How Metagenomics Has Transformed Our Understanding of Bacteriophages in Microbiome Research
Abstract
:1. Introduction
2. A Brief Overview of Metagenomics
3. Curating Metagenomes
4. Issues with Curating Metagenomes
5. Categorising Metagenomes
6. Using AI to Curate Metagenomes
7. Functional Diversity vs. Taxonomic Diversity
8. Phages and the Microbiome
9. Piggyback-the-Winner and Other Hypotheses
10. Prophage Abundances in Different Environments
11. Identifying Prophages
12. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mirsepasi-Lauridsen, H.C.; Vrankx, K.; Engberg, J.; Friis-Møller, A.; Brynskov, J.; Nordgaard-Lassen, I.; Petersen, A.M.; Krogfelt, K.A. Disease-Specific Enteric Microbiome Dysbiosis in Inflammatory Bowel Disease. Frontiers in Medicine. Front. Med. 2018, 5, 304. [Google Scholar] [CrossRef] [PubMed]
- de la Calle, F. Marine microbiome as source of natural products. Microb. Biotechnol. 2017, 10, 1293–1296. [Google Scholar] [CrossRef] [PubMed]
- Staley, J.T.; Konopka, A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 1985, 39, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998, 5, R245–R249. [Google Scholar] [CrossRef]
- DeLong, E.F.; Preston, C.M.; Mincer, T.; Rich, V.; Hallam, S.J.; Frigaard, N.-U.; Martinez, A.; Sullivan, M.B.; Edwards, R.; Brito, B.R.; et al. Community Genomics Among Stratified Microbial Assemblages in the Ocean’s Interior. Science 2006, 311, 496–503. [Google Scholar] [CrossRef]
- Tyson, G.W.; Chapman, J.; Hugenholtz, P.; Allen, E.E.; Ram, R.J.; Richardson, P.M.; Solovyev, V.V.; Rubin, E.M.; Rokhsar, D.S.; Banfield, J.F. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 2004, 428, 37–43. [Google Scholar] [CrossRef]
- Papudeshi, B.; Haggerty, J.M.; Doane, M.; Morris, M.M.; Walsh, K.; Beattie, D.T.; Pande, D.; Zaeri, P.; Silva, G.G.Z.; Thompson, F.; et al. Optimizing and evaluating the reconstruction of Metagenome-assembled microbial genomes. BMC Genom. 2017, 18, 915. [Google Scholar] [CrossRef]
- Abellan-Schneyder, I.; Matchado Monica, S.; Reitmeier, S.; Sommer, A.; Sewald, Z.; Baumbach, J.; List, M.; Neuhaus, K.; Tringe Susannah, G. Primer, Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing. mSphere 2021, 6, e01202-20. [Google Scholar] [CrossRef]
- Thompson, C.C.; Chimetto, L.; Edwards, R.A.; Swings, J.; Stackebrandt, E.; Thompson, F.L. Microbial genomic taxonomy. BMC Genom. 2013, 14, 913. [Google Scholar] [CrossRef]
- Wilson, M.R.; Naccache, S.N.; Samayoa, E.; Biagtan, M.; Bashir, H.; Yu, G.; Salamat, S.M.; Somasekar, S.; Federman, S.; Miller, S.; et al. Actionable Diagnosis of Neuroleptospirosis by Next-Generation Sequencing. N. Engl. J. Med. 2014, 370, 2408–2417. [Google Scholar] [CrossRef]
