Microbiota Succession of Whole and Filleted European Sea Bass (Dicentrarchus labrax) during Storage under Aerobic and MAP Conditions via 16S rRNA Gene High-Throughput Sequencing Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Provision, Storage and Sampling of Samples
2.2. Evaluation of Samples Sensory Rejection
2.3. 16S Metabarcoding Analysis
2.3.1. Samples Preparation and DNA Extraction
2.3.2. Library Preparation, High-Throughput Sequencing and Bioinformatic Analysis
2.4. Statistical Analysis
3. Results
3.1. Sensory Evaluation of Whole and Filleted Sea Bass
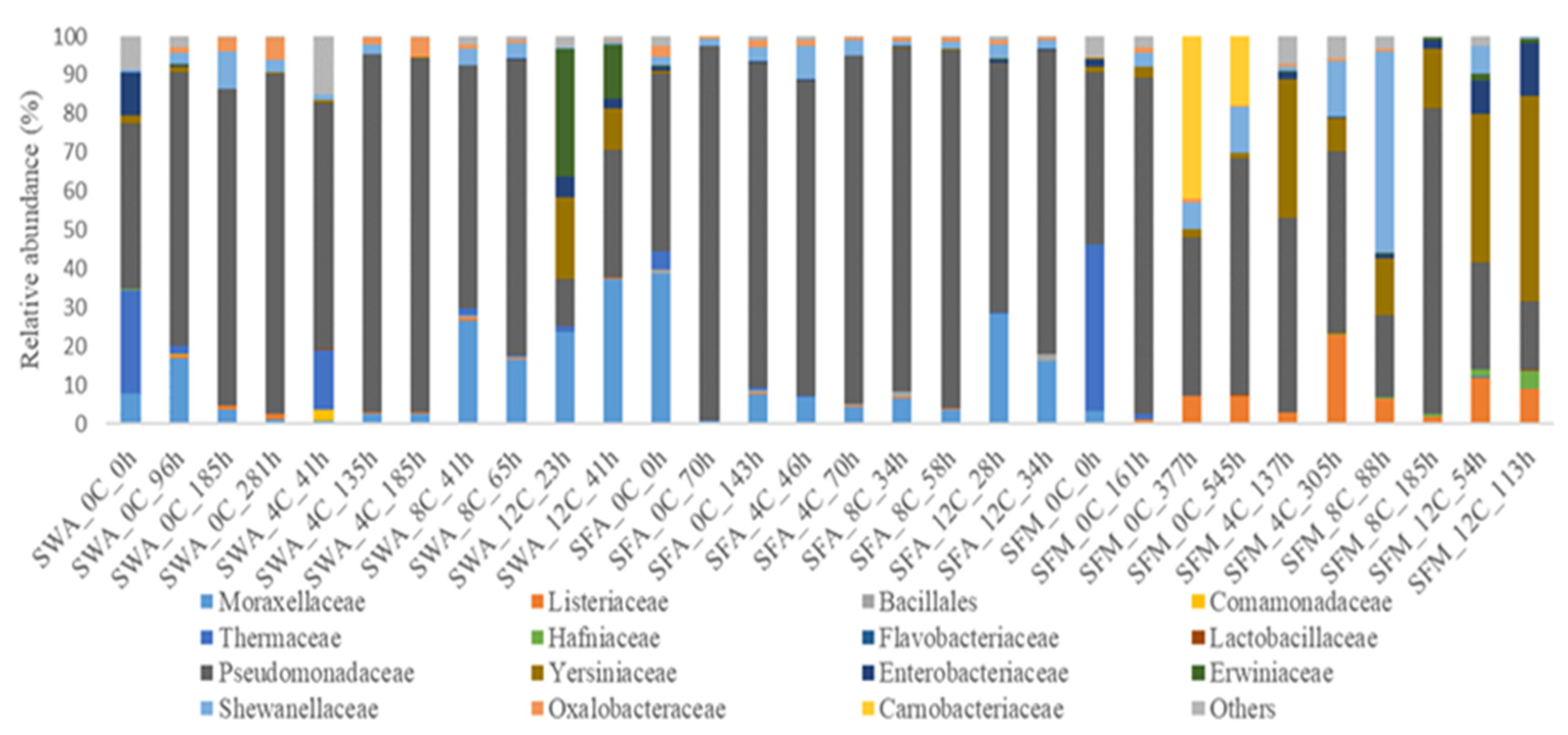
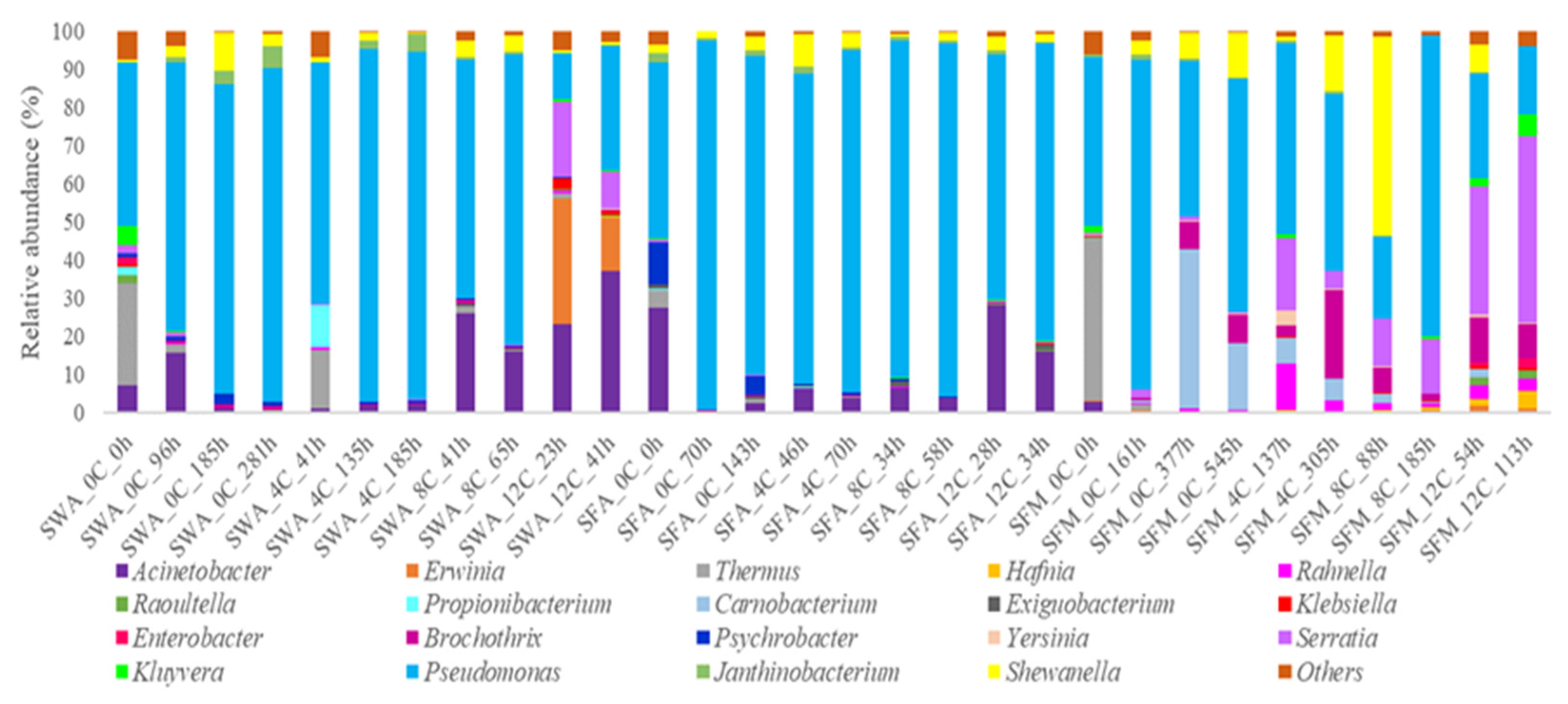
3.2. Microbial Diversity of Whole and Filleted Seabass
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Troell, M.; Jonell, M.; Crona, B. The role of seafood for sustainable and healthy diets. EAT-Lancet Comm. 2019, 24. [Google Scholar]
- Subasinghe, R.; Soto, D.; Jia, J. Global aquaculture and its role in sustainable development. Rev. Aquac. 2009, 1, 2–9. [Google Scholar] [CrossRef]
- FAO. World Fisheries and Aquaculture in Review; FAO: Rome, Italy, 2020; Volume 35, ISBN 9789251326923. [Google Scholar]
- Compassion in Foodbusiness European Seabass (Dicentrarchus labrax) Statistics Summary. 2021, Volume 2, pp. 1–13. Available online: https://www.compassioninfoodbusiness.com/media/7447727/european-seabass-in-numbers.pdf (accessed on 22 July 2022).
