Functional Analysis of Conserved Hypothetical Proteins from the Antarctic Bacterium, Pedobacter cryoconitis Strain BG5 Reveals Protein Cold Adaptation and Thermal Tolerance Strategies
Abstract
1. Introduction
2. Materials and Methods
2.1. Retrieval of Sequence Genome Data
2.2. Sequence Analysis
2.2.1. Physicochemical Characterization
2.2.2. Subcellular Localization
2.2.3. Sequence Comparison
2.2.4. Function Prediction
2.2.5. Determination and Validation of Three-Dimensional Structures
2.3. Cloning of the Genes Coding for the Selected HPs
2.4. Protein Expression and Purification
2.5. Functional Determination
2.5.1. Gene Expression Profile
2.5.2. Enzymatic ATP Hydrolysis
2.5.3. Inhibition of Citrate Synthase Thermal Aggregation
3. Results
3.1. Analysis of the Conserved Hypothetical Proteins from the P. cryoconitis BG5 Genome
3.2. Prediction of Physicochemical Properties and Subcellular Localization
3.3. Predicted Proteins with Adaptational Functions in Response to Temperature Stress
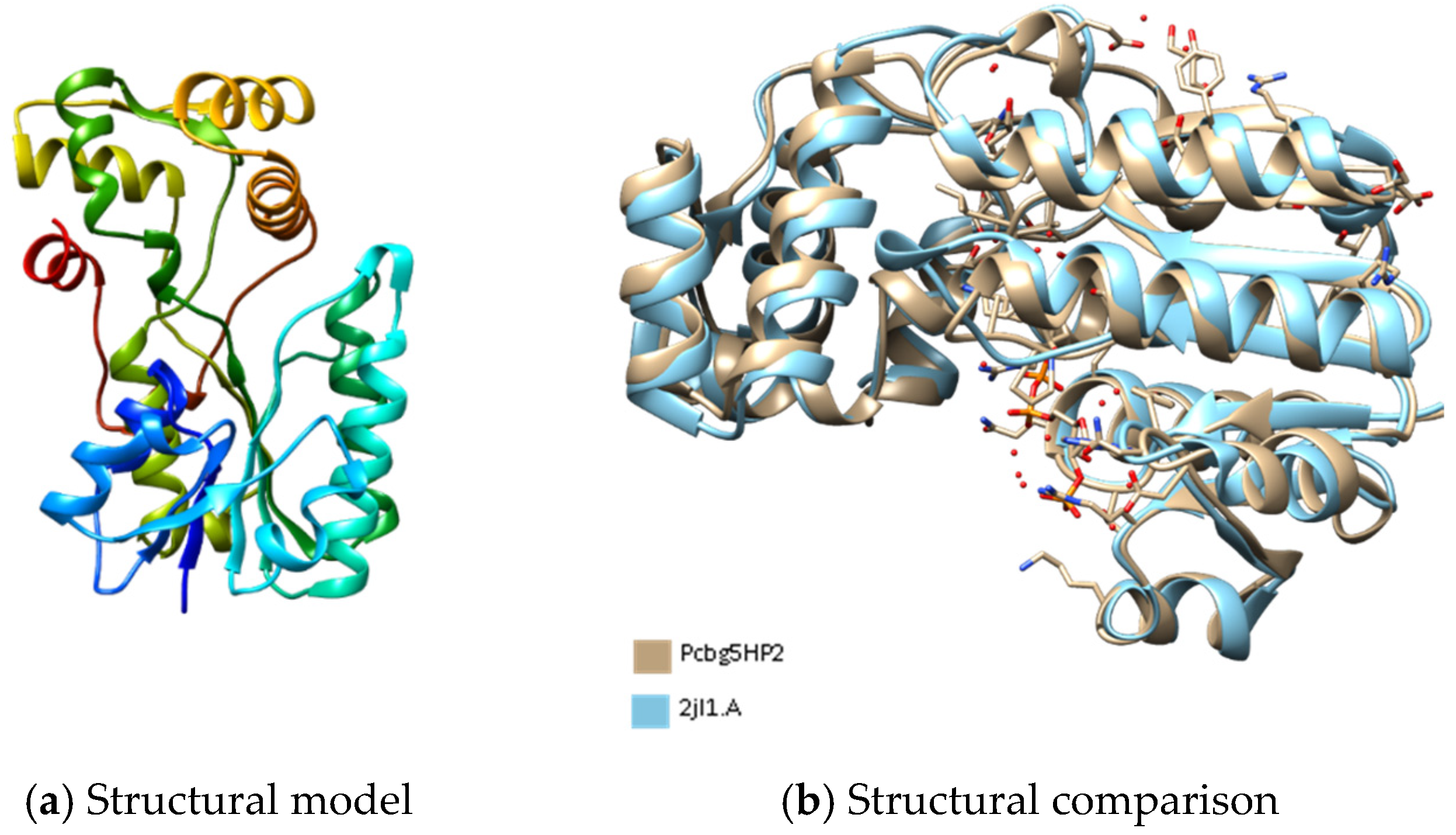
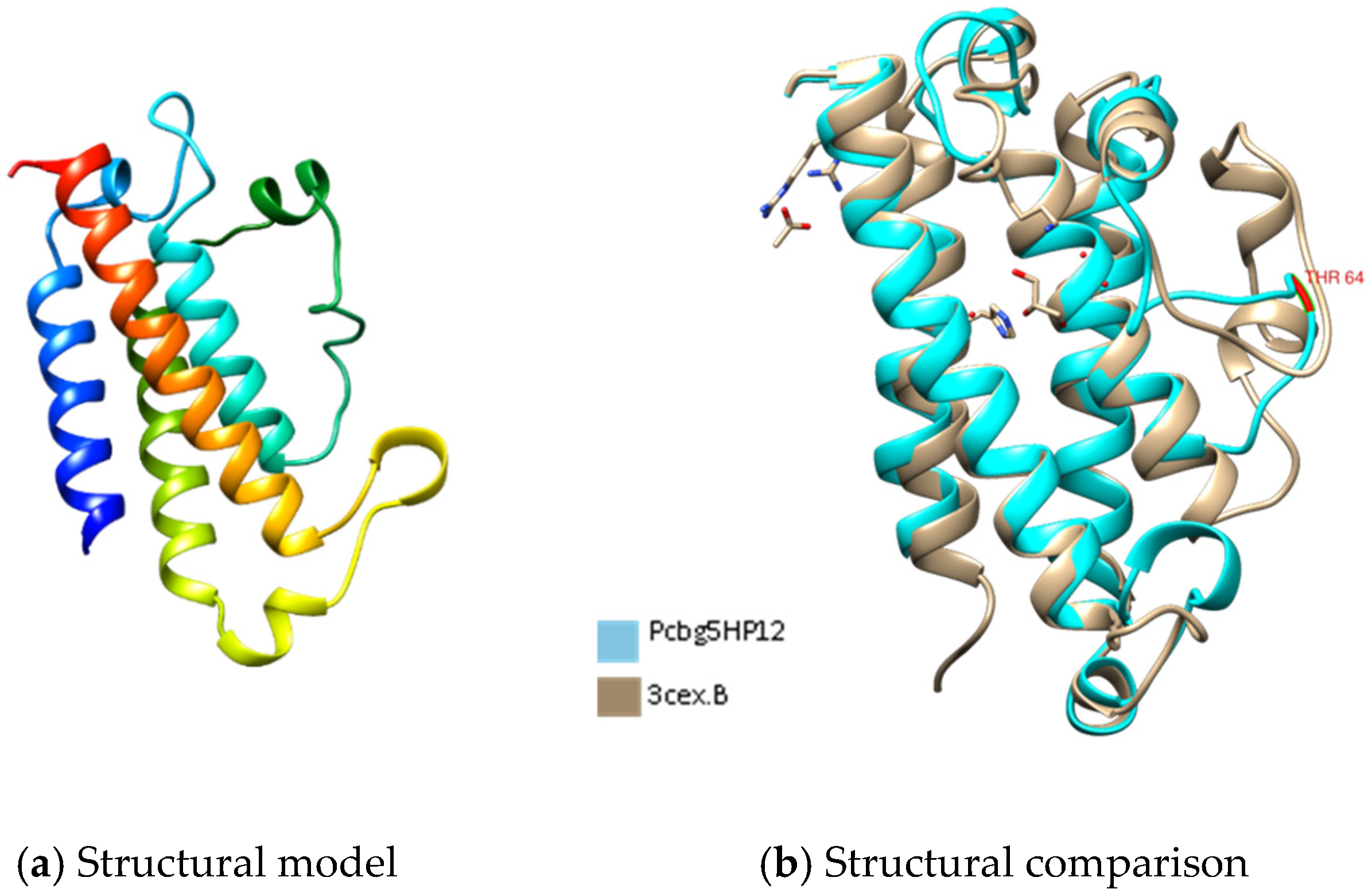
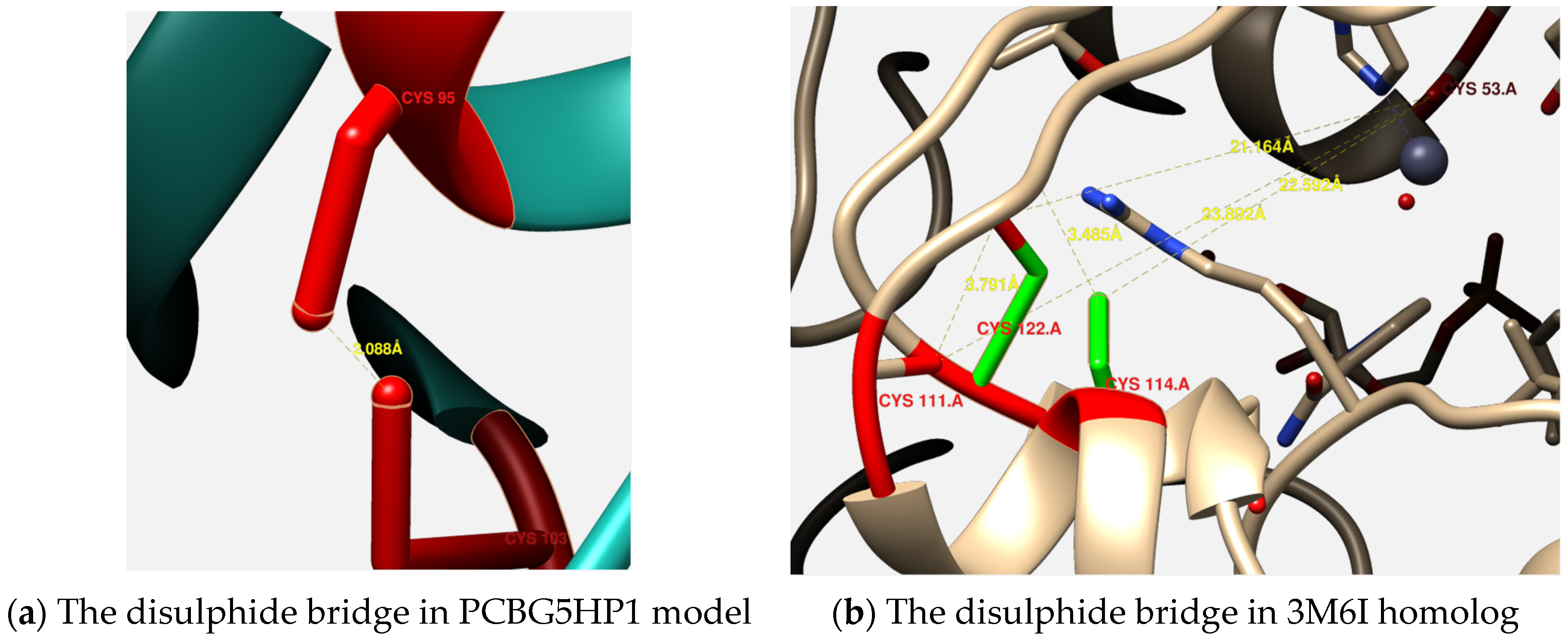
3.4. Three-Dimensional Structures Analysis
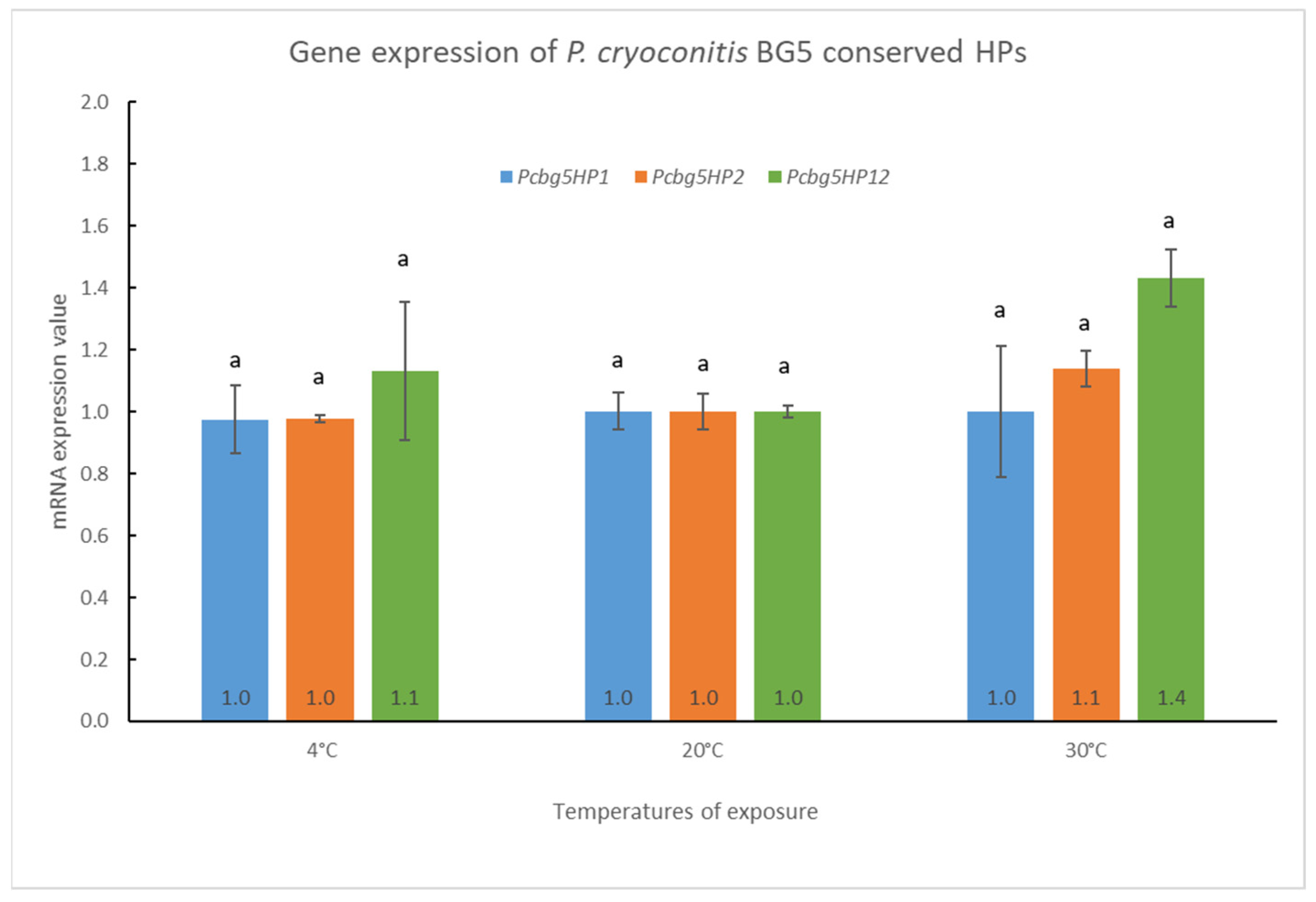
3.5. Pcbg5hp1, Pcbg5hp2, and Pcbg5hp12 mRNA Expression in P. cryoconitis BG5 under Heat-Shock and Cold-Shock Conditions
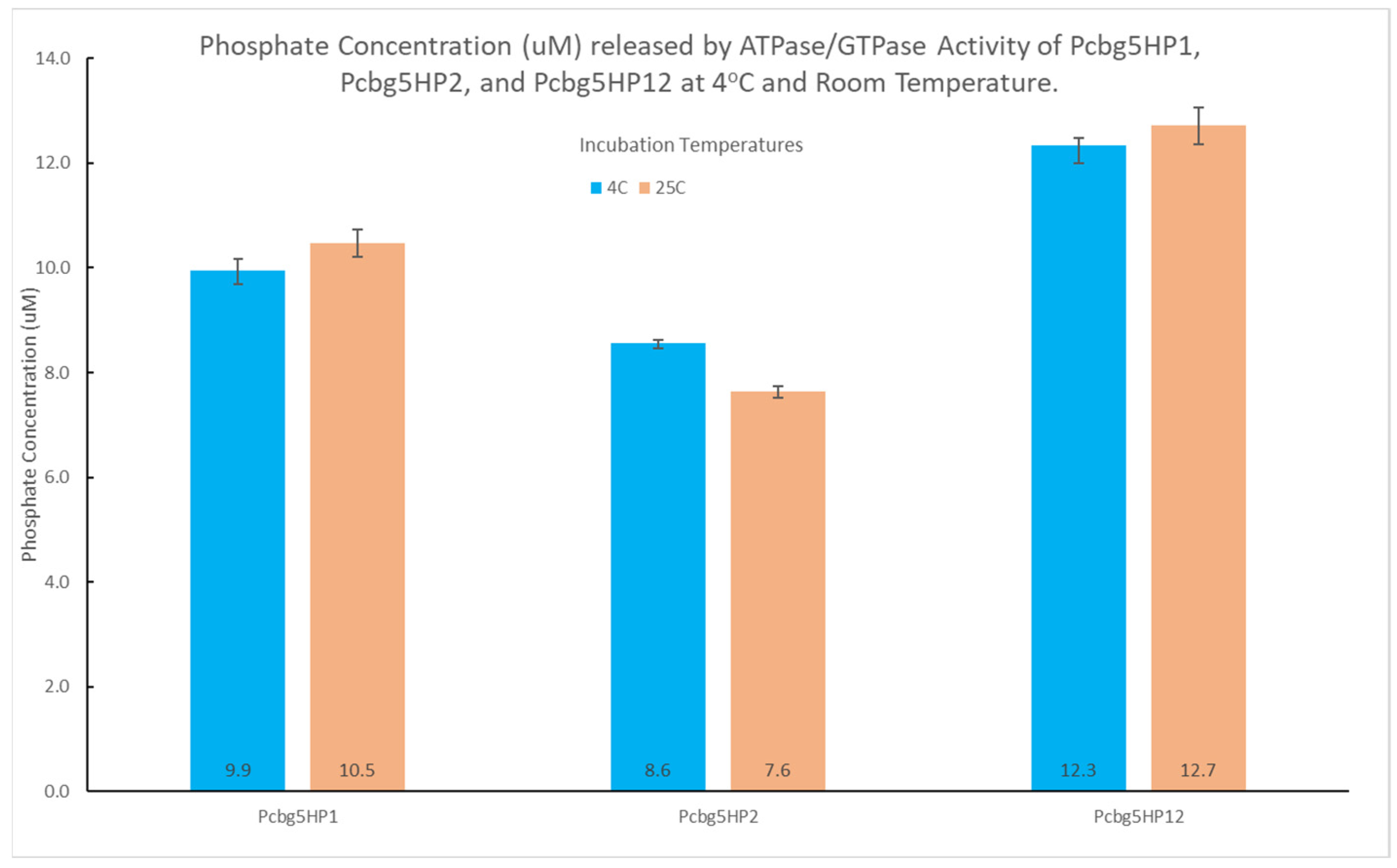
3.6. ATPase Assay Analysis
3.7. Thermal Unfolding Assay Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Song, Y.-S.; Seo, D.-J.; Jung, W.-J. Identification, Purification, and Expression Patterns of Chitinase from Psychrotolerant Pedobacter sp. PR-M6 and Antifungal Activity in vitro. Microb. Pathog. 2017, 107, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.M.V.L.; Tam, H.K.; Ng, W.M.; Boo, S.Y.; González, M. Characterisation of a Cryptic Plasmid from an Antarctic Bacterium Pedobacter cryoconitis strain BG5. Plasmid 2013, 69, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Eisenstein, E.; Gilliland, G.L.; Herzberg, O.; Moult, J.; Orban, J.; Poljak, R.J.; Banerjei, L.; Richardson, D.; Howard, A.J. Biological Function Made Crystal Clear—Annotation of Hypothetical Proteins via Structural Genomics. Curr. Opin. Biotechnol. 