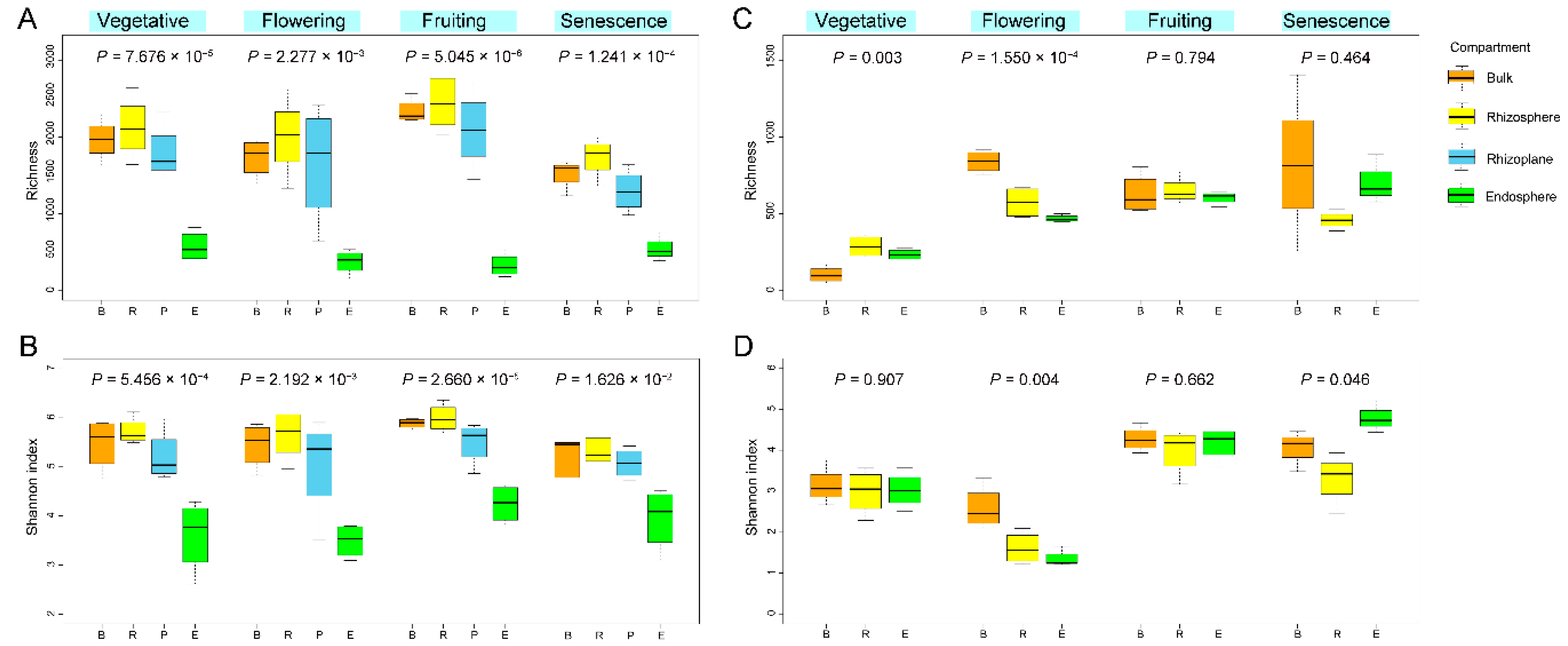
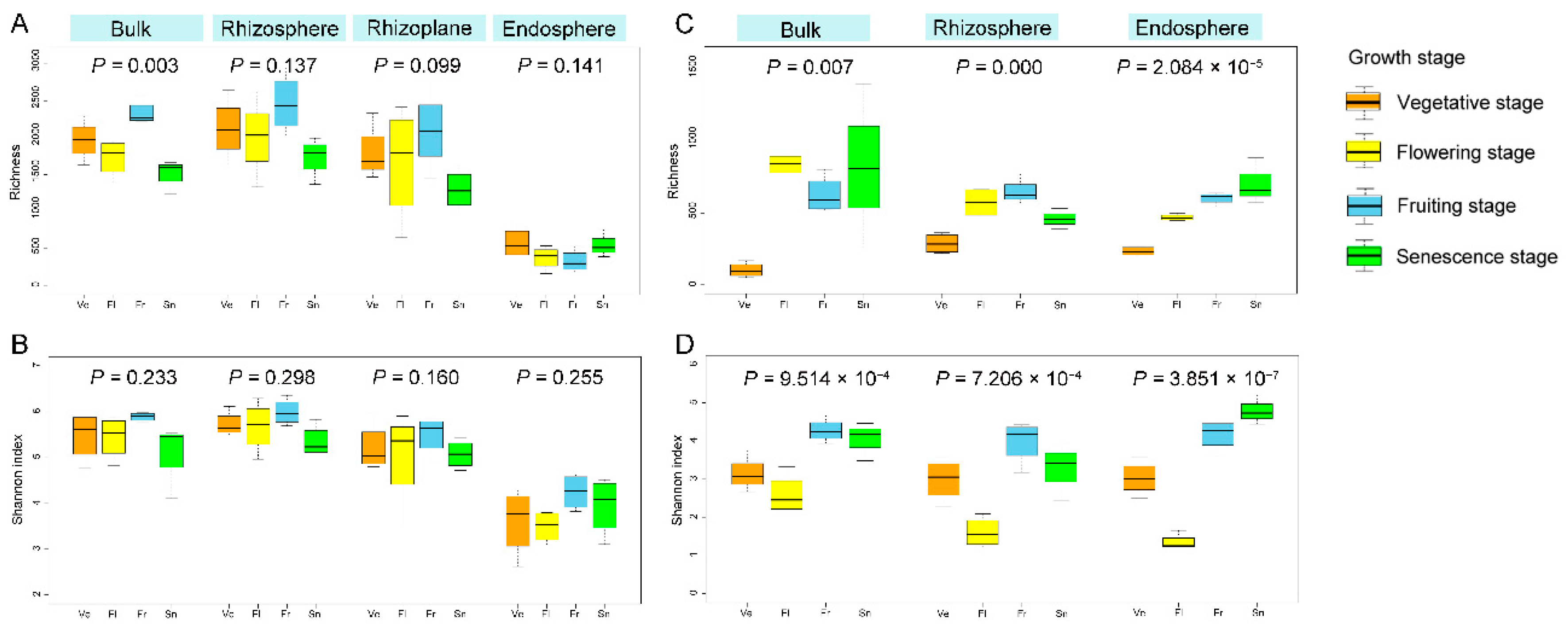
3.6.1. Diversity Dynamics along with Growth Stage
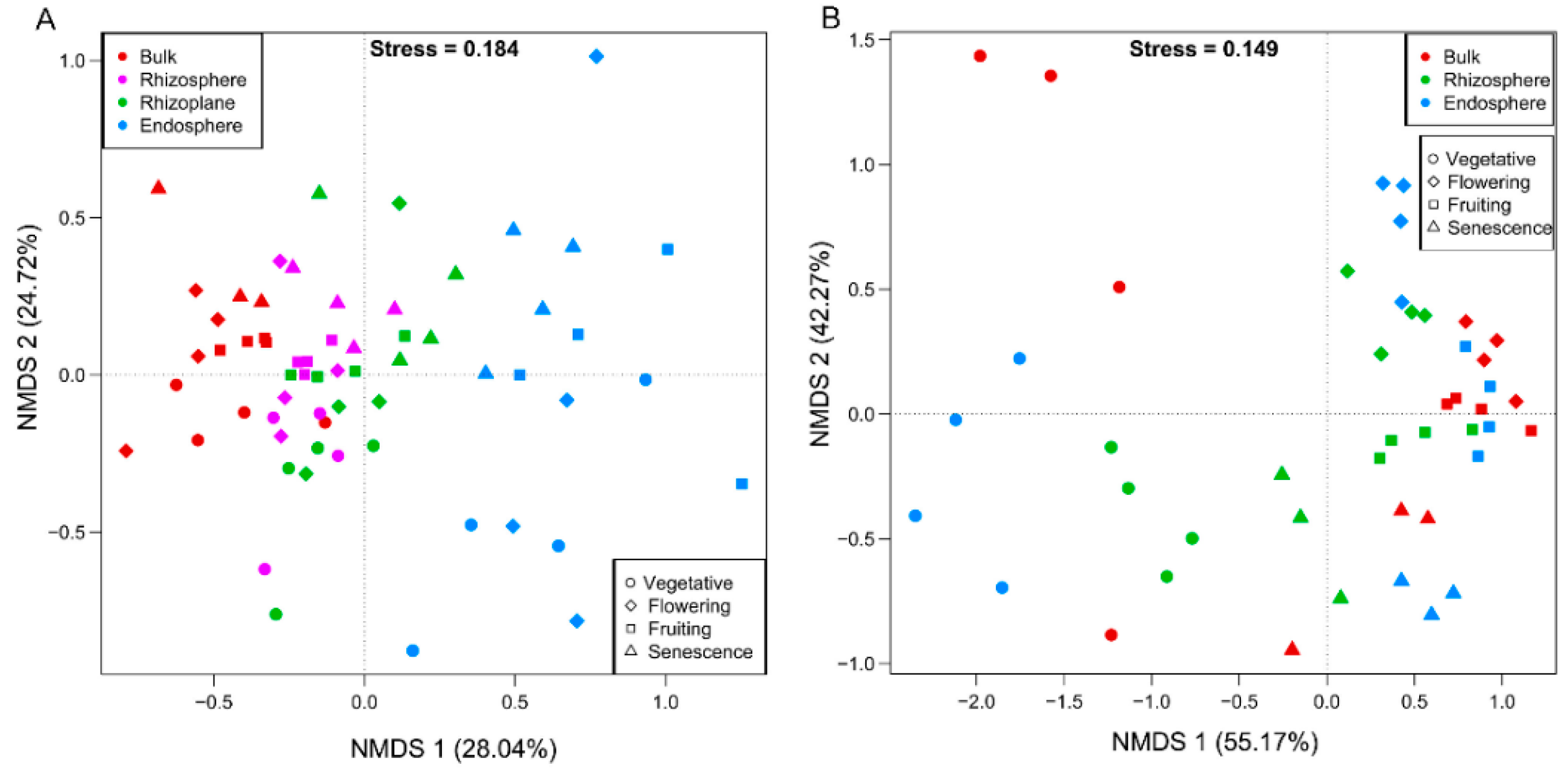
3.6.2. Growth Stage Dynamics of Bacterial Community Structure
3.6.3. Growth Stage Effects on Fungal Community Structure
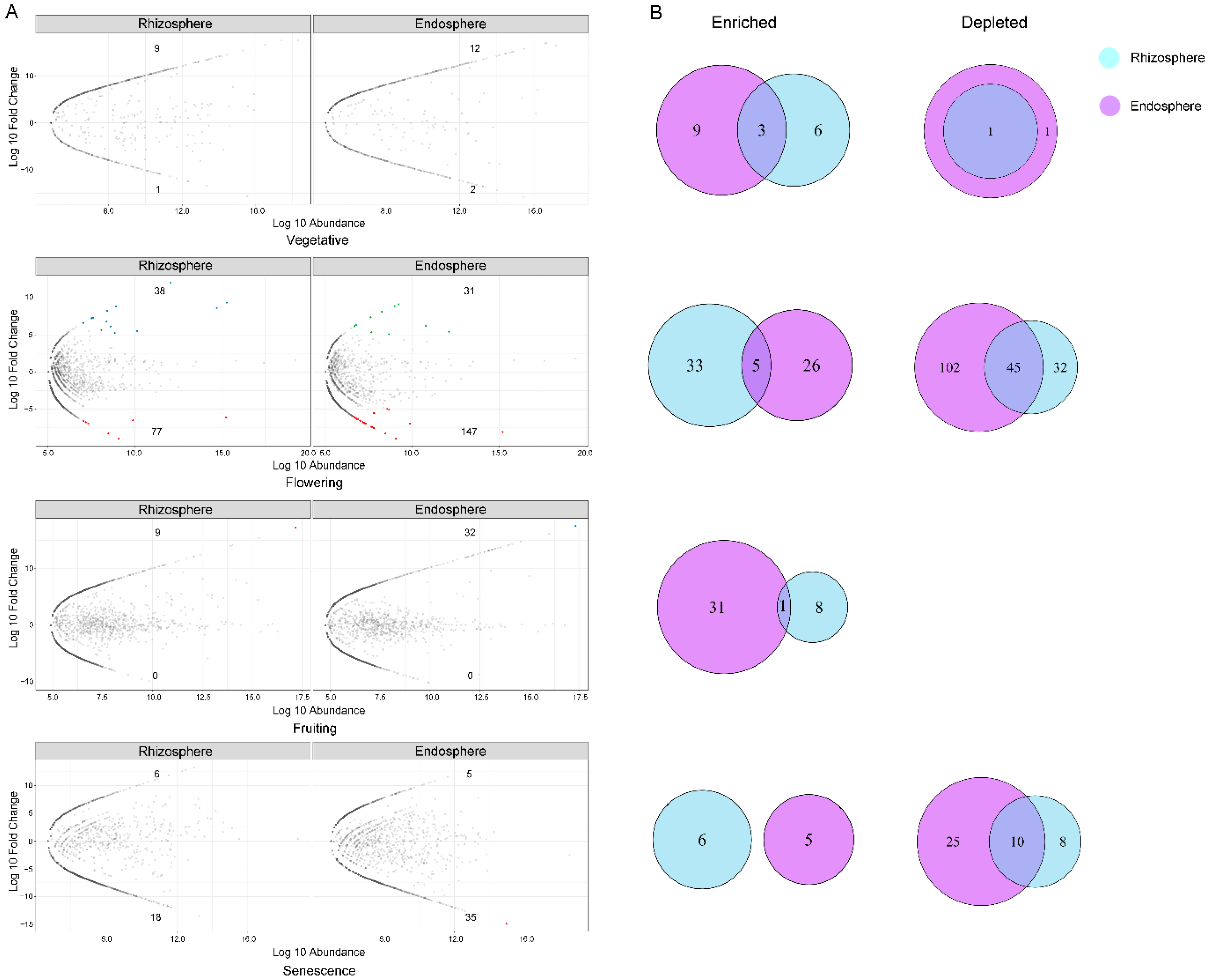
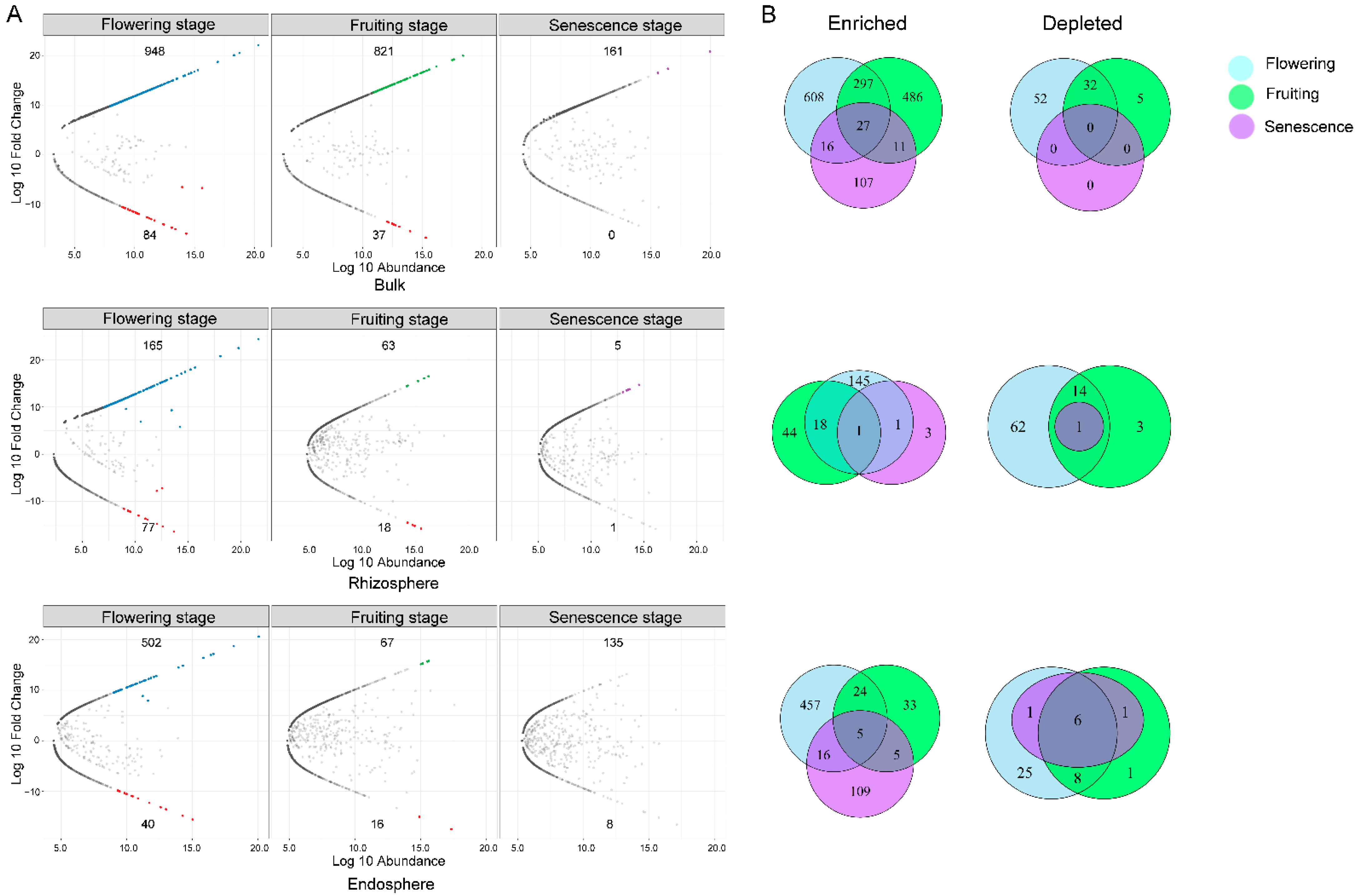
3.7. Growth Stage-Specific Biomarkers Identification in Each Comparment
4. Discussion
4.1. The Dominant Root-Associated Microbial Community Composition
4.2. Bacterial Community Differed between Root Compartments
4.3. Differential Analysis Identified Root Compartment-Specific Enriched or Depleted Bacteria
4.4. Root Compartment-Specific Bacterial Biomarkers Varied with Growth Stage
4.5. Differential Analysis Identified Root Compartment-Specific Enriched or Depleted Fungi
4.6. Root Compartment-Specific Fungal Biomarkers Varied with Growth Stage
4.7. Growth Stage Dynamics of Micobiomes Diversity and Structure in Root Compartment
4.8. Growth Stage Dynamics of Microbial Structure Might Be Induced by Plant Rhizodeposits and Abiotic Factors of Seasonal Changes
4.9. Difference Pattern between Bacteria and Fungi
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; van Themaat, E.V.L.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellin, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.A.; Santos-Medellin, C.M.; Liechty, Z.S.; Nguyen, B.; Lurie, E.; Eason, S.; Phillips, G.; Sundaresan, V. Compositional shifts in root-associated bacterial and archaeal microbiota track the plant life cycle in field-grown rice. PLoS Biol. 2018, 16, e2003862. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.K.; Cui, J.H.; Ren, G.Z.; Wei, S.L.; Yang, P.Y.; Yin, C.P.; Liang, H.K.; Chang, J.H. Changes in the root-associated bacteria of sorghum are driven by the combined effects of salt and sorghum development. Environ. Microbiome 2021, 16, 14. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Davenport, E.R.; Waters, J.L.; Clark, A.G.; Ley, R.E. Cross-species comparisons of host genetic associations with the microbiome. Science 2016, 352, 532–535. [Google Scholar] [CrossRef]
- Li, J.; Luo, Z.Q.; Zhang, C.H.; Qu, X.J.; Chen, M.; Song, T.; Yuan, J. Seasonal variation in the rhizosphere and non-rhizosphere microbial community structures and functions of Camellia yuhsienensis Hu. Microorganisms 2020, 8, 1385. [Google Scholar] [CrossRef]
- Knoth, J.L.; Kim, S.H.; Ettl, G.J.; Doty, S.L. Effects of cross host species inoculation of nitrogen-fixing endophytes on growth and leaf physiology of maize. GCB Bioenergy 2013, 5, 408–418. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.J.; Alqarawi, A.A.; Abd Allah, E.F.; Hashem, A. Phytohormones and beneficial microbes: Essential components for plants to balance stress and fitness. Front. Microbiol. 2017, 8, 2104. [Google Scholar] [CrossRef]