- Brown, J.R.; Bharucha, T.; Breuer, J. Encephalitis diagnosis using metagenomics: Application of next generation sequencing for undiagnosed cases. J. Infect. 2018, 76, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.W.; Evangelista, J.S.; Schmieder, R.; Bailey, B.; Haynes, M.; Furlan, M.; Maughan, H.; Edwards, R.; Rohwer, F.; Conrad, D.; et al. Clinical Insights from Metagenomic Analysis of Sputum Samples from Patients with Cystic Fibrosis. J. Clin. Microbiol. 2014, 52, 425–437. [Google Scholar] [CrossRef] [PubMed]
- de Vries, J.J.C.; Brown, J.R.; Couto, N.; Beer, M.; Le Mercier, P.; Sidorov, I.; Papa, A.; Fischer, N.; Oude Munnink, B.B.; Rodriquez, C.; et al. Recommendations for the introduction of metagenomic next-generation sequencing in clinical virology, part II: Bioinformatic analysis and reporting. J. Clin. Virol. 2021, 138, 104812. [Google Scholar] [CrossRef] [PubMed]
- Winter, A.K.; Hegde, S.T. The important role of serology for COVID-19 control. Lancet Infect. Dis. 2020, 20, 758–759. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Miller, S.A. Clinical metagenomics. Nat. Rev. Genet. 2019, 20, 341–355. [Google Scholar] [CrossRef]
- Gupta, S.; Mortensen, M.S.; Schjørring, S.; Trivedi, U.; Vestergaard, G.; Stokholm, J.; Bisgaard, H.; Krogfelt, K.A.; Sørensen, S.J. Amplicon sequencing provides more accurate microbiome information in healthy children compared to culturing. Commun. Biol. 2019, 2, 291. [Google Scholar] [CrossRef]
- Hematian, A.; Sadeghifard, N.; Mohebi, R.; Taherikalani, M.; Nasrolahi, A.; Amraei, M.; Ghafourian, S. Traditional and Modern Cell Culture in Virus Diagnosis. Osong Public Health Res. Perspect. 2016, 7, 77–82. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Insights into antibiotic resistance through metagenomic approaches. Future Microbiol. 2012, 7, 73–89. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Kantor, R.S.; Olm, M.R.; Whitney, O.N.; Al-Shayeb, B.; Lou, Y.C.; Flamholz, A.; Kennedy, L.C.; Greenwald, H.; Hinkle, A.; et al. Genome Sequencing of Sewage Detects Regionally Prevalent SARS-CoV-2 Variants. mBio 2021, 12, e02703-20. [Google Scholar] [CrossRef]
- COVID-19 Sewage Surveillance Program-COVID-19 (Coronavirus). 2022. Available online: https://www.health.nsw.gov.au/Infectious/covid-19/Pages/sewage-surveillance.aspx (accessed on 1 April 2022).
- Landgraff, C.; Wang, L.Y.R.; Buchanan, C.; Wells, M.; Schonfeld, J.; Bessonov, K.; Ali, J.; Robert, E.; Nadon, C. Metagenomic sequencing of municipal wastewater provides a near-complete SARS-CoV-2 genome sequence identified as the B.1.1.7 variant of concern from a Canadian municipality concurrent with an outbreak. MedRxiv 2021. [Google Scholar] [CrossRef]
- Public Health Alert-New Venues of Concern-News. 2022. Available online: https://www.health.nsw.gov.au/news/Pages/20210813_03.aspx (accessed on 1 April 2022).