2000, 11, 25–30. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Koonin, E.V. “Conserved Hypothetical” Proteins: Prioritization of Targets for Experimental Study. Nucleic Acids Res. 2004, 32, 5452–5463. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jung, Y.J.; Lee, H.K.; Hong, S.G.; Kim, O.S. Complete Genome Sequence of Pedobacter cryoconitis PAMC 27485, a CRISPR-Cas System-Containing Psychrophile Isolated from Antarctica. J. Biotechnol. 2016, 226, 74–75. [Google Scholar] [CrossRef] [PubMed]
- Teoh, C.P.; Lavin, P.; Lee, D.J.H.; González-Aravena, M.; Najimudin, N.; Lee, P.C.; Cheah, Y.K.; Wong, C.M.V.L. Genomics and Transcriptomics Analyses Provide Insights into the Cold Adaptation Strategies of an Antarctic Bacterium, Cryobacterium sp. SO1. Polar Biol. 2021, 44, 1305–1319. [Google Scholar] [CrossRef]
- Koh, H.Y.; Park, H.; Lee, J.H.; Han, S.J.; Sohn, Y.C.; Lee, S.G. Proteomic and Transcriptomic Investigations on Cold-Responsive Properties of the Psychrophilic Antarctic Bacterium Psychrobacter sp. PAMC 21119 at Subzero Temperatures. Environ. Microbiol. 2017, 19, 628–644. [Google Scholar] [CrossRef]
- Wong, C.M.V.L.; Boo, S.Y.; Voo, C.L.Y.; Zainuddin, N.; Najimudin, N. A Comparative Transcriptomic Analysis Provides Insights into the Cold-Adaptation Mechanisms of a Psychrophilic Yeast, Glaciozyma antarctica PI12. Polar Biol. 2019, 42, 541–553. [Google Scholar] [CrossRef]
- Koh, J.S.P.; Wong, C.M.V.L.; Najimudin, N.; Mahadi, N.M. Gene Expression Patterns of Glaciozyma antarctica PI12 in Response to Cold, and Freeze Stress. Polar Sci. 2019, 20, 45–54. [Google Scholar] [CrossRef]
- Margesin, R. Psychrophiles: From Biodiversity to Biotechnology, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2017; ISBN 9783319570570. [Google Scholar]
