Chemotactic Responses of Xanthomonas with Different Host Ranges
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Culture Media, and Growth Conditions
2.2. Carbon Source Use by Xanthomonas Strains
2.3. Chemotactic Response of Xanthomonas Strains to Carbon Compounds
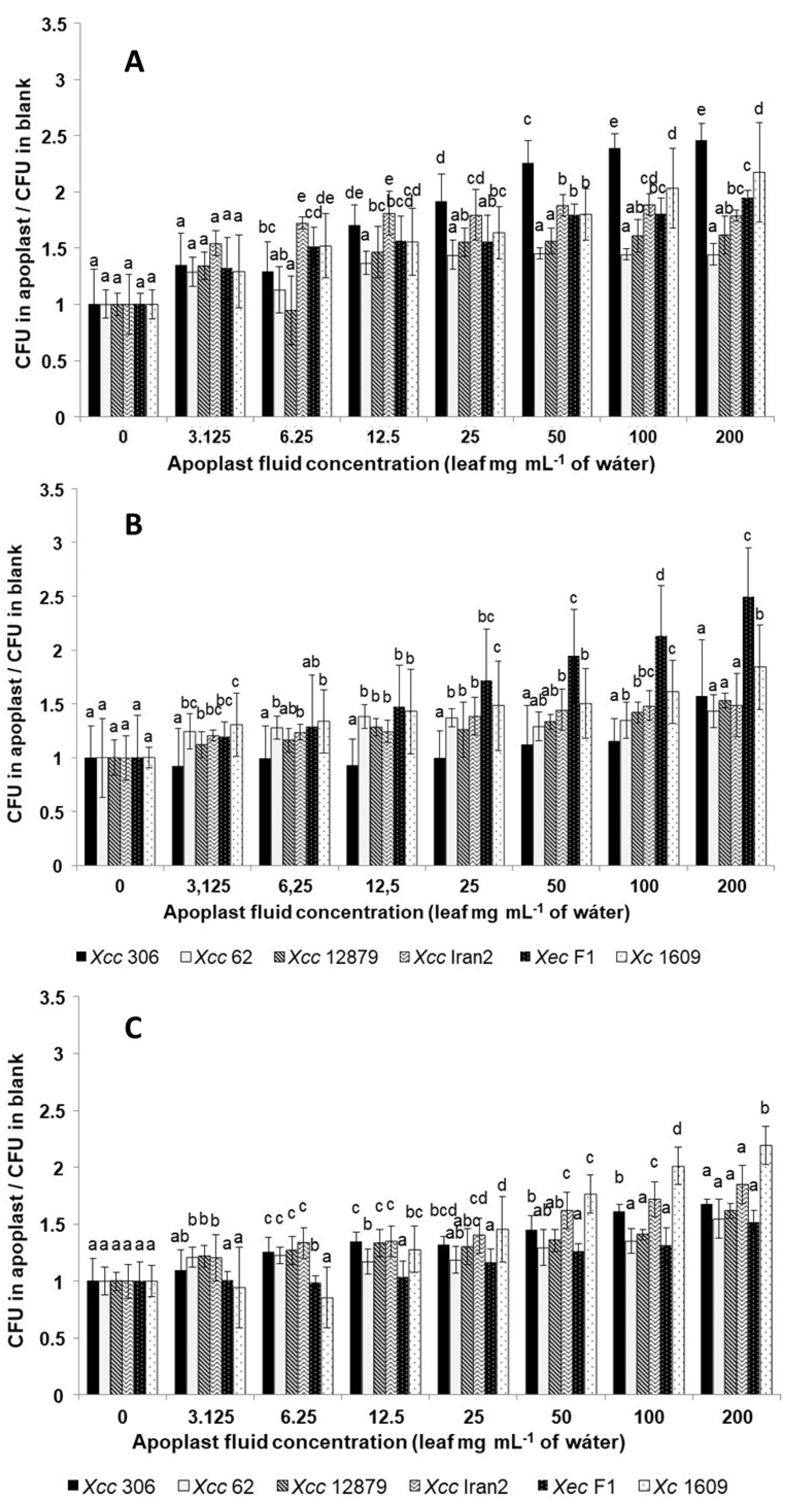
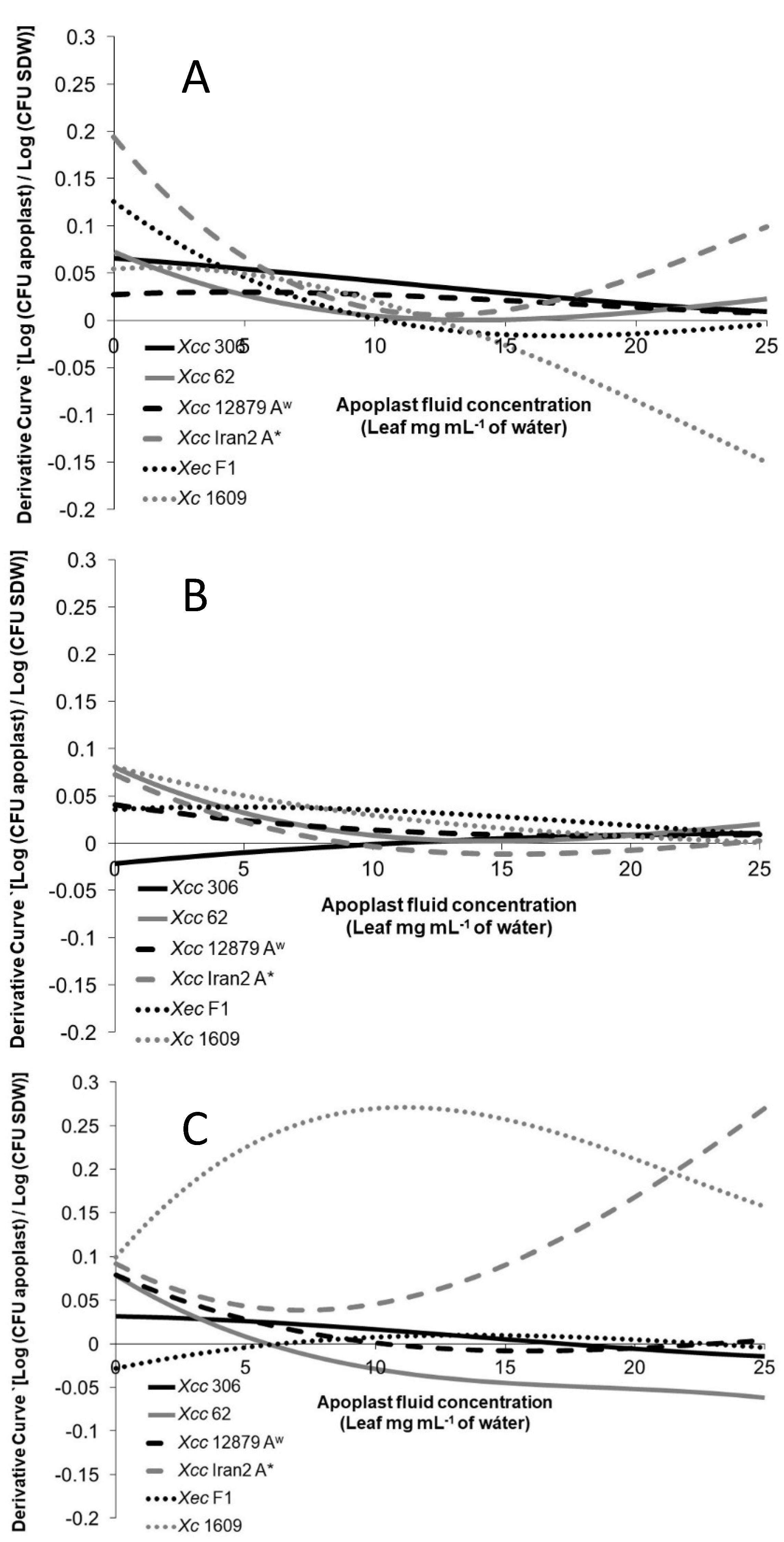
2.4. Chemotactic Response of Xanthomonas Strains to Apoplastic Fluids
2.5. Detection of Methyl-Accepting Chemotaxis Proteins
3. Results
3.1. Carbon Source Use by Xanthomonas Strains
3.2. Chemotactic Response of Xanthomonas Strains to Carbon Compounds
3.3. Identification of MCPs in Xanthomonas Species Used in the Study
3.4. Xanthomonas Strains Are Attracted by Leaf Apoplastic Fluids
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gottwald, T.R.; Hughes, G.; Graham, J.H.; Sun, X.; Riley, T. The Citrus Canker Epidemic in Florida: The Scientific Basis of Regulatory Eradication Policy for an Invasive Species. Phytopathology 2001, 91, 30–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, J.H.; Gottwald, T.R.; Cubero, J.; Achor, D.S. Xanthomonas axonopodis pv. citri: Factors affecting successful eradication of citrus canker. Mol. Plant Pathol. 2004, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Licciardello, G.; Caruso, P.; Bella, P.; Boyer, C.; Smith, M.W.; Pruvost, O.; Robene, I.; Cubero, J.; Catara, V. Pathotyping Citrus Ornamental Relatives with Xanthomonas citri pv. citri and X. citri pv. aurantifolii Refines Our Understanding of Their Susceptibility to These Pathogens. Microorganims 2022, 10, 986. [Google Scholar] [CrossRef]
- Vauterin, L.; Hoste, B.; Kersters, K.; Swings, J. Reclassification of Xanthomonas. Int. J. Syst. Bacteriol. 1995, 45, 472–489. [Google Scholar] [CrossRef] [Green Version]
- Vauterin, L.; Rademaker, J.; Swings, J. Synopsis on the taxonomy of the genus Xanthomonas. Phytopathology 2000, 90, 677–682. [Google Scholar] [CrossRef] [Green Version]
- Schaad, N.W.; Postnikova, E.; Lacy, G.; Sechler, A.; Agarkova, I.; Stromberg, P.E.; Stromberg, V.K.; Vidaver, A.K. Emended classification of xanthomonad pathogens on citrus. Syst. Appl. Microbiol. 2006, 29, 690–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaad, N.W.; Postnikova, E.; Lacy, G.H.; Sechler, A.; Agarkova, I.; Stromberg, P.E.; Stromberg, V.K.; Vidaver, A.K. Reclassification of Xanthomonas campestris pv. citri (ex Hasse 1915) Dye 1978 forms A, B/C/D, and E as X. smithii subsp. citri (ex Hasse) sp. nov. nom. rev. comb. nov., X. fuscans subsp. aurantifolii (ex Gabriel 1989)) sp. nov. nom. r. nom. rev. comb. no. comb. nov., and X. alfalfae subsp. citrumelo (ex Riker and