- Rothman, J.A.; Loveless, T.B.; Griffith, M.L.; Steele, J.A.; Griffith, J.F.; Whiteson, K.L. Metagenomics of Wastewater Influent from Southern California Wastewater Treatment Facilities in the Era of COVID-19. Microbiol. Resour. Announc. 2020, 9, e00907-20. [Google Scholar] [CrossRef] [PubMed]
- Hendriksen, R.S.; Munk, P.; Njage, P.; van Bunnik, B.; McNally, L.; Lukjancenko, O.; Röder, T.; Nieuwenhuijse, D.; Pedersen, S.K.; Kjeldgaard, J.; et al. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 2019, 10, 1124. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Dinsdale, E.A.; Edwards, R.A.; Hall, D.; Angly, F.; Breitbart, M.; Brulc, J.M.; Furlan, M.; Desnues, C.; Haynes, M.; Li, L.; et al. Functional metagenomic profiling of nine biomes. Nature 2008, 452, 629–632. [Google Scholar] [CrossRef]
- Hover, B.M.; Kim, S.-H.; Katz, M.; Charlop-Powers, Z.; Owen, J.G.; Ternei, M.A.; Maniko, J.; Estrela, A.B.; Molina, H.; Park, S.; et al. Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol. 2018, 3, 415–422. [Google Scholar] [CrossRef]
- Tortorella, E.; Tedesco, P.; Palma Esposito, F.; January, G.G.; Fani, R.; Jaspars, M.; De Pascale, D. Antibiotics from Deep-Sea Microorganisms: Current Discoveries and Perspectives. Mar. Drugs 2018, 16, 355. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The Human Microbiome Project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Abu-Ali, G.; Huttenhower, C. The healthy human microbiome. Genome Med. 2016, 8, 51. [Google Scholar] [CrossRef]
- Nelson, W.C.; Tully, B.J.; Mobberley, J.M. Biases in genomes reconstruction from metagenomic data. PeerJ 2020, 8, e10119. [Google Scholar] [CrossRef]
- Welcome to the NCBO BioPortal|NCBO BioPortal. 2022. Available online: https://bioportal.bioontology.org (accessed on 1 April 2022).
- Prestat, E.; David, M.M.; Hultman, J.; Taş, N.; Lamendella, R.; Dvornik, J.; Mackelprang, R.; Myrold, D.D.; Jumpponen, A.; Tringe, S.G.; et al. FOAM (Functional Ontology Assignments for Metagenomes): A Hidden Markov Model (HMM) database with environmental focus. Nucleic Acids Res. 2014, 42, e145. [Google Scholar] [CrossRef] [PubMed]
- Walls, R.L.; Deck, J.; Guralnick, R.; Baskauf, S.; Beaman, R.; Blum, S.; Bowers, S.; Buttigieg, P.L.; Davies, N.; Endresen, D.; et al. Semantics in Support of Biodiversity Knowledge Discovery: An Introduction to the Biological Collections Ontology and Related Ontologies. PLoS ONE 2014, 9, e89606. [Google Scholar] [CrossRef] [PubMed]
- Kushida, T.; Kozaki, K.; Tateisi, Y.; Watanabe, K.; Masuda, T.; Matsumura, K.; Kawamura, T.; Takagi, T. Efficient Construction of a New Ontology for Life Sciences by Sub-classifying Related Terms in the Japan Science, Technology Agency Thesaurus. In Proceedings of the 8th International Conference on Biomedical Ontologies (ICBO), Newcastle upon Tyne, UK, 13–15 September 2017. [Google Scholar]
- Buttigieg, P.L.; Morrison, N.; Smith, B.; Mungall, C.J.; Lewis, S.E.; The ENVO Consortium. The environment ontology: Contextualising biological and biomedical entities. J. Biomed. Semant. 2013, 4, 43. [Google Scholar] [CrossRef] [PubMed]
- Blumberg, K.L.; Ponsero, A.J.; Bomhoff, M.; Wood-Charlson, E.M.; DeLong, E.F.; Hurwitz, B.L. Ontology-Enriched Specifications Enabling Findable, Accessible, Interoperable, and Reusable Marine Metagenomic Datasets in Cyberinfrastructure Systems. Front. Microbiol. 2021, 12, 765268. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Almeida, A.; Beracochea, M.; Boland, M.; Burgin, J.; Cochrane, G.; Crusoe, M.R.; Kale, V.; Potter, S.C.; Richardson, L.J.; et al. MGnify: The microbiome analysis resource in 2020. Nucleic Acids Res. 2020, 48, D570–D578. [Google Scholar] [CrossRef]
- Available online: https://www.ncbi.nlm.nih.gov/sra/docs/sragrowth/ (accessed on 1 April 2022).