- Tomoike, F.; Wakamatsu, T.; Nakagawa, N.; Kuramitsu, S.; Masui, R. Crystal Structure of the Conserved Hypothetical Protein TTHA1606 from Thermus thermophilus HB8. Proteins Struct. Funct. Bioinform. 2009, 76, 244–248. [Google Scholar] [CrossRef]
- Islam, M.S.; Shahik, S.M.; Sohel, M.; Patwary, N.I.A.; Hasan, M.A. In Silico Structural and Functional Annotation of Hypothetical Proteins of Vibrio cholerae O139. Genom. Inform. 2015, 13, 53. [Google Scholar] [CrossRef] [PubMed]
- Sanmukh, S.G.; Paunikar, W.N.; Ghosh, T.K.; Chakrabarti, T. Structure and Function Predictions of Hypothetical Proteins in Vibrio Phages. Int. J. Biom. Bioinform. 2010, 4, 161–175. [Google Scholar]
- Collins, T.; Margesin, R. Psychrophilic Lifestyles: Mechanisms of Adaptation and Biotechnological Tools. Appl. Microbiol. Biotechnol. 2019, 103, 2857–2871. [Google Scholar] [CrossRef] [PubMed]
- Firdaus-Raih, M.; Hashim, N.H.F.; Bharudin, I.; Bakar, M.F.A.; Huang, K.K.; Alias, H.; Lee, B.K.B.; Isa, M.N.M.; Mat-Sharani, S.; Sulaiman, S.; et al. The Glaciozyma antarctica Genome Reveals an Array of Systems That Provide Sustained Responses towards Temperature Variations in a Persistently Cold Habitat. PLoS ONE 2018, 13, e0189947. [Google Scholar] [CrossRef] [PubMed]
- Boo, S.Y.; Wong, C.M.V.L.; Rodrigues, K.F.; Najimudin, N.; Murad, A.M.A.; Mahadi, N.M. Thermal Stress Responses in Antarctic Yeast, Glaciozyma antarctica PI12, Characterized by Real-Time Quantitative PCR. Polar Biol. 2013, 36, 381–389. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. The Proteomics Protocols Handbook; Springer: Berlin/Heidelberg, Germany, 2005; pp. 571–608. [Google Scholar] [CrossRef]
- Chothia, C.; Gough, J. Genomic and Structural Aspects of Protein Evolution. Biochem. J. 2009, 419, 15–28. [Google Scholar] [CrossRef]
- Mohan, R. Computational Structural and Functional Analysis of Hypothetical Proteins of Staphylococcus Aureus. Bioinformation 2012, 8, 722–728. [Google Scholar] [CrossRef]