Jones) Gabriel et al., 1989 sp. nov. nom. r. nom. rev. comb. no. comb. nov.; X. campestris pv malvacearum (ex Smith 1901) Dye 1978 as X. smithii subsp. smithii nov. comb. nov. nom. nov.; X. campestris pv. alfalfae (ex Riker and Jones, 1935) Dye 1978 as X. alfalfae subsp. alfalfae (ex Riker et al., 1935) sp. nov. nom. r. nom. rev.; and ‘‘var. fuscans’ of X. campestris pv. phaseoli (ex Smith, 1987) Dye 1978 as X. fuscans subsp. fuscans sp. nov. Syst. Appl. Microbiol. 2005, 28, 494–518. [Google Scholar] [CrossRef]
- Sun, X.; Stall, R.E.; Jones, J.B.; Cubero, J.; Gottwald, T.R.; Graham, J.H.; Dixon, W.N.; Schubert, T.S.; Chaloux, P.H.; Stromberg, V.K.; et al. Detection and Characterization of a New Strain of Citrus Canker Bacteria from Key/Mexican Lime and Alemow in South Florida. Plant Dis. 2004, 88, 1179–1188. [Google Scholar] [CrossRef] [Green Version]
- Vernière, C.; Hartung, J.; Pruvost, O.; Civerolo, E.; Alvarez, A.; Maestri, P.; Luisetti, J. Characterization of phenotypically distinct strains of Xanthomonas axonopodis pv. citri from Southwest Asia. Eur. J. Plant Pathol. 1998, 104, 477–487. [Google Scholar] [CrossRef]
- Pruvost, O.; Goodarzi, T.; Boyer, K.; Soltaninejad, H.; Escalon, A.; Alavi, S.M.; Javegny, S.; Boyer, C.; Cottyn, B.; Gagnevin, L.; et al. Genetic structure analysis of strains causing citrus canker in Iran reveals the presence of two different lineages of Xanthomonas citri pv. citri pathotype A*. Plant Pathol. 2015, 64, 776–784. [Google Scholar] [CrossRef]
- Gordon, J.L.; Lefeuvre, P.; Escalon, A.; Barbe, V.; Cruveiller, S.; Gagnevin, L.; Pruvost, O. Comparative genomics of 43 strains of Xanthomonas citri pv. citri reveals the evolutionary events giving rise to pathotypes with different host ranges. BMC Genomics 2015, 16, 1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adler, J. Chemotaxis in bacteria. J. Supramol. Struct. 1976, 4, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Río-Álvarez, I.; Muñoz-Gómez, C.; Navas-Vásquez, M.; Martínez-García, P.M.; Antúnez-Lamas, M.; Rodríguez-Palenzuela, P.; López-Solanilla, E. Role of Dickeya dadantii 3937 chemoreceptors in the entry to Arabidopsis leaves through wounds. Mol. Plant Pathol. 2015, 16, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Antunez-Lamas, M.; Cabrera, E.; Lopez-Solanilla, E.; Solano, R.; González-Melendi, P.; Chico, J.M.; Toth, I.; Birch, P.; Pritchard, L.; Liu, H.; et al. Bacterial chemoattraction towards jasmonate plays a role in the entry of Dickeya dadantii through wounded tissues. Mol. Microbiol. 2009, 74, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Lund, S.P.; Scott, R.A.; Greenwald, J.W.; Records, A.H.; Nettleton, D.; Lindow, S.E.; Gross, D.C.; Beattie, G.A. Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc. Natl. Acad. Sci. USA 2013, 110, E425–E434. [Google Scholar] [CrossRef] [Green Version]