- Torres, P.J.; Edwards, R.A.; McNair, K.A. PARTIE: A partition engine to separate metagenomic and amplicon projects in the Sequence Read Archive. Bioinformatics 2017, 33, 2389–2391. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef]
- Yi, H.; Yong, D.; Lee, K.; Cho, Y.-J.; Chun, J. Profiling bacterial community in upper respiratory tracts. BMC Infect. Dis. 2014, 14, 583. [Google Scholar] [CrossRef]
- Lassalle, F.; Spagnoletti, M.; Fumagalli, M.; Shaw, L.; Dyble, M.; Walker, C.; Thomas, M.G.; Bamberg Migliano, A.; Balloux, F. Oral microbiomes from hunter-gatherers and traditional farmers reveal shifts in commensal balance and pathogen load linked to diet. Mol. Ecol. 2018, 27, 182–195. [Google Scholar] [CrossRef]
- Man, W.H.; de Steenhuijsen Piters, W.A.A.; Bogaert, D. The microbiota of the respiratory tract: Gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017, 15, 259–270. [Google Scholar] [CrossRef]
- Fernández, A.; Huang, S.; Seston, S.; Xing, J.; Hickey, R.; Criddle, C.; Tiedje, J. How Stable Is Stable? Function versus Community Composition. Appl. Environ. Microbiol. 1999, 65, 3697–3704. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, A.K.; Yin, C.; Hulbert, S.H. Community Structure, Species Variation, and Potential Functions of Rhizosphere-Associated Bacteria of Different Winter Wheat (Triticum aestivum) Cultivars. Front. Plant Sci. 2017, 8, 132. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.T. Machine-Learning Based Identification of Metagenomic Source Environments. Master’s Thesis, San Diego State University, San Diego, CA, USA, 2019. Available online: https://digitallibrarydev.sdsu.edu/islandora/object/sdsu%3A27581 (accessed on 1 April 2022).
- Cuthbertson, L.; Walker, A.W.; Oliver, A.E.; Rogers, G.B.; Rivett, D.W.; Hampton, T.H.; Ashare, A.; Elborn, J.S.; De Soyza, A.; Carroll, M.P.; et al. Lung function and microbiota diversity in cystic fibrosis. Microbiome 2020, 8, 45. [Google Scholar] [CrossRef]
- Yatera, K.; Noguchi, S.; Mukae, H. The microbiome in the lower respiratory tract. Respir. Investig. 2018, 56, 432–439. [Google Scholar] [CrossRef]
- Lim, Y.W.; Schmieder, R.; Haynes, M.; Furlan, M.; Matthews, T.D.; Whiteson, K.; Poole, S.J.; Hayes, C.S.; Low, D.A.; Maughan, H.; et al. Mechanistic Model of Rothia mucilaginosa Adaptation toward Persistence in the CF Lung, Based on a Genome Reconstructed from Metagenomic Data. PLoS ONE 2013, 8, e64285. [Google Scholar] [CrossRef]