- Qu, Q.; Zhang, Z.Y.; Peijnenburg, W.J.G.M.; Liu, W.Y.; Lu, T.; Hu, B.L.; Chen, J.M.; Chen, J.; Lin, Z.F.; Qian, H.F. Rhizosphere microbiome assembly and its impact on plant growth. J. Agric. Food Chem. 2020, 68, 5024–5038. [Google Scholar] [CrossRef]
- Mathur, P.; Roy, S. Insights into the plant responses to drought and decoding the potential of root associated microbiome for inducing drought tolerance. Physiol. Plant. 2021, 172, 1016–1029. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Chakraborty, A.P.; Chakraborty, R. Understanding the potential of root microbiome influencing salt-tolerance in plants and mechanisms involved at the transcriptional and translational level. Physiol. Plant. 2021, 173, 1657–1681. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.L.; Cheng, R.Y.; Xiao, L.N.; Wei, F.G.; Wei, G.F.; Xu, J.; Wang, Y.; Guo, X.T.; Chen, Z.J.; Chen, S.L. Diversity and composition of bacterial endophytes among plant parts of Panax notoginseng. Chin. Med. 2018, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Ibekwe, A.M.; Ors, S.; Ferreira, J.F.S.; Liu, X.; Suarez, D.L. Seasonal induced changes in spinach rhizosphere microbial community structure with varying salinity and drought. Sci. Total Environ. 2017, 579, 1485–1495. [Google Scholar] [CrossRef]
- Wu, Z.Y.; Lin, W.X.; Li, J.J.; Liu, J.F.; Li, B.L.; Wu, L.K.; Fang, C.X.; Zhang, Z.X. Effects of seasonal variations on soil microbial community composition of two typical zonal vegetation types in the Wuyi Mountains. J. Mt. Sci. 2016, 13, 1056–1065. [Google Scholar] [CrossRef]
- Peng, F.; Huang, C.H.; You, Q.G.; Gao, T.P. Effects of plantation of Lycium ruthenicum on the soil salt distribution in the minqin basin. J. Desert Res. 2013, 33, 1406–1412. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Zhang, J.J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-supported software for describing and comparing microbial communities. Appl. Environ. Microb. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Moriwaki, T.; Miyazawa, Y.; Kobayashi, A.; Uchida, M.; Watanabe, C.; Fujii, N.; Takahashi, H. Hormonal regulation of lateral root development in Arabidopsis modulated by MIZ1 and requirement of GNOM activity for MIZ1 function. Plant Physiol. 2011, 157, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef]
- Ai, C.; Liang, G.Q.; Sun, J.W.; Wang, X.B.; He, P.; Zhou, W.; He, X.H. Reduced dependence of rhizosphere microbiome on plant-derived carbon in 32-year long-term inorganic and organic fertilized soils. Soil Biol. Biochem. 2015, 80, 70–78. [Google Scholar] [CrossRef]