- Indiana, A. Rôles du Chimiotactisme et de la Mobilité Flagellaire dans la Fitness des Xanthomonas. Ph.D. Thesis, Université d’Angers, Angers, France, 2014. [Google Scholar]
- Lacal, J.; García-Fontana, C.; Muñoz-Martínez, F.; Ramos, J.-L.; Krell, T. Sensing of environmental signals: Classification of chemoreceptors according to the size of their ligand binding regions. Environ. Microbiol. 2010, 12, 2873–2884. [Google Scholar] [CrossRef]
- Ud-Din, A.I.M.S.; Roujeinikova, A. Methyl-accepting chemotaxis proteins: A core sensing element in prokaryotes and archaea. Cell Mol. Life Sci. 2017, 74, 3293–3303. [Google Scholar] [CrossRef]
- Mhedbi-Hajri, N.; Darrasse, A.; Pigné, S.; Durand, K.; Fouteau, S.; Barbe, V.; Manceau, C.; Lemaire, C.; Jacques, M.-A. Sensing and adhesion are adaptive functions in the plant pathogenic xanthomonads. BMC Evol. Biol. 2011, 11, 67. [Google Scholar] [CrossRef] [Green Version]
- Garita-Cambronero, J.; Sena-Vélez, M.; Ferragud, E.; Sabuquillo, P.; Redondo, C.; Cubero, J. Xanthomonas citri subsp. citri and Xanthomonas arboricola pv. pruni: Comparative analysis of two pathogens producing similar symptoms in different host plants. Alcaraz LD, editor. PLoS ONE 2019, 14, e0219797. [Google Scholar] [CrossRef] [Green Version]
- Yaryura, P.M.; Conforte, V.P.; Malamud, F.; Roeschlin, R.; de Pino, V.; Castagnaro, A.P.; McCarthy, Y.; Dow, J.M.; Marano, M.R.; Vojnov, A.A. XbmR, a new transcription factor involved in the regulation of chemotaxis, biofilm formation and virulence in Xanthomonas citri subsp. citri. Environ. Microbiol. 2015, 17, 4164–4176. [Google Scholar] [CrossRef]
- Graham, J.H. Susceptibility of Citrus Fruit to Bacterial Spot and Citrus Canker. Phytopathology 1992, 82, 452–457. [Google Scholar] [CrossRef]
- Bock, C.H.; Graham, J.H.; Gottwald, T.R.; Cook, A.Z.; Parker, P.E. Wind speed and wind-associated leaf injury affect severity of citrus canker on Swingle citrumelo. Eur. J. Plant Pathol. 2010, 128, 21–38. [Google Scholar] [CrossRef]
- Ference, C.M.; Gochez, A.M.; Behlau, F.; Wang, N.; Graham, J.H.; Jones, J.B. Recent advances in the understanding of Xanthomonas citri ssp. citri pathogenesis and citrus canker disease management. Mol. Plant Pathol. 2018, 19, 1302–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cubero, J.; Gell, I.; Johnson, E.G.; Redondo, A.; Graham, J.H. Unstable green fluorescent protein for study of Xanthomonas citri subsp. citri survival on citrus. Plant Pathol. 2011, 60, 977–985. [Google Scholar] [CrossRef]