- Willis, J.R.; Saus, E.; Iraola-Guzmán, S.; Cabello-Yeves, E.; Ksiezopolska, E.; Cozzuto, L.; Bejarano, L.A.; Andreu-Somavilla, N.; Alloza-Trabado, M.; Blanco, A.; et al. Citizen-science based study of the oral microbiome in Cystic fibrosis and matched controls reveals major differences in diversity and abundance of bacterial and fungal species. J. Oral Microbiol. 2021, 13, 1897328. [Google Scholar] [CrossRef]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-level analysis of gut microbiome variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef]
- Camarillo-Guerrero, L.F.; Almeida, A.; Rangel-Pineros, G.; Finn, R.D.; Lawley, T.D. Massive expansion of human gut bacteriophage diversity. Cell 2021, 184, 1098–1109.e9. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Brito, B.; Li, L.; Wegley, L.; Furlan, M.; Angly, E.F.; Breitbart, M.; Buchanan, J.; Desnues, C.; Dinsdale, E.; Edwards, R.; et al. Viral and microbial community dynamics in four aquatic environments. ISME J. 2010, 4, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Dwivedi, B.; Akhter, S.; Breitbart, M.; Edwards, R.A. Multidimensional metrics for estimating phage abundance, distribution, gene density, and sequence coverage in metagenomes. Front. Microbiol. 2015, 6, 381. [Google Scholar] [CrossRef]
- Howard-Varona, C.; Hargreaves, K.R.; Abedon, S.T.; Sullivan, M.B. Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME J. 2017, 11, 1511–1520. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Genet. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Romero-Calle, D.; Guimarães Benevides, R.; Góes-Neto, A.; Billington, C. Bacteriophages as Alternatives to Antibiotics in Clinical Care. Antibiotics 2019, 8, 138. [Google Scholar] [CrossRef] [PubMed]
- Tagliaferri, T.L.; Jansen, M.; Horz, H.-P. Fighting Pathogenic Bacteria on Two Fronts: Phages and Antibiotics as Combined Strategy. Front. Cell. Infect. Microbiol. 2019, 9, 22. [Google Scholar] [CrossRef]
- Hatful, G.F.; Dedrick, R.M.; Schooley, R.T. Phage Therapy for Antibiotic-Resistant Bacterial Infections. Annu. Rev. Med. 2022, 73, 197–211. [Google Scholar] [CrossRef]
- Khalid, A.; Lin, R.C.Y.; Iredell, J.R. A Phage Therapy Guide for Clinicians and Basic Scientists: Background and Highlighting Applications for Developing Countries. Front. Microbiol. 2020, 11, 599906. [Google Scholar] [CrossRef]
- Luong, T.; Salabarria, A.-C.; Edwards, R.A.; Roach, D.R. Standardized bacteriophage purification for personalized phage therapy. Nat. Protoc. 2020, 15, 2867–2890. [Google Scholar] [CrossRef]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 2017, 61, e00954-17. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a dis-seminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Hay, I.D.; Lithgow, T. Filamentous phages: Masters of a microbial sharing economy. EMBO Rep. 2019, 20, e47247. [Google Scholar] [CrossRef]