- FAO. Text by Makino, M. Fisheries and Aquaculture Division; FAO: Rome, Italy, 2021. [Google Scholar]
- Anagnostopoulos, D.A.; Parlapani, F.F.; Boziaris, I.S. The evolution of knowledge on seafood spoilage microbiota from the 20th to the 21st century: Have we finished or just begun? Trends Food Sci. Technol. 2022, 120, 236–247. [Google Scholar] [CrossRef]
- Gram, L.; Dalgaard, P. Fish spoilage bacteria—Problems and solutions. Curr. Opin. Biotechnol. 2002, 13, 262–266. [Google Scholar] [CrossRef]
- Boziaris, I.S.; Parlapani, F.F. Specific Spoilage Organisms (SSOs) in Fish. Microbiol. Qual. Food Foodborne Spoilers 2017, 5, 61–98. [Google Scholar] [CrossRef]
- Parlapani, F.F.; Michailidou, S.; Pasentsis, K.; Argiriou, A.; Krey, G.; Boziaris, I.S. A meta-barcoding approach to assess and compare the storage temperature-dependent bacterial diversity of gilt-head sea bream (Sparus aurata) originating from fish farms from two geographically distinct areas of Greece. Int. J. Food Microbiol. 2018, 278, 36–43. [Google Scholar] [CrossRef]
- Parlapani, F.F.; Michailidou, S.; Anagnostopoulos, D.A.; Sakellariou, A.K.; Pasentsis, K.; Psomopoulos, F.; Argiriou, A.; Haroutounian, S.A.; Boziaris, I.S. Microbial spoilage investigation of thawed common cuttlefish (Sepia officinalis) stored at 2 °C using next generation sequencing and volatilome analysis. Food Microbiol. 2018, 76, 518–525. [Google Scholar] [CrossRef]
- Zhuang, S.; Hong, H.; Zhang, L.; Luo, Y. Spoilage-related microbiota in fish and crustaceans during storage: Research progress and future trends. Compr. Rev. Food Sci. Food Saf. 2021, 20, 252–288. [Google Scholar] [CrossRef]
- Nychas, G.J.E.; Skandamis, P.N.; Tassou, C.C.; Koutsoumanis, K.P. Meat spoilage during distribution. Meat Sci. 2008, 78, 77–89. [Google Scholar] [CrossRef]
- Doulgeraki, A.I.; Ercolini, D.; Villani, F.; Nychas, G.J.E. Spoilage microbiota associated to the storage of raw meat in different conditions. Int. J. Food Microbiol. 2012, 157, 130–141. [Google Scholar] [CrossRef]
- Parlapani, F.F.; Kormas, K.A.; Boziaris, I.S. Microbiological changes, shelf life and identification of initial and spoilage microbiota of sea bream fillets stored under various conditions using 16S rRNA gene analysis. J. Sci. Food Agric. 2015, 95, 2386–2394. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Mataragas, M.; Bourdichon, F.; Doulgeraki, A.; Pilet, M.F.; Jagadeesan, B.; Rantsiou, K.; Phister, T. Next generation microbiological risk assessment meta-omics: The next need for integration. Int. J. Food Microbiol. 2018, 287, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Maillet, A.; Bouju-Albert, A.; Roblin, S.; Vaissié, P.; Leuillet, S.; Dousset, X.; Jaffrès, E.; Combrisson, J.; Prévost, H. Impact of DNA extraction and sampling methods on bacterial communities monitored by 16S rDNA metabarcoding in cold-smoked salmon and processing plant surfaces. Food Microbiol. 2021, 95, 103705. [Google Scholar] [CrossRef]
- Mayo, B.; Rachid, C.; Alegria, A.; Leite, A.; Peixoto, R.; Delgado, S. Impact of Next Generation Sequencing Techniques in Food Microbiology. Curr. Genom. 2014, 15, 293–309. [Google Scholar] [CrossRef] [PubMed]