- Sena-Vélez, M.; Redondo, C.; Gell, I.; Ferragud, E.; Johnson, E.; Graham, J.H.; Cubero, J. Biofilm formation and motility of Xanthomonas strains with different citrus host range. Plant Pathol. 2015, 64, 767–775. [Google Scholar] [CrossRef]
- Al-Saadi, A.; Reddy, J.D.; Duan, Y.P.; Brunings, A.M.; Yuan, Q.; Gabriel, D.W. All Five Host-Range Variants of Xanthomonas citri Carry One pthA Homolog With 17.5 Repeats That Determines Pathogenicity on Citrus, but None Determine Host-Range Variation. Mol. Plant-Microbe Interact. 2007, 20, 934–943. [Google Scholar] [CrossRef] [Green Version]
- Rybak, M.; Minsavage, G.V.; Stall, R.E.; Jones, J.B. Identification of Xanthomonas citri ssp. citri host specificity genes in a heterologous expression host. Mol. Plant Pathol. 2009, 10, 249–262. [Google Scholar] [CrossRef]
- Escalon, A.; Javegny, S.; Vernière, C.; Noël, L.D.; Vital, K.; Poussier, S.; Hajri, A.; Boureau, T.; Pruvost, O.; Arlat, M.; et al. Variations in type III effector repertoires, pathological phenotypes and host range of Xanthomonas citri pv. citri pathotypes. Mol. Plant Pathol. 2013, 14, 483–496. [Google Scholar] [CrossRef] [Green Version]
- Teper, D.; Xu, J.; Pandey, S.S.; Wang, N. PthAW1, a Transcription Activator-Like Effector of Xanthomonas citri subsp. citri, Promotes Host-Specific Immune Responses. Mol. Plant-Microbe Interact. 2021, 34, 1033–1047. [Google Scholar] [CrossRef]
- An, S.-Q.; Potnis, N.; Dow, M.; Vorhölter, F.-J.; He, Y.-Q.; Becker, A.; Teper, D.; Li, Y.; Wang, N.; Bleris, L.; et al. Mechanistic insights into host adaptation, virulence and epidemiology of the phytopathogen Xanthomonas. FEMS Microbiol. Rev. 2019, 44, 1–32. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D.; Ryan, D.D.; Ryan, P.D. Past: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electron 2001, 4, 9. [Google Scholar]
- Han, G.; Cooney, J.J. A modified capillary assay for chemotaxis. J. Ind. Microbiol. 1993, 12, 396–398. [Google Scholar] [CrossRef]
- Antúnez-Lamas, M.; Cabrera-Ordóñez, E.; López-Solanilla, E.; Raposo, R.; Trelles-Salazar, O.; Rodríguez-Moreno, A.; Rodríguez-Palenzuela, P. Role of motility and chemotaxis in the pathogenesis of Dickeya dadantii 3937 (ex Erwinia chrysanthemi 3937). Microbiology 2009, 155, 434–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rico, A.; Preston, G.M. Pseudomonas syringae pv. tomato DC3000 Uses Constitutive and Apoplast-Induced Nutrient Assimilation Pathways to Catabolize Nutrients That Are Abundant in the Tomato Apoplast. Mol. Plant Microbe Interact. 2008, 21, 269–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, T.-Y.; Kuo, T.-T. Bacterial leaf blight of rice plant. VI. Chemotactic responses of Xanthomonas oryzae to water droplets exudated from water pores on the leaf of rice plants. Bot. Bull. Acad. Sin. 1975, 16, 126–136. [Google Scholar]