- Rice, S.A.; Tan, C.H.; Mikkelsen, P.J.; Kung, V.; Woo, J.; Tay, M.; Hauser, A.; McDougald, D.; Webb, J.S.; Kjelleberg, S. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 2009, 3, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Zinder, N.D. Lysogenization and superinfection immunity in Salmonella. Virology 1958, 5, 291–326. [Google Scholar] [CrossRef]
- Mavrich, T.N.; Hatfull, G.F. Evolution of Superinfection Immunity in Cluster A Mycobacteriophages. mBio 2019, 10, e00971-19. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.; Kauffman, K.; Hussain, F.; Kalatzis, P.; Rørbo, N.; Polz, M.F.; Middelboe, M. Widespread distribution of prophage-encoded virulence factors in marine Vibrio communities. Sci. Rep. 2018, 8, 9973. [Google Scholar] [CrossRef] [PubMed]
- Sweere, J.M.; Van Belleghem, J.D.; Ishak, H.; Bach, M.S.; Popescu, M.; Sunkari, V.; Kaber, G.; Manasherob, R.; Suh, G.A.; Cao, X.; et al. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 2019, 363, aat9691. [Google Scholar] [CrossRef]
- Bielaszewska, M.; Idelevich Evgeny, A.; Zhang, W.; Bauwens, A.; Schaumburg, F.; Mellmann, A.; Peters, G.; Karch, H. Effects of Antibiotics on Shiga Toxin 2 Production and Bacteriophage Induction by Epidemic Escherichia coli O104:H4 Strain. Antimicrob. Agents Chemother. 2012, 56, 3277–3282. [Google Scholar] [CrossRef]
- von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef]
- Waldor, M.K.; Mekalanos, J.J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 1996, 272, 1910–1914. [Google Scholar] [CrossRef]
- Argen, J.A.; Clark, A.G. Selfish genetic elements. PLoS Genet. 2018, 14, e1007700. [Google Scholar] [CrossRef]
- Ptashne, M. A Genetic Switch: Phage Lambda Revisited; CSHL Press: Cold Spring Harbor, NY, USA, 2004. [Google Scholar]
- Knowles, B.; Silveira, C.B.; Bailey, B.A.; Barott, K.; Cantu, V.A.; Cobián-Güemes, A.G.; Coutinho, F.H.; Dinsdale, E.A.; Felts, B.; Furby, K.A.; et al. Lytic to temperate switching of viral communities. Nature 2016, 531, 466–470 . [Google Scholar] [CrossRef] [PubMed]
- Thingstad, T.F. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 2000, 45, 1320–1328. [Google Scholar] [CrossRef]
- Silveira, C.B.; Luque, A.; Rohwer, F. The landscape of lysogeny across microbial community density, diversity and energetics. Environ. Microbiol. 2021, 23, 4098–4111. [Google Scholar] [CrossRef] [PubMed]
- Paterson, J.S.; Smith, R.J.; McKerral, J.C.; Dann, L.M.; Launer, E.; Goonan, P.; Kleinig, T.; Fuhrman, J.A.; Mitchell, J.G. A hydrocarbon-contaminated aquifer reveals a Piggyback-the-Persistent viral strategy. FEMS Microbiol. Ecol. 2019, 95, fiz116. [Google Scholar] [CrossRef]