- Rangwala, H.; Karypis, G. Introduction to Protein Structure Prediction: Methods and Algorithms; John Wiley & Sons: New York, NY, USA, 2011; Volume 18, ISBN 111809946X. [Google Scholar]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete Genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Sonnhammer, E.L.L.; Krogh, A. A Hidden Markov Model for Predicting Transmembrane Helices in Protein Sequence. In Proceedings of the 6th International Conference on Intelligent Systems for Molecular Biology; ACM Digital Library: New York, NY, USA, 2008; p. 8. [Google Scholar]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT Protein Knowledgebase and Its Supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving Coverage, Classification and Access to Protein Sequence Annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein Domains Identifier. Nucleic Acids Res. 2005, 33, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods 2014, 12, 7–8. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Tina, K.G.; Bhadra, R.; Srinivasan, N. PIC: Protein Interactions Calculator. Nucleic Acids Res. 2007, 35, 473–476. [Google Scholar] [CrossRef]
- Wong, C.M.V.L.; Tam, H.; Alias, S.; González, M.; González-Rocha, G.; Domínguez-Yévenes, M. Pseudomonas and Pedobacter Isolates from King George Island Inhibited the Growth of Foodborne Pathogens. Pol. Polar Res. 2011, 32, 3–14. [Google Scholar] [CrossRef]
- Yusof, N.A.; Bakar, F.D.A.; Beddoe, T.; Murad, A.M.A. Cloning, Expression and Crystallisation of SGT1 Co-Chaperone Protein from Glaciozyma antarctica. AIP Conf. Proc. 2013, 1571, 292–297. [Google Scholar] [CrossRef]
- Bharudin, I.; Zaki, N.Z.; Bakar, F.D.A.; Mahadi, N.M.; Najmudin, N.; Illias, R.M.; Murad, A.M.A. Comparison of RNA Extraction Methods for Transcript Analysis from the Psychrophilic Yeast, Glaciozyma antarctica. Malays. Appl. Biol. 2014, 43, 71–79. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Villeneuve, T.S.; Garant, K.A.; Amons, R.; MacRae, T.H. Functional Characterization of Artemin, a Ferritin Homolog Synthesized in Artemia Embryos during Encystment and Diapause. FEBS J. 2007, 274, 1093–1101. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Havlíček, V. Protein Extraction and Precipitation. In Proteomic Profiling and Analytical Chemistry: The Crossroads, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 52–62. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Gazi, M.A.; Mahmud, S.; Fahim, S.M.; Islam, M.R.; Das, S.; Mahfuz, M.; Ahmed, T. Questing Functions and Structures of Hypothetical Proteins from Campylobacter jejuni: A Computer-Aided Approach. Biosci. Rep. 2020, 40, BSR20193939. [Google Scholar] [CrossRef]
- De Maayer, P.; Anderson, D.; Cary, C.; Cowan, D.A. Some like It Cold: Understanding the Survival Strategies of Psychrophiles. EMBO Rep. 2014, 15, 508–517. [Google Scholar] [CrossRef]
- Michetti, D.; Brandsdal, B.O.; Bon, D.; Isaksen, G.V.; Tiberti, M.; Papaleo, E. A Comparative Study of Cold- and Warm-Adapted Endonucleases A Using Sequence Analyses and Molecular Dynamics Simulations. PLoS ONE 2017, 12, e0169586. [Google Scholar]
- Siddiqui, K.S.; Williams, T.J.; Wilkins, D.; Yau, S.; Allen, M.A.; Brown, M.V.; Lauro, F.M.; Cavicchioli, R. Psychrophiles. Annu. Rev. Earth Planet. Sci. 2013, 41, 87–115. [Google Scholar] [CrossRef]
- Pauly, T.A.; Ekstrom, J.L.; Beebe, D.A.; Chrunyk, B.; Cunningham, D.; Griffor, M.; Kamath, A.; Lee, S.E.; Madura, R.; Mcguire, D.; et al. X-Ray Crystallographic and Kinetic Studies of Human Sorbitol Dehydrogenase. Structure 2003, 11, 1071–1085. [Google Scholar] [CrossRef]
- Kim, M.H.; Kim, Y.; Park, H.-J.; Lee, J.S.; Kwak, S.-N.; Jung, W.-H.; Lee, S.-G.; Kim, D.; Lee, Y.-C.; Oh, T.-K. Structural Insight into Bioremediation of Triphenylmethane Dyes by Citrobacter Sp. Triphenylmethane Reductase. J. Biol. Chem. 2008, 283, 31981–31990. [Google Scholar] [CrossRef]
- RCSB PDB—3CEX: Crystal Structure of the Conserved Protein of Locus EF_3021 from Enterococcus Faecalis. Available online: https://www.rcsb.org/structure/3CEX (accessed on 17 April 2022).