- Palleroni, N.J. Chamber for Bacterial Chemotaxis Experiments. Appl. Environ. Microbiol. 1976, 32, 729–730. [Google Scholar] [CrossRef] [Green Version]
- Killiny, N. Metabolite signature of the phloem sap of fourteen citrus varieties with different degrees of tolerance to Candidatus Liberibacter asiaticus. Physiol. Mol. Plant Pathol. 2017, 97, 20–29. [Google Scholar] [CrossRef]
- Kamoun, S.; Kado, C.I. Phenotypic Switching Affecting Chemotaxis, Xanthan Production, and Virulence in Xanthomonas campestris. Appl. Environ. Microbiol. 1990, 56, 3855–3860. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Allen, C. Chemotaxis Is Required for Virulence and Competitive Fitness of the Bacterial Wilt Pathogen Ralstonia solanacearum. J. Bacteriol. 2006, 188, 3697–3708. [Google Scholar] [CrossRef] [Green Version]
- Merritt, P.M.; Danhorn, T.; Fuqua, C. Motility and Chemotaxis in Agrobacterium tumefaciens Surface Attachment and Biofilm Formation. J. Bacteriol. 2007, 189, 8005–8014. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, J.; Müsken, M.; Becker, T.; Magnowska, Z.; Bertinetti, D.; Möller, S.; Zimmermann, B.; Herberg, F.W.; Jänsch, L.; Häussler, S. The Pseudomonas aeruginosa Chemotaxis Methyltransferase CheR1 Impacts on Bacterial Surface Sampling. PLoS ONE 2011, 6, e18184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostakioti, M.; Hadjifrangiskou, M.; Hultgren, S.J. Bacterial Biofilms: Development, Dispersal, and Therapeutic Strategies in the Dawn of the Postantibiotic Era. Cold Spring Harb. Perspect. Med. 2013, 3, a010306. [Google Scholar] [CrossRef] [PubMed]
- Moreira, L.M.; Facincani, A.P.; Ferreira, C.B.; Ferreira, R.M.; Ferro, M.I.T.; Gozzo, F.C.; De Oliveira, J.C.F.; Ferro, J.A.; Soares, M.R. Chemotactic signal transduction and phosphate metabolism as adaptive strategies during citrus canker induction by Xanthomonas citri. Funct. Integr. Genom. 2015, 15, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Yang, X.; Ye, X.; Feng, J.; Cheng, T.; Zhou, X.; Liu, D.X.; Xu, L.; Wang, J. The Methyltransferase HemK Regulates the Virulence and Nutrient Utilization of the Phytopathogenic Bacterium Xanthomonas citri subsp. citri. Int. J. Mol. Sci. 2022, 23, 3931. [Google Scholar] [CrossRef] [PubMed]
- Rigano, L.A.; Siciliano, F.; Enrique, R.; Sendín, L.; Filippone, P.; Torres, P.S.; Qüesta, J.; Dow, J.M.; Castagnaro, A.P.; Vojnov, A.A.; et al. Biofilm Formation, Epiphytic Fitness, and Canker Development in Xanthomonas axonopodis pv. citri. Mol. Plant Microbe Interact. 2007, 20, 1222–1230. [Google Scholar] [CrossRef] [Green Version]