- Brum, J.R.; Hurwitz, B.L.; Schofield, O.; Ducklow, H.W.; Sullivan, M.B. Seasonal time bombs: Dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J. 2016, 10, 437–449. [Google Scholar] [CrossRef]
- Silveira, C.B.; Rohwer, F.L. Piggyback-the-Winner in host-associated microbial communities. NPJ Biofilms Microbiomes 2016, 2, 16010. [Google Scholar] [CrossRef]
- Cheng, H.H.; Muhlrad, P.J.; Hoyt, M.A.; Echols, H. Cleavage of the cII protein of phage lambda by purified HflA protease: Control of the switch between lysis and lysogeny. Proc. Natl. Acad. Sci. USA 1988, 85, 7882–7886. [Google Scholar] [CrossRef]
- Kim, M.S.; Bae, J.W. Lysogeny is prevalent and widely distributed in the murine gut microbiota. ISME J. 2018, 12, 1127–1141. [Google Scholar] [CrossRef]
- Cazares, D.; Cazares, A.; Figueroa, W.; Guarneros, G.; Edwards Robert, A.; Vinuesa, P.; Bordenstein, S.; Svircev, A. A Novel Group of Promiscuous Podophages Infecting Diverse Gammaproteobacteria from River Communities Exhibits Dynamic Intergenus Host Adaptation. mSystems 2021, 6, e00773-20. [Google Scholar] [CrossRef]
- Dutilh, B.E.; Cassman, N.; McNair, K.; Sanchez, S.E.; Silva, G.G.Z.; Boling, L.; Barr, J.; Speth, D.; Seguritan, V.; Aziz, R.; et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 2014, 5, 4498. [Google Scholar] [CrossRef]
- Edwards, R.A.; Vega, A.A.; Norman, H.M.; Ohaeri, M.; Levi, K.; Dinsdale, E.A.; Cinek, O.; Aziz, R.K.; McNair, K.; Barr, J.J.; et al. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat. Microbiol. 2019, 4, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.-H.; Lin, X.B.; Zhang, S.; Tollenaar Stephanie, L.; Özçam, M.; Dunphy, C.; Walter, J.; van Pijkeren, J.-P.; Kivisaar, M. Prophages in Lactobacillus reuteri Are Associated with Fitness Trade-Offs but Can Increase Competitiveness in the Gut Ecosystem. Appl. Environ. Microbiol. 2019, 86, e01922-19. [Google Scholar] [CrossRef] [PubMed]
- Motlagh, A.M.; Bhattacharjee, A.S.; Coutinho, F.H.; Dutilh, B.E.; Casjens, S.R.; Goel, R.K. Insights of Phage-Host Interaction in Hypersaline Ecosystem through Metagenomics Analyses. Front. Microbiol. 2017, 8, 352. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, L.; Breitbart, M.; Mobberley, J.; Long, A.; Haynes, M.; Rohwer, F.; Paul, J.H. Metagenomic Analysis of Lysogeny in Tampa Bay: Implications for Prophage Gene Expression. PLoS ONE 2008, 3, e3263. [Google Scholar] [CrossRef]
- McDaniel, L.D.; Rosario, K.; Breitbart, M.; Paul, J.H. Comparative metagenomics: Natural populations of induced prophages demonstrate highly unique, lower diversity viral sequences. Environ. Microbiol. 2014, 16, 570–585. [Google Scholar] [CrossRef]