- Bertone, P. SPINE: An Integrated Tracking Database and Data Mining Approach for Identifying Feasible Targets in High-Throughput Structural Proteomics. Nucleic Acids Res. 2001, 29, 2884–2898. [Google Scholar] [CrossRef]
- Gupta, D.; Banerjee, S.; Pailan, S.; Saha, P. In Silico Identification and Characterization of a Hypothetical Protein of Mycobacterium tuberculosis EAI5 as a Potential Virulent Factor. Bioinformation 2016, 12, 182–191. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hawkins, T.; Kihara, D. Function Prediction of Uncharacterized Proteins. J. Bioinform. Comput. Biol. 2007, 5, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Dönnes, P.; Höglund, A. Predicting Protein Subcellular Localization: Past, Present, and Future. Genom. Proteom. Bioinform. 2004, 2, 209–215. [Google Scholar] [CrossRef]
- Shen, Y.; Tang, J.; Guo, F. Identification of Protein Subcellular Localization via Integrating Evolutionary and Physicochemical Information into Chou’s General PseAAC. J. Theor. Biol. 2019, 462, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.A.T.; Shahbaaz, M.; Ahmad, F.; Hassan, M.I. Identification of Functional Candidates amongst Hypothetical Proteins of Treponema pallidum ssp. pallidum. PLoS ONE 2015, 10, e0124177. [Google Scholar] [CrossRef]
- Andrade, M.A.; O’Donoghue, S.I.; Rost, B. Adaptation of Protein Surfaces to Subcellular Location. J. Mol. Biol. 1998, 276, 517–525. [Google Scholar] [CrossRef]
- Hua, S.; Sun, Z. Support Vector Machine Approach for Protein Subcellular Localization Prediction. Bioinformatics 2001, 17, 721–728. [Google Scholar] [CrossRef]
- Nakashima, H.; Nishikawa, K. Discrimination of Intracellular and Extracellular Proteins Using Amino Acid Composition and Residue-Pair Frequencies. J. Mol. Biol. 1994, 238, 54–61. [Google Scholar] [CrossRef]
- Raymond-Bouchard, I.; Goordial, J.; Zolotarov, Y.; Ronholm, J.; Stromvik, M.; Bakermans, C.; Whyte, L.G. Conserved Genomic and Amino Acid Traits of Cold Adaptation in Subzero-Growing Arctic Permafrost Bacteria. FEMS Microbiol. Ecol. 2018, 94, 1–13. [Google Scholar] [CrossRef]
- D’Amico, S.; Collins, T.; Marx, J.C.; Feller, G.; Gerday, C. Psychrophilic Microorganisms: Challenges for Life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef]
- Anastasiou, R.; Leverrier, P.; Krestas, I.; Rouault, A.; Kalantzopoulos, G.; Boyaval, P.; Tsakalidou, E.; Jan, G. Changes in Protein Synthesis during Thermal Adaptation of Propionibacterium freudenreichii subsp. shermanii. Int. J. Food Microbiol. 2006, 108, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.; Barrand, N.E.; Bracegirdle, T.J.; Convey, P.; Hodgson, D.A.; Jarvis, M.; Jenkins, A.; Marshall, G.; Meredith, M.P.; Roscoe, H.; et al. Antarctic Climate Change and the Environment: An Update. Polar Rec. 2014, 50, 237–259. [Google Scholar] [CrossRef]
- Chong, G.L.; Chu, W.L.; Othman, R.Y.; Phang, S.M. Differential Gene Expression of an Antarctic Chlorella in Response to Temperature Stress. Polar Biol. 2011, 34, 637–645. [Google Scholar] [CrossRef]
- Geisel, N. Constitutive versus Responsive Gene Expression Strategies for Growth in Changing Environments. PLoS ONE 2011, 6, e27033. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Mo, A.; Liu, S.; Yang, L.; Li, L. Constitutive Expression of a Peanut Ubiquitin-Conjugating Enzyme Gene in Arabidopsis Confers Improved Water-Stress Tolerance through Regulation of Stress-Responsive Gene Expression. J. Biosci. Bioeng. 2011, 111, 478–484. [Google Scholar] [CrossRef]
- Hu, Z.; Song, N.; Zheng, M.; Liu, X.; Liu, Z.; Xing, J.; Ma, J.; Guo, W.; Yao, Y.; Peng, H. Histone Acetyltransferase GCN 5 Is Essential for Heat Stress-responsive Gene Activation and Thermotolerance in Arabidopsis. Plant J. 2015, 84, 1178–1191. [Google Scholar] [CrossRef]
- Fortuna, A.; Collalto, D.; Schiaffi, V.; Pastore, V.; Visca, P.; Ascenzioni, F.; Rampioni, G.; Leoni, L. The Pseudomonas aeruginosa DksA1 Protein Is Involved in H2O2 Tolerance and Within-Macrophages Survival and Can Be Replaced by DksA2. Sci. Rep. 2022, 12, 1–11. [Google Scholar] [CrossRef]