- Schlaeppi, K.; Dombrowski, N.; Oter, R.G.; van Themaat, E.V.L.; Schulze-Lefert, P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc. Natl. Acad. Sci. USA 2014, 111, 585–592. [Google Scholar] [CrossRef]
- Mukhtar, S.; Mehnaz, S.; Malik, K.A. Comparative study of the rhizosphere and root endosphere microbiomes of Cholistan desert plants. Front. Microbiol. 2021, 12, 618742. [Google Scholar] [CrossRef]
- Deng, S.S.; Chang, X.L.; Zhang, Y.M.; Ren, L.Z.; Jiang, F.; Qu, Z.H.; Peng, F. Nocardioides antarcticus sp nova, isolated from marine sediment. Int. J. Syst. Evol. Micr. 2015, 65, 2615–2621. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Z.F.; Zhang, Y.L.; Ding, M.J.; Li, L.H. Identification of traffic-related metals and the effects of different environments on their enrichment in roadside soils along the Qinghai-Tibet highway. Sci. Total Environ. 2015, 521, 160–172. [Google Scholar] [CrossRef]
- Kielak, A.M.; Cipriano, M.A.P.; Kuramae, E.E. Acidobacteria strains from subdivision 1 act as plant growth-promoting bacteria. Arch. Microbiol. 2016, 198, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M.; Mirza, M.S.; Hameed, S.; Dimitrov, M.R.; Smidt, H. Cultivation-based and molecular assessment of bacterial diversity in the rhizosheath of wheat under different crop rotations. PLoS ONE 2015, 10, e0130030. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.L.; Sun, Y.; Wu, B.; Zhao, S.; Yuan, B.; Qin, S.; Jiang, J.H.; Huang, Y. Actinophytocola glycyrrhizae sp nov isolated from the rhizosphere of Glycyrrhiza inflata. Int. J. Syst. Evol. Micr. 2018, 68, 2504–2508. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, B.; Meng, H.Y.; Xing, Z.B.; Lai, Q.L.; Yuan, L.J. Actinophytocola xanthii sp nov.; an actinomycete isolated from rhizosphere soil of the plant Xanthium sibiricum. Int. J. Syst. Evol. Microbiol. 2017, 67, 1152–1157. [Google Scholar] [CrossRef]
- Cabanás, C.G.L.; Fernández-González, A.J.; Cardoni, M.; Valverde-Corredor, A.; López-Cepero, J.; Fernández-López, M.; Mercado-Blanco, J. The Banana root endophytome: Differences between mother plants and suckers and evaluation of selected bacteria to control Fusarium oxysporum f. sp. Cubense. J. Fungi 2021, 7, 194. [Google Scholar] [CrossRef] [PubMed]
- Fernández-González, A.J.; Ramirez-Tejero, J.A.; Nevado-Berzosa, M.P.; Luque, F.; Fernández-López, M.; Mercado-Blanco, J. Coupling the endophytic microbiome with the host transcriptome in olive roots. Comput. Struct. Biotechnol. J. 2021, 19, 4777–4789. [Google Scholar] [CrossRef] [PubMed]
- McGuinness, M.; Dowling, D. Plant-associated bacterial degradation of toxic organic compounds in soil. Int. J. Environ. Res. Public Health 2009, 6, 2226–2247. [Google Scholar] [CrossRef]
- Kim, M.K.; Srinivasan, S.; Park, M.J.; Sathiyaraj, G.; Kim, Y.J.; Yang, D.C. Nocardioides humi sp nov.; A beta-glucosidase-producing bacterium isolated from soil of a ginseng field. Int. J. Syst. Evol. Microbiol. 2009, 59, 2724–2728. [Google Scholar] [CrossRef]