- Syropoulou, F.; Parlapani, F.F.; Anagnostopoulos, D.A.; Stamatiou, A.; Mallouchos, A.; Boziaris, I.S. Spoilage Investigation of Chill Stored Meagre (Argyrosomus regius) Using Modern Microbiological and Analytical Techniques. Foods 2021, 10, 3109. [Google Scholar] [CrossRef]
- Howgate, P.; Association, W.E.F.T.; Johnston, A.; Whittle, K.J.; Torry Research Station, A. Multilingual Guide to EC Freshness Grades for Fishery Products, December 1992; Scientific Research Publishing: Wuhan, China, 1992. [Google Scholar]
- Anagnostopoulos, D.A.; Parlapani, F.F.; Mallouchos, A.; Angelidou, A.; Syropoulou, F.; Minos, G.; Boziaris, I.S. Volatile Organic Compounds and 16S Metabarcoding in Ice-Stored Red Seabream Pagrus major. Foods 2022, 11, 666. [Google Scholar] [CrossRef]
- Dowd, S.E.; Callaway, T.R.; Wolcott, R.D.; Sun, Y.; McKeehan, T.; Hagevoort, R.G.; Edrington, T.S. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 2008, 8, 125. [Google Scholar] [CrossRef]
- Edgar, R.C.; Bateman, A. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Eren, A.M.; Zozaya, M.; Taylor, C.M.; Dowd, S.E.; Martin, D.H.; Ferris, M.J. Exploring the Diversity of Gardnerella vaginalis in the Genitourinary Tract Microbiota of Monogamous Couples Through Subtle Nucleotide Variation. PLoS ONE 2011, 6, e26732. [Google Scholar] [CrossRef]
- Swanson, K.S.; Dowd, S.E.; Suchodolski, J.S.; Middelbos, I.S.; Vester, B.M.; Barry, K.A.; Nelson, K.E.; Torralba, M.; Henrissat, B.; Coutinho, P.M.; et al. Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J. 2010, 5, 639–649. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ 2018. [Google Scholar] [CrossRef] [PubMed]
- Gram, L.; Huss, H.H. Microbiological spoilage of fish and fish products. Int. J. Food Microbiol. 1996, 33, 121–137. [Google Scholar] [CrossRef]
- Duan, S.; Zhou, X.; Miao, J.; Duan, X. Succession of bacterial microbiota in tilapia fillets at 4 °C and in situ investigation of spoilers. World J. Microbiol. Biotechnol. 2018, 34, 69. [Google Scholar] [CrossRef] [PubMed]
- Parlapani, F.F.; Haroutounian, S.A.; Nychas, G.J.E.; Boziaris, I.S. Microbiological spoilage and volatiles production of gutted European sea bass stored under air and commercial modified atmosphere package at 2 °C. Food Microbiol. 2015, 50, 44–53. [Google Scholar] [CrossRef]
- Syropoulou, F.; Parlapani, F.F.; Bosmali, I.; Madesis, P.; Boziaris, I.S. HRM and 16S rRNA gene sequencing reveal the cultivable microbiota of the European sea bass during ice storage. Int. J. Food Microbiol. 2020, 327, 108658. [Google Scholar] [CrossRef]
- Syropoulou, F.; Parlapani, F.F.; Kakasis, S.; Nychas, G.J.E.; Boziaris, I.S. Primary Processing and Storage Affect the Dominant Microbiota of Fresh and Chill-Stored Sea Bass Products. Foods 2021, 10, 671. [Google Scholar] [CrossRef] [PubMed]
- Parlapani, F.F. Microbial diversity of seafood. Curr. Opin. Food Sci. 2021, 37, 45–51. [Google Scholar] [CrossRef]
- Tian, B.; Hua, Y. Carotenoid biosynthesis in extremophilic Deinococcus-Thermus bacteria. Trends Microbiol. 2010, 18, 512–520. [Google Scholar] [CrossRef]