- Jakobsson, M.E.; Moen, A.; Bousset, L.; Egge-Jacobsen, W.; Kernstock, S.; Melki, R.; Falnes, P. Identification and Characterization of a Novel Human Methyltransferase Modulating Hsp70 Protein Function through Lysine Methylation. J. Biol. Chem. 2013, 288, 27752–27763. [Google Scholar] [CrossRef]
- Czapski, T.R.; Trun, N. Expression of Csp Genes in E. Coli K-12 in Defined Rich and Defined Minimal Media during Normal Growth, and after Cold-Shock. Gene 2014, 547, 91–97. [Google Scholar] [CrossRef]
- Georlette, D.; Damien, B.; Blaise, V.; Depiereux, E.; Uversky, V.N.; Gerday, C.; Feller, G. Structural and Functional Adaptations to Extreme Temperatures in Psychrophilic, Mesophilic, and Thermophilic DNA Ligases. J. Biol. Chem. 2003, 278, 37015–37023. [Google Scholar] [CrossRef]
- Fields, P.A.; Somero, G.N. Hot Spots in Cold Adaptation: Localized Increases in Conformational Flexibility in Lactate Dehydrogenase A4 Orthologs of Antarctic Notothenioid Fishes. Proc. Natl. Acad. Sci. USA 1998, 95, 11476–11481. [Google Scholar] [CrossRef] [PubMed]
- Marx, J.C.; Collins, T.; D’Amico, S.; Feller, G.; Gerday, C. Cold-Adapted Enzymes from Marine Antarctic Microorganisms. Mar. Biotechnol. 2007, 9, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.L.; Bushman, D.; Wang, F.; Li, P.W.-L.; Walker, A.; Cummiskey, J.; Branciforte, D.; Williams, M.C. A Single Amino Acid Substitution in ORF1 Dramatically Decreases L1 Retrotransposition and Provides Insight into Nucleic Acid Chaperone Activity. Nucleic Acids Res. 2008, 36, 5845–5854. [Google Scholar] [CrossRef] [PubMed]
- Maikova, A.; Zalutskaya, Z.; Lapina, T.; Ermilova, E. The HSP70 Chaperone Machines of Chlamydomonas Are Induced by Cold Stress. J. Plant Physiol. 2016, 204, 85–91. [Google Scholar] [CrossRef]
- Li, D.C.; Yang, F.; Lu, B.; Chen, D.F.; Yang, W.J. Thermotolerance and Molecular Chaperone Function of the Small Heat Shock Protein HSP20 Fromhyperthermophilic Archaeon, Sulfolobus solfataricus P2. Cell Stress Chaperones 2012, 17, 103–108. [Google Scholar] [CrossRef]
- Feller, G. Psychrophilic Enzymes: From Folding to Function and Biotechnology. Scientifica 2013, 2013, 1–28. [Google Scholar] [CrossRef]
- Collins, T.; Meuwis, M.A.; Gerday, C.; Feller, G. Activity, Stability and Flexibility in Glycosidases Adapted to Extreme Thermal Environments. J. Mol. Biol. 2003, 328, 419–428. [Google Scholar] [CrossRef]
- Rutkiewicz, M.; Bujacz, A.; Bujacz, G. Structural Features of Cold-Adapted Dimeric GH2 β-D-Galactosidase from Arthrobacter sp. 32cB. Biochim. Et Biophys. Acta-Proteins Proteom. 2019, 1867, 776–786. [Google Scholar] [CrossRef]
- Yusuf, S.N.H.M.; Bakar, F.D.A.; Mahadi, N.M.; Murad, A.M.A. In Silico Analysis of Glucoamylase from a Psychrophilic Yeast, Glaciozyma antarctica PI12. AIP Conf. Proc. 2015, 1678, 030032. [Google Scholar]
- Lonhienne, T.; Zoidakis, J.; Vorgias, C.E.; Feller, G.; Gerday, C.; Bouriotis, V. Modular Structure, Local Flexibility and Cold-Activity of a Novel Chitobiase from a Psychrophilic Antarctic Bacterium. J. Mol. Biol. 2001, 310, 291–297. [Google Scholar] [CrossRef]
- Davey, L.; Ng, C.K.W.; Halperin, S.A.; Lee, S.F. Functional Analysis of Paralogous Thiol-Disulfide Oxidoreductases in Streptococcus gordonii. J. Biol. Chem. 2013, 288, 16416–16429. [Google Scholar] [CrossRef] [PubMed]
- Georlette, D.; Blaise, V.; Collins, T.; D’Amico, S.; Gratia, E.; Hoyoux, A.; Marx, J.C.; Sonan, G.; Feller, G.; Gerday, C. Some like It Cold: Biocatalysis at Low Temperatures. FEMS Microbiol. Rev. 2004, 28, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Mavromatis, K.; Feller, G.; Kokkinidis, M.; Bouriotis, V. Cold Adaptation of a Psychrophilic Chitinase: A Mutagenesis Study. Protein Eng. 2003, 16, 497–503. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Feller, G.; Narinx, E.; Arpigny, J.L.; Aittaleb, M.; Baise, E.; Genicot, S.; Gerday, C. Enzymes from Psychrophilic Organisms. FEMS Microbiol. Rev. 1996, 18, 189–202. [Google Scholar] [CrossRef]