- King, W.L.; Yates, C.F.; Guo, J.; Fleishman, S.M.; Trexler, R.V.; Centinari, M.; Bell, T.H.; Eissenstat, D.M. The hierarchy of root branching order determines bacterial composition, microbial carrying capacity and microbial filtering. Commun. Biol. 2021, 4, 483. [Google Scholar] [CrossRef]
- Dai, Y.Y.; Liu, R.; Zhou, Y.M.; Li, N.; Hou, L.Q.; Ma, Q.; Gao, B. Fire Phoenix facilitates phytoremediation of PAH-Cd co-contaminated soil through promotion of beneficial rhizosphere bacterial communities. Environ. Int. 2020, 136, 105421. [Google Scholar] [CrossRef]
- Hu, H.Y.; Li, H.; Hao, M.M.; Ren, Y.N.; Zhang, M.K.; Liu, R.Y.; Zhang, Y.; Li, G.; Chen, J.S.; Ning, T.Y.; et al. Nitrogen fixation and crop productivity enhancements co-driven by intercrop root exudates and key rhizosphere bacteria. J. Appl. Ecol. 2021, 58, 2243–2255. [Google Scholar] [CrossRef]
- Jin, K.M.; Li, H.B.; Li, X.Q.; Li, H.G.; Dodd, I.C.; Belimov, A.A.; Davies, W.J.; Shen, J.B. Rhizosphere bacteria containing ACC deaminase decrease root ethylene emission and improve maize root growth with localized nutrient supply. Food Energy Secur. 2021, 10, e278. [Google Scholar] [CrossRef]
- Lemanceau, P.; Blouin, M.; Muller, D.; Moenne-Loccoz, Y. Let the core microbiota be functional. Trends Plant Sci. 2017, 22, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, Y.; Zhang, P.F.; Trivedi, P.; Riera, N.; Wang, Y.Y.; Liu, X.; Fan, G.Y.; Tang, J.L.; Coletta, H.D.; et al. The structure and function of the global citrus rhizosphere microbiome. Nat. Commun. 2018, 9, 4894. [Google Scholar] [CrossRef]
- Chaluvadi, S.; Bennetzen, J.L. Species-associated differences in the below-ground microbiomes of wild and domesticated Setaria. Front. Plant Sci. 2018, 9, 1183. [Google Scholar] [CrossRef]
- Li, M.; Jain, S.; Baker, B.J.; Taylor, C.; Dick, G.J. Novel hydrocarbon monooxygenase genes in the metatranscriptome of a natural deep-sea hydrocarbon plume. Environ. Microbiol. 2014, 16, 60–71. [Google Scholar] [CrossRef]
- Maarastawi, S.A.; Frindte, K.; Geer, R.; Krober, E.; Knief, C. Temporal dynamics and compartment specific rice straw degradation in bulk soil and the rhizosphere of maize. Soil Biol. Biochem. 2018, 127, 200–212. [Google Scholar] [CrossRef]
- Yaish, M.W.; Al-Lawati, A.; Jana, G.A.; Patankar, H.V.; Glick, B.R. Impact of soil salinity on the structure of the bacterial endophytic community identified from the roots of caliph medic (Medicago truncatula). PLoS ONE 2016, 11, e0159007. [Google Scholar] [CrossRef]
- Yang, H.; Hu, J.X.; Long, X.H.; Liu, Z.P.; Rengel, Z. Salinity altered root distribution and increased diversity of bacterial communities in the rhizosphere soil of Jerusalem artichoke. Sci. Rep. 2016, 6, 20687. [Google Scholar] [CrossRef]