- Parlapani, F.F.; Meziti, A.; Kormas, K.A.; Boziaris, I.S. Indigenous and spoilage microbiota of farmed sea bream stored in ice identified by phenotypic and 16S rRNA gene analysis. Food Microbiol. 2013, 33, 85–89. [Google Scholar] [CrossRef]
- Kozińska, A.; Paździor, E.; Pȩkala, A.; Niemczuk, W. Acinetobacter johnsonii and Acinetobacter lwoffii—The emerging fish pathogens. Bull. Vet. Inst. Pulawy 2014, 58, 193–199. [Google Scholar] [CrossRef]
- Dalgaard, P. Freshness, Quality and Safety in Seafoods Flair-Flow Europe Technical Manual; DK-2800 Kgs; Danish Institute for Fisheries Research: Lyngby, Denmark, 2000. [Google Scholar]
- Koutsoumanis, K.; Nychas, G.J.E. Application of a systematic experimental procedure to develop a microbial model for rapid fish shelf life predictions. Int. J. Food Microbiol. 2000, 60, 171–184. [Google Scholar] [CrossRef]
- Tryfinopoulou, P.; Tsakalidou, E.; Nychas, G.J.E. Characterization of Pseudomonas spp. associated with spoilage of gilt-head sea bream stored under various conditions. Appl. Environ. Microbiol. 2002, 68, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, V.; Chouliara, I.; Badeka, A.; Savvaidis, I.N.; Kontominas, M.G. Effect of gutting on microbiological, chemical, and sensory properties of aquacultured sea bass (Dicentrarchus labrax) stored in ice. Food Microbiol. 2003, 20, 411–420. [Google Scholar] [CrossRef]
- Paleologos, E.K.; Savvaidis, I.N.; Kontominas, M.G. Biogenic amines formation and its relation to microbiological and sensory attributes in ice-stored whole, gutted and filleted Mediterranean Sea bass (Dicentrarchus labrax). Food Microbiol. 2004, 21, 549–557. [Google Scholar] [CrossRef]
- Zotta, T.; Parente, E.; Ianniello, R.G.; De Filippis, F.; Ricciardi, A. Dynamics of bacterial communities and interaction networks in thawed fish fillets during chilled storage in air. Int. J. Food Microbiol. 2019, 293, 102–113. [Google Scholar] [CrossRef]
- Kuuliala, L.; Al Hage, Y.; Ioannidis, A.G.; Sader, M.; Kerckhof, F.M.; Vanderroost, M.; Boon, N.; De Baets, B.; De Meulenaer, B.; Ragaert, P.; et al. Microbiological, chemical and sensory spoilage analysis of raw Atlantic cod (Gadus morhua) stored under modified atmospheres. Food Microbiol. 2018, 70, 232–244. [Google Scholar] [CrossRef]
- Jääskeläinen, E.; Jakobsen, L.M.A.; Hultman, J.; Eggers, N.; Bertram, H.C.; Björkroth, J. Metabolomics and bacterial diversity of packaged yellowfin tuna (Thunnus albacares) and salmon (Salmo salar) show fish species-specific spoilage development during chilled storage. Int. J. Food Microbiol. 2019, 293, 44–52. [Google Scholar] [CrossRef]
- Chinivasagam, H.N.; Bremner, H.A.; Thrower, S.J.; Nottingham, S.M. Spoilage pattern of five species of australian prawns: Deterioration is influenced by environment of capture and mode of storage. J. Aquat. Food Prod. Technol. 1996, 5, 25–50. [Google Scholar] [CrossRef]
- Aru, V.; Pisano, M.B.; Savorani, F.; Engelsen, S.B.; Cosentino, S.; Cesare Marincola, F. Data on the changes of the mussels’ metabolic profile under different cold storage conditions. Data Br. 2016, 7, 951–957. [Google Scholar] [CrossRef]