- Alexandre, G. Coupling metabolism and chemotaxis-dependent behaviours by energy taxis receptors. Microbiology 2010, 156, 2283–2293. [Google Scholar] [CrossRef] [Green Version]
- Alexandre, G.; Zhulin, I.B. More than one way to sense chemicals. J. Bacteriol. 2001, 183, 4681–4686. [Google Scholar] [CrossRef] [Green Version]
- Egbert, M.D.; Barandiaran, X.E.; Di Paolo, E.A. A minimal model of metabolism-based chemotaxis. PLoS Comput. Biol. 2010, 6, e1001004. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.-C.; Leu, Y.-W.; Chang-Chien, H.-C.; Hu, R.-M. Flagellar Biogenesis of Xanthomonas campestris Requires the Alternative Sigma Factors RpoN2 and FliA and Is Temporally Regulated by FlhA, FlhB, and FlgM. J. Bacteriol. 2009, 191, 2266–2275. [Google Scholar] [CrossRef]
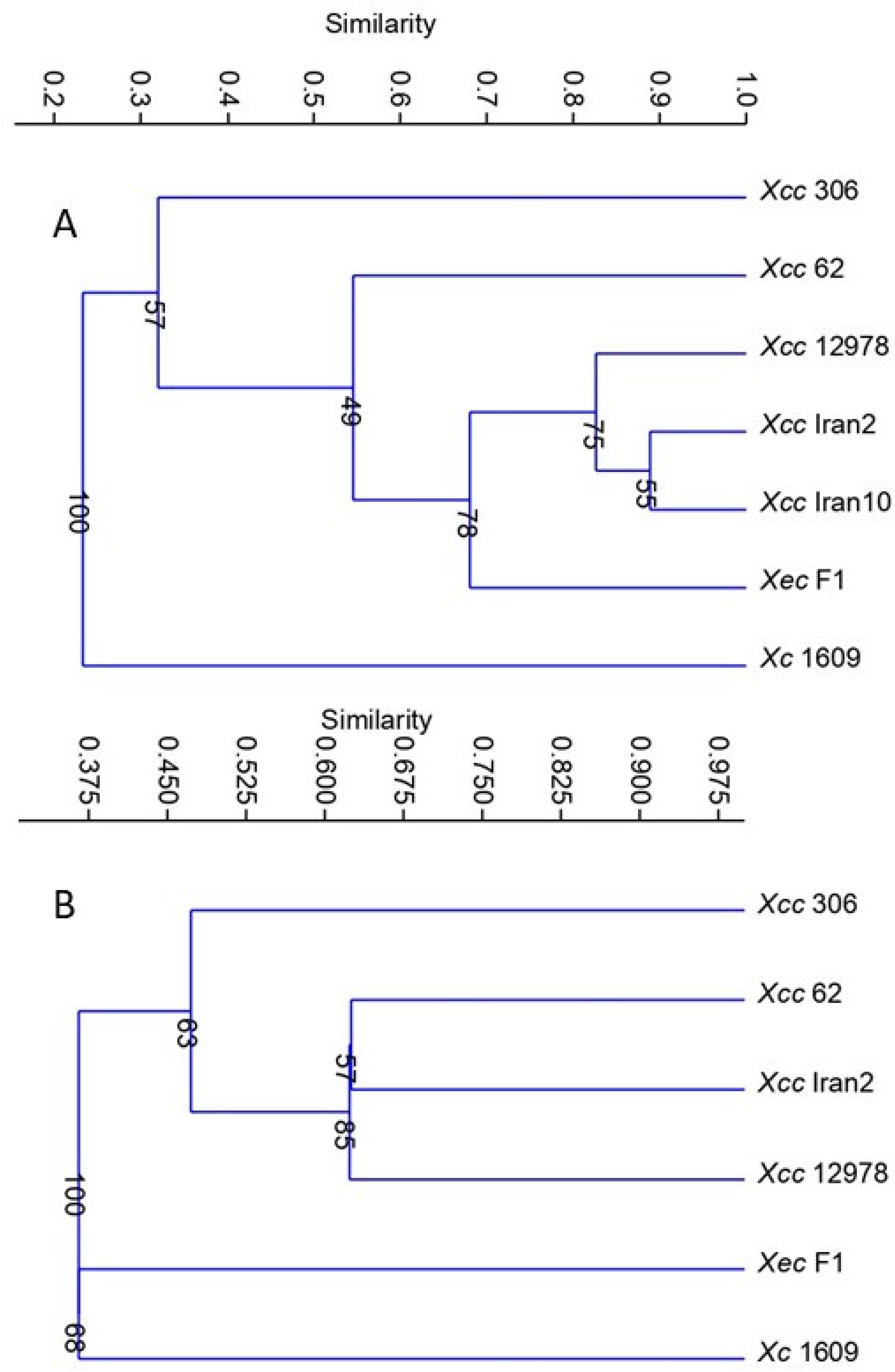
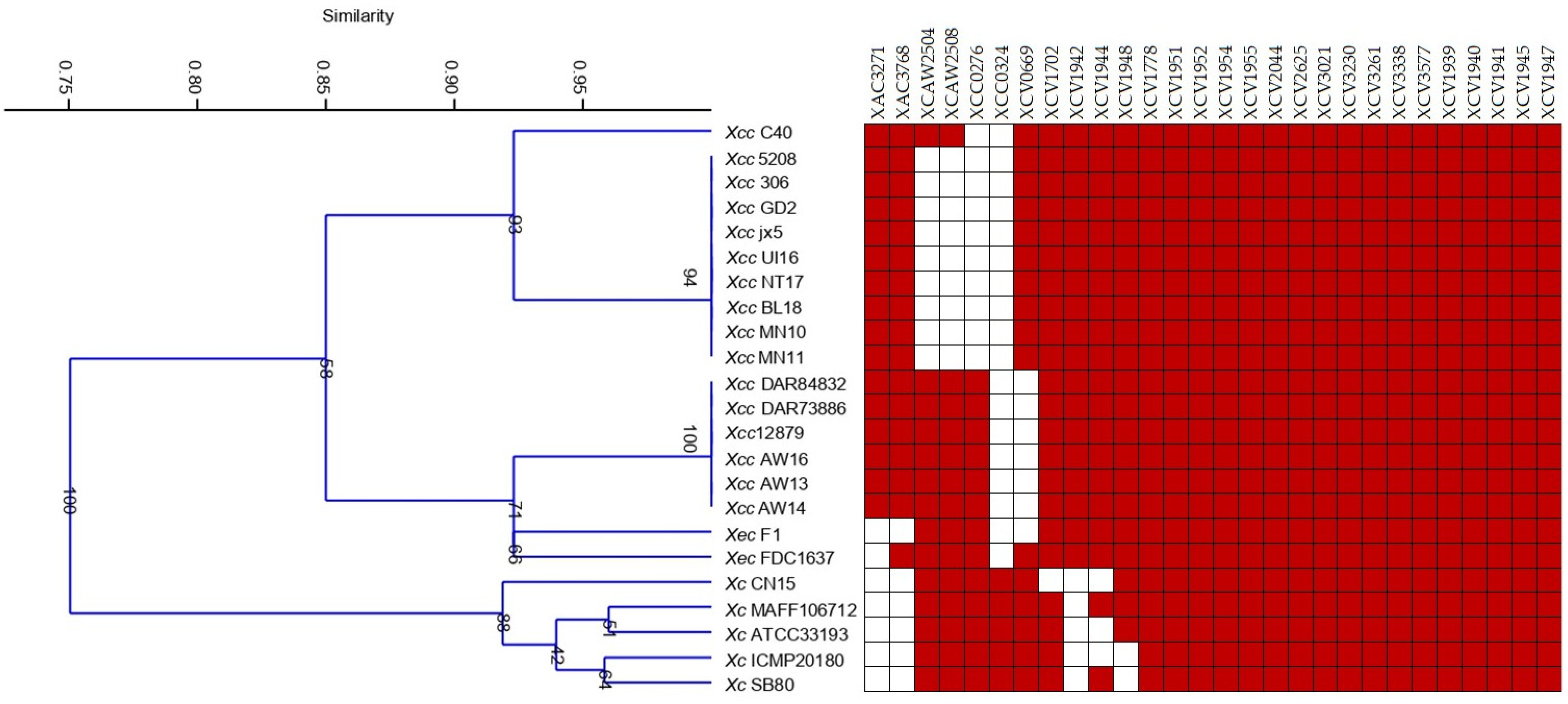
| Strain | Taxon, Disease and Disease Type | Natural Host |
|---|---|---|
| Xcc 306 | Xanthomonas citri pv. citri, CBC a A | Citrus sinensis |
| Xcc 62 | Xanthomonas citri pv. citri, CBC A | Citrus paradisi |
| Xcc Iran2 | Xanthomonas citri pv. citri, CBC A* | Citrus aurantifolia |
| Xcc Iran10 | Xanthomonas citri pv. citri, CBC A* | Citrus aurantifolia |
| Xcc 12879 | Xanthomonas citri pv. citri, CBC Aw | Citrus aurantifolia |
| Xec F1 | Xanthomonas euvesicatoria pv. citrumelonis, CBS b | Citrus spp. |
| Xc 1609 | Xanthomonas campestris pv. campestris, CBR c | Brassica spp. |
| Strain | Species/Pathovar | Type | Accession/Assembly |
|---|---|---|---|
| Xcc C40 | X. citri pv. citri | A | CCWX01 |
| Xcc 5208 | X. citri pv. citri | A | NZ_CP009028.1 |
| Xcc 306 | X. citri pv. citri | A | NC_003919.1 |
| Xcc gd2 | X. citri pv. citri | A | NZ_CP009019.1 |
| Xcc jx5 | X. citri pv. citri | A | NZ_CP009010.1 |
| Xcc UI6 | X. citri pv. citri | A | NZ_CP008990.1 |
| Xcc NT17 | X. citri pv. citri | A | NZ_CP008993.1 |
| Xcc BL18 | X. citri pv. citri | A | NZ_CP009023.1 |
| Xcc MN10 | X. citri pv. citri | A | NZ_CP009002.1 |
| Xcc MN11 | X. citri pv. citri | A | NZ_CP008999.1 |
| Xcc DAR73886 | X. citri pv. citri | A* | GCA_016801635.1 |
| Xcc DAR84832 | X. citri pv. citri | A* | GCA_016801615.1 |
| Xcc 12879 | X. citri pv. citri | Aw | NC_020815.1 |
| Xcc AW13 | X. citri pv. citri | Aw | NZ_CP009031.1 |
| Xcc AW14 | X. citri pv. citri | Aw | NZ_CP009034.1 |
| Xcc AW16 | X. citri pv. citri | Aw | NZ_CP009040.1 |
| Xec F1 | X. euvesicatoria pv. citrumelonis | NAb | GCA_000225915.1 |