- Engelbrecht, G.; Claassens, S.; Mienie, C.M.S.; Fourie, H. Screening of rhizosphere bacteria and nematode populations associated with soybean roots in the Mpumalanga Highveld of South Africa. Microorganisms 2021, 9, 1813. [Google Scholar] [CrossRef]
- Ferreira, S.D.; Nakasone, A.K.; do Nascimento, S.M.C.; de Oliveira, D.A.; Siqueira, A.S.; Cunha, E.F.M.; de Castro, G.L.S.; de Souza, C.R.B. Isolation and characterization of cassava root endophytic bacteria with the ability to promote plant growth and control the in vitro and in vivo growth of Phytopythium sp. Physiol. Mol. Plant Prod. 2021, 116, 101709. [Google Scholar] [CrossRef]
- Kang, S.M.; Joo, G.J.; Hamayun, M.; Na, C.I.; Shin, D.H.; Kim, H.Y.; Hong, J.K.; Lee, I.J. Gibberellin production and phosphate solubilization by newly isolated strain of Acinetobacter calcoaceticus and its effect on plant growth. Biotechnol. Lett. 2009, 31, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Moslehi, S.; Pourmehr, S.; Shirzad, A.; Khakvar, R. Potential of some endophytic bacteria in biological control of root-knot nematode Meloidogyne incognita. Egypt. J. Biol. Pest Control 2021, 31, 50. [Google Scholar] [CrossRef]
- Singh, K.; Gera, R.; Sharma, R.; Maithani, D.; Chandra, D.; Bhat, M.A.; Kumar, R.; Bhatt, P. Mechanism and application of Sesbania root-nodulating bacteria: An alternative for chemical fertilizers and sustainable development. Arch. Microbiol. 2021, 203, 1259–1270. [Google Scholar] [CrossRef]
- Vives-Peris, V.; de Ollas, C.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Root exudates: From plant to rhizosphere and beyond. Plant Cell Rep. 2020, 39, 3–17. [Google Scholar] [CrossRef]
- Bai, Y.N.; Eijsink, V.G.H.; Kielak, A.M.; van Veen, J.A.; de Boer, W. Genomic comparison of chitinolytic enzyme systems from terrestrial and aquatic bacteria. Environ. Microbiol. 2016, 18, 38–49. [Google Scholar] [CrossRef]
- Worsley, S.F.; Newitt, J.; Rassbach, J.; Batey, S.F.D.; Holmes, N.A.; Murrell, J.C.; Wilkinson, B.; Hutchings, M.I. Streptomyces endophytes promote host health and enhance growth across plant species. Appl. Environ. Microbiol. 2020, 86, e01053-20. [Google Scholar] [CrossRef]
- Worsley, S.F.; Macey, M.; Prudence, S.; Wilkinson, B.; Murrell, J.C.; Hutchings, M.I. Investigating the role of root exudates in recruiting streptomyces bacteria to the Arabidopsis thaliana microbiome. Front. Mol. Biosci. 2021, 8, 686110. [Google Scholar] [CrossRef]
- Newitt, J.T.; Prudence, S.M.M.; Hutchings, M.I.; Worsley, S.F. Biocontrol of cereal crop diseases using Streptomycetes. Pathogens 2019, 8, 78. [Google Scholar] [CrossRef]