- Parlapani, F.F.; Michailidou, S.; Anagnostopoulos, D.A.; Koromilas, S.; Kios, K.; Pasentsis, K.; Psomopoulos, F.; Argiriou, A.; Haroutounian, S.A.; Boziaris, I.S. Bacterial communities and potential spoilage markers of whole blue crab (Callinectes sapidus) stored under commercial simulated conditions. Food Microbiol. 2019, 82, 325–333. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, X.; Ding, Y.; Ke, Z.; Zhou, X.; Zhang, J. Diversity and succession of the microbial community and its correlation with lipid oxidation in dry-cured black carp (Mylopharyngodon piceus) during storage. Food Microbiol. 2021, 98, 103686. [Google Scholar] [CrossRef] [PubMed]
- Du, G.; Gai, Y.; Zhou, H.; Fu, S.; Zhang, D. Assessment of Spoilage Microbiota of Rainbow Trout (Oncorhynchus mykiss) during Storage by 16S rDNA Sequencing. J. Food Qual. 2022, 2022, 5367984. [Google Scholar] [CrossRef]
- Rawat, S. Food Spoilage: Microorganisms and their prevention Microbial Food Spoilage-Losses and Control Strategies A Brief Review of t he Lit erat ure. Asian J. Plant Sci. Res. 2015, 5, 47–56. [Google Scholar]
- Zhu, S.; Wu, H.; Zhang, C.; Jie, J.; Liu, Z.; Zeng, M.; Wang, C. Spoilage of refrigerated Litopenaeus vannamei: Eavesdropping on Acinetobacter acyl-homoserine lactones promotes the spoilage potential of Shewanella baltica. J. Food Sci. Technol. 2018, 55, 1903–1912. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, Z.; Jia, S.; Zhang, J.; Li, K.; Luo, Y. The roles of bacteria in the biochemical changes of chill-stored bighead carp (Aristichthys nobilis): Proteins degradation, biogenic amines accumulation, volatiles production, and nucleotides catabolism. Food Chem. 2018, 255, 174–181. [Google Scholar] [CrossRef]
- Devlieghere, F.; Debevere, J. Influence of Dissolved Carbon Dioxide on the Growth of Spoilage Bacteria. LWT Food Sci. Technol. 2000, 33, 531–537. [Google Scholar] [CrossRef]
- Powell, S.M.; Tamplin, M.L. Microbial communities on Australian modified atmosphere packaged Atlantic salmon. Food Microbiol. 2012, 30, 226–232. [Google Scholar] [CrossRef]
- Macé, S.; Joffraud, J.J.; Cardinal, M.; Malcheva, M.; Cornet, J.; Lalanne, V.; Chevalier, F.; Sérot, T.; Pilet, M.F.; Dousset, X. Evaluation of the spoilage potential of bacteria isolated from spoiled raw salmon (Salmo salar) fillets stored under modified atmosphere packaging. Int. J. Food Microbiol. 2013, 160, 227–238. [Google Scholar] [CrossRef]
- Chaillou, S.; Chaulot-Talmon, A.; Caekebeke, H.; Cardinal, M.; Christieans, S.; Denis, C.; Hélène Desmonts, M.; Dousset, X.; Feurer, C.; Hamon, E.; et al. Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. ISME J. 2015, 9, 1105–1118. [Google Scholar] [CrossRef]
- McMillin, K.W. Where is MAP Going? A review and future potential of modified atmosphere packaging for meat. Meat Sci. 2008, 80, 43–65. [Google Scholar] [CrossRef]