- Parmar, K.; Dafale, N.; Pal, R.; Tikariha, H.; Purohit, H. An Insight into Phage Diversity at Environmental Habitats using Comparative Metagenomics Approach. Curr. Microbiol. 2018, 75, 132–141. [Google Scholar] [CrossRef]
- Stokar-Avihail, A.; Tal, N.; Erez, Z.; Lopatina, A.; Sorek, R. Widespread Utilization of Peptide Communication in Phages Infecting Soil and Pathogenic Bacteria. Cell Host Microbe 2019, 25, 746–755.e5. [Google Scholar] [CrossRef]
- Gómez, P.; Bennie, J.; Gaston, K.J.; Buckling, A. The Impact of Resource Availability on Bacterial Resistance to Phages in Soil. PLoS ONE 2015, 10, e0123752. [Google Scholar] [CrossRef]
- Braga, L.P.; Spor, A.; Kot, W.; Breuil, M.-C.; Hansen, L.H.; Setubal, J.C.; Philippot, L. Impact of phages on soil bacterial communities and nitrogen availability under different assembly scenarios. Microbiome 2020, 8, 52. [Google Scholar] [CrossRef]
- Rezaei Javan, R.; Ramos-Sevillano, E.; Akter, A.; Brown, J.; Brueggemann, A.B. Prophages and satellite prophages are widespread in Streptococcus and may play a role in pneumococcal pathogenesis. Nat. Commun. 2019, 10, 4852. [Google Scholar] [CrossRef]
- Roach, M.; McNair, K.; Giles, S.; Inglis, L.; Pargin, E.; Decewicz, P.; Edwards, R. Philympics 2021: Prophage Predictions Perplex Programs. bioRxiv 2021. [Google Scholar] [CrossRef]
- Akhter, S.; Aziz, R.K.; Edwards, R.A. PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012, 40, e126. [Google Scholar] [CrossRef] [PubMed]
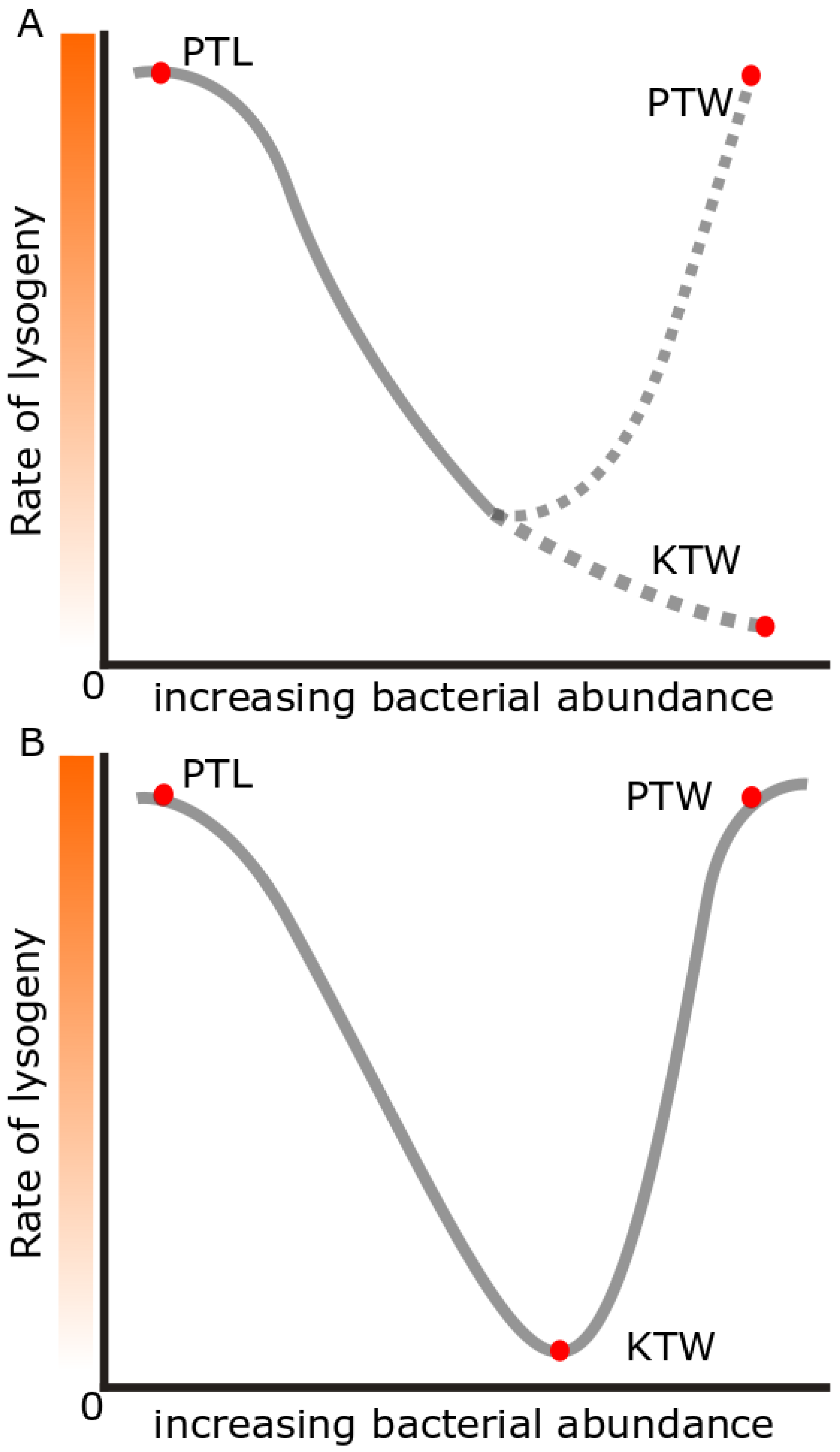
| Lysogeny Hypothesis | Environmental Conditions | Level of Lysogeny |
|---|---|---|
| Kill-the-Winner | High energy, cells growing | Low |
| Piggyback-the-Winner | High energy, cells growing | High |
| Piggyback-the-Loser | Low energy, little growth | High |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inglis, L.K.; Edwards, R.A. How Metagenomics Has Transformed Our Understanding of Bacteriophages in Microbiome Research. Microorganisms 2022, 10, 1671. https://doi.org/10.3390/microorganisms10081671
Inglis LK, Edwards RA. How Metagenomics Has Transformed Our Understanding of Bacteriophages in Microbiome Research. Microorganisms. 2022; 10(8):1671. https://doi.org/10.3390/microorganisms10081671
Chicago/Turabian StyleInglis, Laura K., and Robert A. Edwards. 2022. "How Metagenomics Has Transformed Our Understanding of Bacteriophages in Microbiome Research" Microorganisms 10, no. 8: 1671. https://doi.org/10.3390/microorganisms10081671
APA StyleInglis, L. K., & Edwards, R. A. (2022). How Metagenomics Has Transformed Our Understanding of Bacteriophages in Microbiome Research. Microorganisms, 10(8), 1671. https://doi.org/10.3390/microorganisms10081671