- Aliyu, I.A.; Yusuf, A.A.; Uyovbisere, E.O.; Masso, C.; Sanders, I.R. Effect of co-application of phosphorus fertilizer and in vitro-produced mycorrhizal fungal inoculants on yield and leaf nutrient concentration of cassava. PLoS ONE 2019, 14, e0218969. [Google Scholar] [CrossRef]
- van der Heijden, M.G.A.; Martin, F.M.; Selosse, M.A.; Sanders, I.R. Mycorrhizal ecology and evolution: The past, the present, and the future. New Phytol. 2015, 205, 1406–1423. [Google Scholar] [CrossRef] [PubMed]
- Vanegas-León, M.L.; Sulzbacher, M.A.; Rinaldi, A.C.; Roy, M.; Selosse, M.A.; Neves, M.A. Are Trechisporales ectomycorrhizal or non-mycorrhizal root endophytes? Mycol. Prog. 2019, 18, 1231–1240. [Google Scholar] [CrossRef]
- Huang, Z.J.; Cai, X.L.; Shao, C.L.; She, Z.G.; Xia, X.K.; Chen, Y.G.; Yang, J.X.; Zhou, S.N.; Lin, Y.C. Chemistry and weak antimicrobial activities of phomopsins produced by mangrove endophytic fungus Phomopsis sp. ZSU-H76. Phytochemistry 2008, 69, 1604–1608. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chen, L.; Redmile-Gordon, M.; Zhang, J.B.; Zhang, C.Z.; Ning, Q.; Li, W. Mortierella elongata’s roles in organic agriculture and crop growth promotion in a mineral soil. Land Degrad. Dev. 2018, 29, 1642–1651. [Google Scholar] [CrossRef]
- Tamayo-Velez, A.; Osorio, N.W. Soil fertility improvement by litter decomposition and inoculation with the fungus Mortierella sp in avocado plantations of Colombia. Commun. Soil Sci. Plan. 2018, 49, 139–147. [Google Scholar] [CrossRef]
- Zhang, H.S.; Wu, X.H.; Li, G.; Qin, P. Interactions between arbuscular mycorrhizal fungi and phosphate-solubilizing fungus (Mortierella sp.) and their effects on Kostelelzkya virginica growth and enzyme activities of rhizosphere and bulk soils at different salinities. Biol. Fert. Soils 2011, 47, 543–554. [Google Scholar] [CrossRef]
- Qiu, R.; Bai, J.K.; Li, C.J.; Li, S.J.; Li, X.J.; Chen, Y.G.; Hu, Y.J.; Liu, D.S. Molecular identification and pathogenicity analysis of tobacco Fusarium spp. in Henan. Acta Tab. Sin. 2018, 24, 129–134. [Google Scholar] [CrossRef]
- Schlatter, D.; Kinkel, L.; Thomashow, L.; Weller, D.; Paulitz, T. Disease suppressive soils: New insights from the soil microbiome. Phytopathology 2017, 107, 1284–1297. [Google Scholar] [CrossRef]
- Behie, S.W.; Bidochka, M.J. Nutrient transfer in plant-fungal symbioses. Trends Plant Sci. 2014, 19, 734–740. [Google Scholar] [CrossRef]
- Bencherif, K.; Boutekrabt, A.; Dalpe, Y.; Sahraoui, A.L.H. Soil and seasons affect arbuscular mycorrhizal fungi associated with Tamarix rhizosphere in arid and semi-arid steppes. Appl. Soil Ecol. 2016, 107, 182–190. [Google Scholar] [CrossRef]
- Liu, M.; Yue, Y.J.; Wang, Z.H.; Li, L.; Duan, G.Z.; Bai, S.L.; Li, T. Composition of the arbuscular mycorrhizal fungal community and changes in diversity of the rhizosphere of Clematis fruticosa over three seasons across different elevations. Eur. J. Soil Sci. 2020, 71, 511–523. [Google Scholar] [CrossRef]