- Sarmiento, F.; Peralta, R.; Blamey, J.M. Cold and Hot Extremozymes: Industrial Relevance and Current Trends. Front. Bioeng. Biotechnol. 2015, 3, 148. [Google Scholar] [CrossRef]
- D’Amico, S.; Gerday, C.; Feller, G. Temperature Adaptation of Proteins: Engineering Mesophilic-like Activity and Stability in a Cold-Adapted α-Amylase. J. Mol. Biol. 2003, 332, 981–988. [Google Scholar] [CrossRef]
- Cotugno, R.; Ruocco, M.R.; Marco, S.; Falasca, P.; Evangelista, G.; Raimo, G.; Chambery, A.; di Maro, A.; Masullo, M.; de Vendittis, E. Differential Cold–Adaptation among Protein Components of the Thioredoxin System in the Psychrophilic Eubacterium Pseudoalteromonas haloplanktis TAC 125. Mol. BioSyst. 2009, 5, 519–528. [Google Scholar] [CrossRef]
- Yusof, N.A.; Kamaruddin, S.; Abu Bakar, F.D.; Mahadi, N.M.; Abdul Murad, A.M. Structural and Functional Insights into TRiC Chaperonin from a Psychrophilic Yeast, Glaciozyma antarctica. Cell Stress Chaperones 2019, 24, 351–368. [Google Scholar] [CrossRef]


| Gene Target | Direction | Primer DNA Sequence | Amplicon Length (Bp) |
|---|---|---|---|
| pcbghp1 | Forward | 5′-GGATATCCAGCTCAAAGAAAG-3′ | 96 |
| Reverse | 5′-AGGTCTGTTCCGCAAATA-3′ | ||
| pcbghp2 | Forward | 5′-CAGAGTAGCGGTACAAATAAG-3, | 119 |
| Reverse | 5′-GACTAAGATCAACAACTCTGG-3, | ||
| pcbghp12 | Forward | 5′-AAAGGATTGGCGTAACCGGG-3′ | 165 |
| Reverse | 5′-GAGTGCCCCTGATTCCTGATG-3′ |
| ID | Physicochemical Properties | ||
|---|---|---|---|
| Number of Amino Acids | Molecular Weight (Da) | Isoelectric Point (pI) | |
| PCBG5HP1 | 340 | 37,301.82 | 5.56 |
| PCBG5HP2 | 293 | 31,926.54 | 5.78 |
| PCBG5HP12 | 190 | 22,542.34 | 5.77 |
| Protein ID | Sequence Identity | e Value | Description |
|---|---|---|---|
| PCBG5HP1 | 28% | 5 × 10−29 | O35045.1 Uncharacterized zinc-type alcohol dehydrogenase-like protein YjmD (Bacillus subtilis subsp. subtilis str. 168) |
| PCBG5HP2 | 25% | 2 × 10−8 | Q54LW0.2 Prestalk A differentiation protein A (Dictyostelium discoideum) |
| PCBG5HP12 | - | - | No significant similarity found |
| Protein ID | Homologous Superfamily | Domains | Biological Process | Molecular Function | Cellular Component |
|---|---|---|---|---|---|
| PCBG5HP1 | GroES-like superfamily (IPR011032) NAD(P)-binding domain superfamily (IPR036291) | Alcohol dehydrogenase, N-terminal (IPR013154) Glucose dehydrogenase, C-terminal (IPR031640) | GO:0055114 oxidation-reduction process | GO:0008270 zinc ion binding GO:0016491oxidoreductase activity | None predicted |
| PCBG5HP2 | NAD(P)-binding domain superfamily (IPR036291) | NAD(P)-binding domain (IPR016040) | None predicted | None predicted | None predicted |
| PCBG5HP12 | DinB/YfiT-like putative metalloenzymes (IPR034660) | None predicted | None predicted | None predicted | None predicted |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masnoddin, M.; Ling, C.M.W.V.; Yusof, N.A. Functional Analysis of Conserved Hypothetical Proteins from the Antarctic Bacterium, Pedobacter cryoconitis Strain BG5 Reveals Protein Cold Adaptation and Thermal Tolerance Strategies. Microorganisms 2022, 10, 1654. https://doi.org/10.3390/microorganisms10081654
Masnoddin M, Ling CMWV, Yusof NA. Functional Analysis of Conserved Hypothetical Proteins from the Antarctic Bacterium, Pedobacter cryoconitis Strain BG5 Reveals Protein Cold Adaptation and Thermal Tolerance Strategies. Microorganisms. 2022; 10(8):1654. https://doi.org/10.3390/microorganisms10081654
Chicago/Turabian StyleMasnoddin, Makdi, Clemente Michael Wong Vui Ling, and Nur Athirah Yusof. 2022. "Functional Analysis of Conserved Hypothetical Proteins from the Antarctic Bacterium, Pedobacter cryoconitis Strain BG5 Reveals Protein Cold Adaptation and Thermal Tolerance Strategies" Microorganisms 10, no. 8: 1654. https://doi.org/10.3390/microorganisms10081654
APA StyleMasnoddin, M., Ling, C. M. W. V., & Yusof, N. A. (2022). Functional Analysis of Conserved Hypothetical Proteins from the Antarctic Bacterium, Pedobacter cryoconitis Strain BG5 Reveals Protein Cold Adaptation and Thermal Tolerance Strategies. Microorganisms, 10(8), 1654. https://doi.org/10.3390/microorganisms10081654