- Antunes-rohling, A.; Calero, S.; Halaihel, N.; Marquina, P.; Raso, J.; Calanche, J.; Beltr, A.; Ignacio, Á.; Cebri, G. Characterization of the Spoilage Microbiota of Hake Di ff erent Temperatures. Foods 2019, 2, 489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Y.; Liu, X.; Lei, Y.; Regenstein, J.M.; Luo, Y. Characterization of the microbial composition and quality of lightly salted grass carp (Ctenopharyngodon idellus) fillets with vacuum or modified atmosphere packaging. Int. J. Food Microbiol. 2019, 293, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, J.S.; Bøknæs, N.; Mejlholm, O.; Dalgaard, P. Superchilling in combination with modified atmosphere packaging resulted in long shelf-life and limited microbial growth in Atlantic cod (Gadus morhua L.) from capture-based-aquaculture in Greenland. Food Microbiol. 2020, 88, 103405. [Google Scholar] [CrossRef]
- Schuerger, A.C.; Ulrich, R.; Berry, B.J.; Nicholson, W.L. Growth of serratia liquefaciens under 7 mbar, 0 C, and CO 2-enriched anoxic atmospheres. Astrobiology 2013, 13, 115–131. [Google Scholar] [CrossRef]
- González-Rodríguez, M.N.; Sanz, J.J.; Santos, J.Á.; Otero, A.; García-López, M.L. Numbers and types of microorganisms in vacuum-packed cold-smoked freshwater fish at the retail level. Int. J. Food Microbiol. 2002, 77, 161–168. [Google Scholar] [CrossRef]
- Joffraud, J.J.; Cardinal, M.; Cornet, J.; Chasles, J.S.; Léon, S.; Gigout, F.; Leroi, F. Effect of bacterial interactions on the spoilage of cold-smoked salmon. Int. J. Food Microbiol. 2006, 112, 51–61. [Google Scholar] [CrossRef]
- Macé, S.; Cornet, J.; Chevalier, F.; Cardinal, M.; Pilet, M.F.; Dousset, X.; Joffraud, J.J. Characterisation of the spoilage microbiota in raw salmon (Salmo salar) steaks stored under vacuum or modified atmosphere packaging combining conventional methods and PCR-TTGE. Food Microbiol. 2012, 30, 164–172. [Google Scholar] [CrossRef]
- Stohr, V.; Joffraud, J.J.; Cardinal, M.; Leroi, F. Spoilage potential and sensory profile associated with bacteria isolated from cold-smoked salmon. Food Res. Int. 2001, 34, 797–806. [Google Scholar] [CrossRef]
- Alfaro, B.; Hernandez, I. Evolution of the indigenous microbiota in modified atmosphere packaged Atlantic horse mackerel (Trachurus trachurus) identified by conventional and molecular methods. Int. J. Food Microbiol. 2013, 167, 117–123. [Google Scholar] [CrossRef]
- Jaffrès, E.; Lalanne, V.; Macé, S.; Cornet, J.; Cardinal, M.; Sérot, T.; Dousset, X.; Joffraud, J.J. Sensory characteristics of spoilage and volatile compounds associated with bacteria isolated from cooked and peeled tropical shrimps using SPME-GC-MS analysis. Int. J. Food Microbiol. 2011, 147, 195–202. [Google Scholar] [CrossRef]
- Begrem, S.; Jérôme, M.; Leroi, F.; Delbarre-Ladrat, C.; Grovel, O.; Passerini, D. Genomic diversity of Serratia proteamaculans and Serratia liquefaciens predominant in seafood products and spoilage potential analyses. Int. J. Food Microbiol. 2021, 354. [Google Scholar] [CrossRef] [PubMed]
| Sample 2 | Atmosphere | Temperature (°C) |
|---|---|---|
| Whole Seabass | Air | 0, 4, 8, 12 |
| Filleted Seabass | Air | 0, 4, 8, 12 |
| Filleted Seabass | MAP 1 | 0, 4, 8, 12 |
| Sample | Atmosphere | Temperature (°C) | End of Shelf-Life (h) |
|---|---|---|---|
| Whole Sea bass | Air | 0 | 281 |
| Whole Sea bass | Air | 4 | 185 |