- Wang, H.H.; Chu, H.L.; Dou, Q.; Feng, H.; Tang, M.; Zhang, S.X.; Wang, C.Y. Seasonal changes in Pinus tabuliformis root-associated fungal microbiota drive N and P cycling in terrestrial ecosystem. Front. Microbiol. 2021, 11, 526898. [Google Scholar] [CrossRef] [PubMed]
- Buckeridge, K.M.; Banerjee, S.; Siciliano, S.D.; Grogan, P. The seasonal pattern of soil microbial community structure in mesic low arctic tundra. Soil Biol. Biochem. 2013, 65, 338–347. [Google Scholar] [CrossRef]
- Carbone, M.J.; Alaniz, S.; Mondino, P.; Gelabert, M.; Eichmeier, A.; Tekielska, D.; Bujanda, R.; Gramaje, D. Drought influences fungal community dynamics in the grapevine rhizosphere and root microbiome. J. Fungi 2021, 7, 686. [Google Scholar] [CrossRef] [PubMed]
- Naylor, D.; Coleman-Derr, D. Drought stress and root-associated bacterial communities. Front. Plant Sci. 2018, 8, 2223. [Google Scholar] [CrossRef] [PubMed]
- Cantó, C.D.; Simonin, M.; King, E.; Moulin, L.; Bennett, M.J.; Castrillo, G.; Laplaze, L. An extended root phenotype: The rhizosphere, its formation and impacts on plant fitness. Plant J. 2020, 103, 951–964. [Google Scholar] [CrossRef] [PubMed]
- Chaparro, J.M.; Badri, D.V.; Bakker, M.G.; Sugiyama, A.; Manter, D.K.; Vivanco, J.M. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE 2013, 8, e55731. [Google Scholar] [CrossRef]
- Williams, A.; de Vries, F.T. Plant root exudation under drought: Implications for ecosystem functioning. New Phytol. 2020, 225, 1899–1905. [Google Scholar] [CrossRef]
- Sugiyama, A.; Ueda, Y.; Takase, H.; Yazaki, K. Pyrosequencing assessment of rhizosphere fungal communities from a soybean field. Can. J. Microbiol. 2014, 60, 687–690. [Google Scholar] [CrossRef]
- Zimudzi, J.; van der Waals, J.E.; Coutinho, T.A.; Cowan, D.A.; Valverde, A. Temporal shifts of fungal communities in the rhizosphere and on tubers in potato fields. Fungal Biol. 2018, 122, 928–934. [Google Scholar] [CrossRef]
- Taketani, R.G.; Lanconi, M.D.; Kavamura, V.N.; Durrer, A.; Andreote, F.D.; Melo, I.S. Dry season constrains bacterial phylogenetic diversity in a semi-arid rhizosphere system. Microb. Ecol. 2017, 73, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Coleman-Derr, D.; Desgarennes, D.; Fonseca-Garcia, C.; Gross, S.; Clingenpeel, S.; Woyke, T.; North, G.; Visel, A.; Partida-Martinez, L.P.; Tringe, S.G. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016, 209, 798–811. [Google Scholar] [CrossRef] [PubMed]
- Desgarennes, D.; Garrido, E.; Torres-Gomez, M.J.; Pena-Cabriales, J.J.; Partida-Martinez, L.P. Diazotrophic potential among bacterial communities associated with wild and cultivated Agave species. FEMS Microbiol. Ecol. 2014, 90, 844–857. [Google Scholar] [CrossRef]