| Xec FDC1637 a | X. euvesicatoria pv. citrumelonis | NA | GCA_005059795.1 |
| Xc CN15 | X. campestris pv. campestris | NA | GCA_000403575.2 |
| Xc MAFF302021 | X. campestris pv. campestris | NA | GCA_009177345.1 |
| Xc ATCC33193 | X. campestris pv. campestris | NA | GCA_000007145.1 |
| Xc ICMP20180 | X. campestris pv. campestris | NA | GCA_001186415.1 |
| Xc SB80 | X. campestris pv. campestris | NA | GCA_021459985.1 |
| Strains/Additive a | Xcc 306 | Xcc 62 | Xcc 12879 | Xcc Iran2 | Xcc Iran10 | Xec F1 | Xc 1609 |
|---|---|---|---|---|---|---|---|
| Dextrin | − | + | + | + | + | + | + |
| Glycogen | + a | + | NI | + | + | + | NI |
| Tween 80 | − b | − | − | − | − | − | + |
| L-Arabinose | − | − | − | − | NI | NI | − |
| D-Arabitol | − | − | − | − | − | NI | − |
| L-Fucose | NI c | + | + | + | + | + | + |
| α-D-Lactose | − | − | − | + | + | NI | − |
| Lactulose | NI | + | + | + | + | + | + |
| D-Melobiose | − | NI | − | + | + | + | + |
| D-Raffinose | − | − | − | NI | NI | NI | NI |
| Sucrose | + | + | + | + | + | NI | + |
| Turanose | − | − | − | + | + | NI | NI |
| Succinic Acid Mono-Methyl-Ester | NI | + | + | + | + | + | + |
| Cis-Aconitic Acid | + | + | + | + | + | + | − |
| D-Gluconic Acid | − | − | − | − | NI | − | − |
| α-Hidroxybutyric Acid | − | − | NI | + | + | + | + |
| β-Hidroxybutyric Acid | − | − | − | − | NI | NI | NI |
| α-Keto Butyric Acid | − | + | + | + | + | + | + |
| D,L-Lactic Acid | − | − | NI | + | + | + | + |
| Malonic Acid | NI | NI | + | + | + | + | + |
| Propionic Acid | − | − | + | + | + | − | + |
| D-Saccharic Acid | − | − | − | − | − | − | + |
| Succinamic Acid | NI | + | + | + | + | + | NI |
| L-Alaninamide | + | + | + | + | + | + | NI |
| D-Alanine | − | − | + | + | + | + | − |
| L-Alanine | NI | − | + | + | + | + | NI |
| L-Alanyl-Glicine | + | + | + | + | + | + | − |
| L-Asparagine | − | − | NI | − | NI | − | − |
| L-Aspartic Acid | − | − | NI | + | + | − | NI |
| Glycyl-L-Aspartic Acid | − | − | + | + | + | − | − |
| Glycyl-L-Glutamic Acid | + | + | + | + | + | + | NI |
| Hydroxy-L-Proline | − | − | − | − | NI | − | NI |
| Urocanic Acid | − | − | − | − | NI | − | − |
| Uridine | − | − | − | − | + | − | + |
| D,L-α-Glycerol Phosphate | − | − | NI | + | + | + | − |
| α-D-Glucose-1-Phosphate | − | NI | − | + | + | NI | − |
| D-Glucose-6-Phosphate | − | − | − | + | + | + | − |
| Additive | Xcc 306 | Xcc 62 | Xcc Iran2 A* | Xcc 12879 Aw | Xec F1 | Xc 1609 |
|---|---|---|---|---|---|---|
| Sodium Citrate 10 mM | + a | 0 b | + | + | 0 | 0 |
| Fructose 10 mM | 0 | + | 0 | + | − b | 0 |
| Galactose 10 mM | 0 | + | 0 | + | 0 | + |
| Glucose 10 mM | 0 | 0 | 0 | + | − | 0 |
| Maltose 10 mM | 0 | + | + | + | 0 | + |