| Whole Sea bass | Air | 8 | 65 |
| Whole Sea bass | Air | 12 | 41 |
| Filleted Sea bass | Air | 0 | 143 |
| Filleted Sea bass | Air | 4 | 70 |
| Filleted Sea bass | Air | 8 | 58 |
| Filleted Sea bass | Air | 12 | 34 |
| Filleted Sea bass | MAP | 0 | 545 |
| Filleted Sea bass | MAP | 4 | 305 |
| Filleted Sea bass | MAP | 8 | 185 |
| Filleted Sea bass | MAP | 12 | 113 |
| Sample | Raw Reads | Filtered Passing Reads | Observed Features 1 | Shannon |
|---|---|---|---|---|
| SWA_0C_0h | 4994 | 3685 | 76 | 4.89 |
| SWA_0C_96h | 20,085 | 13,930 | 135 | 5.82 |
| SWA_0C_185h | 20,709 | 9664 | 45 | 4.90 |
| SWA_0C_281h | 27,096 | 13,238 | 47 | 4.55 |
| SWA_4C_41h | 7255 | 4692 | 47 | 4.30 |
| SWA_4C_135h | 18,099 | 8600 | 35 | 4.49 |
| SWA_4C_185h | 15,564 | 6544 | 29 | 4.01 |
| SWA_8C_41h | 24,288 | 13,319 | 117 | 5.62 |
| SWA_8C_65h | 21,130 | 10,526 | 57 | 4.90 |
| SWA_12C_23h | 25,780 | 13,013 | 81 | 4.77 |
| SWA_12C_41h | 29,266 | 14,590 | 74 | 5.44 |
| SFA_0C_0h | 13,708 | 9355 | 114 | 5.75 |
| SFA_0C_70h | 58,857 | 32,042 | 50 | 4.00 |
| SFA_0C_143h | 23,662 | 15,583 | 93 | 4.87 |
| SFA_4C_46h | 30,998 | 15,236 | 67 | 4.96 |
| SFA_4C_70h | 33,789 | 17,742 | 65 | 4.63 |
| SFA_8C_34h | 16,466 | 7169 | 41 | 4.35 |
| SFA_8C_58h | 25,308 | 13,846 | 39 | 4.32 |
| SFA_12C_28h | 10,658 | 4227 | 25 | 4.04 |
| SFA_12C_34h | 22,962 | 12,020 | 49 | 4.97 |
| SFM_0C_0h | 7089 | 5845 | 56 | 4.16 |
| SFM_0C_161h | 19,552 | 9812 | 64 | 5.01 |
| SFM_0C_377h | 11,336 | 5188 | 24 | 3.57 |
| SFM_0C_545h | 23,868 | 12,248 | 30 | 3.66 |
| SFM_4C_137h | 36,548 | 15,649 | 50 | 4.78 |
| SFM_4C_305h | 37,086 | 19,233 | 53 | 4.56 |
| SFM_8C_88h | 37,098 | 15,620 | 53 | 4.84 |
| SFM_8C_185h | 37,291 | 20,632 | 46 | 4.55 |
| SFM_12C_54h | 34,161 | 14,363 | 52 | 5.13 |
| SFM_12C_113h | 46,307 | 17,838 | 50 | 4.87 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Syropoulou, F.; Anagnostopoulos, D.A.; Parlapani, F.F.; Karamani, E.; Stamatiou, A.; Tzokas, K.; Nychas, G.-J.E.; Boziaris, I.S. Microbiota Succession of Whole and Filleted European Sea Bass (Dicentrarchus labrax) during Storage under Aerobic and MAP Conditions via 16S rRNA Gene High-Throughput Sequencing Approach. Microorganisms 2022, 10, 1870. https://doi.org/10.3390/microorganisms10091870
Syropoulou F, Anagnostopoulos DA, Parlapani FF, Karamani E, Stamatiou A, Tzokas K, Nychas G-JE, Boziaris IS. Microbiota Succession of Whole and Filleted European Sea Bass (Dicentrarchus labrax) during Storage under Aerobic and MAP Conditions via 16S rRNA Gene High-Throughput Sequencing Approach. Microorganisms. 2022; 10(9):1870. https://doi.org/10.3390/microorganisms10091870
Chicago/Turabian StyleSyropoulou, Faidra, Dimitrios A. Anagnostopoulos, Foteini F. Parlapani, Evangelia Karamani, Anastasios Stamatiou, Kostas Tzokas, George-John E. Nychas, and Ioannis S. Boziaris. 2022. "Microbiota Succession of Whole and Filleted European Sea Bass (Dicentrarchus labrax) during Storage under Aerobic and MAP Conditions via 16S rRNA Gene High-Throughput Sequencing Approach" Microorganisms 10, no. 9: 1870. https://doi.org/10.3390/microorganisms10091870