- Fonseca-Garcia, C.; Coleman-Derr, D.; Garrido, E.; Visel, A.; Tringe, S.G.; Partida-Martinez, L.P. The cacti microbiome: Interplay between habitat-filtering and host-specificity. Front. Microbiol. 2016, 7, 150. [Google Scholar] [CrossRef] [PubMed]
- Shakya, M.; Gottel, N.; Castro, H.; Yang, Z.M.K.; Gunter, L.; Labbe, J.; Muchero, W.; Bonito, G.; Vilgalys, R.; Tuskan, G.; et al. A multifactor analysis of fungal and bacterial community structure in the root microbiome of mature Populus deltoides trees. PLoS ONE 2013, 8, e76382. [Google Scholar] [CrossRef]



| Microbial Community | Growth Stage | Rhizocompartment | PERMANOVA | ANOSIM (Genus Level) | ||
|---|---|---|---|---|---|---|
| Statistic (R2) | p Value | Statistic (R) | p Value | |||
| Bacteria/Archaea 16S | Between growth stages | Bulk | 0.268 | 0.106 | 0.222 | 0.019 |
| Rhizosphere | 0.554 | 0.002 | 0.568 | 0.001 | ||
| Rhizoplane | 0.274 | 0.14 | 0.122 | 0.123 | ||
| Endosphere | 0.339 | 0.047 | 0.247 | 0.021 | ||
| Between rhizocompartment | Vegetative | 0.526 | 0.001 | 0.373 | 0.001 | |
| flowering | 0.527 | 0.001 | 0.456 | 0.001 | ||
| fruiting | 0.637 | 0.001 | 0.483 | 0.001 | ||
| Senescence | 0.637 | 0.001 | 0.449 | 0.001 | ||
| Fungi 18S | Between growth stages | Bulk | 0.663 | 0.001 | 0.717 | 0.001 |
| Rhizosphere | 0.755 | 0.001 | 0.804 | 0.001 | ||
| Endosphere | 0.807 | 0.001 | 0.852 | 0.001 | ||
| Between rhizocompartment | Vegetative | 0.451 | 0.009 | 0.308 | 0.032 | |
| flowering | 0.695 | 0.007 | 0.361 | 0.012 | ||
| fruiting | 0.398 | 0.027 | 0.144 | 0.079 | ||
| Senescence | 0.188 | 0.718 | −0.021 | 0.503 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; He, X.; Yuan, H.; Lv, G. Differed Growth Stage Dynamics of Root-Associated Bacterial and Fungal Community Structure Associated with Halophytic Plant Lycium ruthenicum. Microorganisms 2022, 10, 1644. https://doi.org/10.3390/microorganisms10081644
Li Y, He X, Yuan H, Lv G. Differed Growth Stage Dynamics of Root-Associated Bacterial and Fungal Community Structure Associated with Halophytic Plant Lycium ruthenicum. Microorganisms. 2022; 10(8):1644. https://doi.org/10.3390/microorganisms10081644
Chicago/Turabian StyleLi, Yan, Xuemin He, Hongfei Yuan, and Guanghui Lv. 2022. "Differed Growth Stage Dynamics of Root-Associated Bacterial and Fungal Community Structure Associated with Halophytic Plant Lycium ruthenicum" Microorganisms 10, no. 8: 1644. https://doi.org/10.3390/microorganisms10081644
APA StyleLi, Y., He, X., Yuan, H., & Lv, G. (2022). Differed Growth Stage Dynamics of Root-Associated Bacterial and Fungal Community Structure Associated with Halophytic Plant Lycium ruthenicum. Microorganisms, 10(8), 1644. https://doi.org/10.3390/microorganisms10081644