| Sucrose 10 mM | + | + | + | + | + | + |
| Xylose 10 mM | 0 | 0 | 0 | 0 | − | − |
| Arginine 10 mM | 0 | + | 0 | 0 | 0 | 0 |
| Arginine 100 mM | 0 | + | + | + | + | + |
| Alanine 10 mM | 0 | + | + | 0 | 0 | − |
| Alanine 250 mM | + | + | + | + | 0 | + |
| Cysteine 10 mM | − c | − | − | − | − | − |
| Leucine 10 mM | + | 0 | 0 | 0 | 0 | − |
| Leucine 150 mM | + | + | + | + | − | 0 |
| Serine 10 mM | 0 | 0 | 0 | 0 | − | − |
| Serine 200 mM | + | + | + | + | + | + |
| Glycerol 0.2% | + | + | + | + | + | + |
| Mannitol 0.2% | + | + | + | + | − | 0 |
| Galacturonic Acid 10 mM | + | + | 0 | 0 | + | 0 |
| Glucuronic Acid 10 mM | 0 | 0 | 0 | 0 | 0 | + |
| Citric Acid 10 mM | − | 0 | 0 | − | 0 | 0 |
| Succinic Acid 10 mM | 0 | + | + | 0 | + | 0 |
| Cumaric Acid 10 mM | 0 | + | 0 | + | + | 0 |
| Strains/Primers | XAC3271 | XAC3768 | XCCAW2504 | XCCAW2508 | XCV1702 | XCV1778 | XCV1942 | XCV1944 | XCV1947 | XCV1951 | XCC0324 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CBC a A type | Xcc 306 | |||||||||||
| Xcc 62 | ||||||||||||
| CBC Aw type | Xcc 12879 | |||||||||||
| CBC A* type | Xcc Iran2 | |||||||||||
| Xcc Iran10 | ||||||||||||
| CBS b | Xec F1 | |||||||||||
| CBR c | Xc 1609 | |||||||||||
| Strain/Concentration (mg mL−1) | 3.12 | 6.25 | 12.5 | 25 | 50 | 100 | 200 | |
|---|---|---|---|---|---|---|---|---|
| Xcc 306 A | Sweet orange | + a | + | + | + | + | + | + |
| Xcc 62 A | + | + | + | + | + | + | + | |
| Xcc 12879 Aw | + | 0 | + | + | + | + | + | |
| Xcc Iran2 A* | + | + | + | + | + | + | + | |
| Xec F1 | + | + | + | + | + | + | + | |
| Xc 1609 | 0 b | + | + | + | + | + | + | |
| Xcc 306 A | Mexican lime | 0 | 0 | 0 | 0 | 0 | 0 | + |
| Xcc 62 A | + | + | + | + | + | + | + | |
| Xcc 12879 Aw | 0 | + | + | + | + | + | + | |
| Xcc Iran2 A* | + | + | + | + | + | + | + | |
| Xec F1 | 0 | 0 | 0 | + | + | + | + | |
| Xc 1609 | + | + | + | + | + | + | + | |
| Xcc 306 A | Chinese cabbage | 0 | + | + | + | + | + | + |
| Xcc 62 A | + | + | + | + | + | + | + | |
| Xcc 12879 Aw | + | + | + | + | + | + | + | |
| Xcc Iran2 A* | 0 | + | + | + | + | + | + | |
| Xec F1 | 0 | 0 | 0 | + | + | + | + | |
| Xc 1609 | 0 | 0 | + | + | + | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sena-Vélez, M.; Ferragud, E.; Redondo, C.; Graham, J.H.; Cubero, J. Chemotactic Responses of Xanthomonas with Different Host Ranges. Microorganisms 2023, 11, 43. https://doi.org/10.3390/microorganisms11010043
Sena-Vélez M, Ferragud E, Redondo C, Graham JH, Cubero J. Chemotactic Responses of Xanthomonas with Different Host Ranges. Microorganisms. 2023; 11(1):43. https://doi.org/10.3390/microorganisms11010043
Chicago/Turabian StyleSena-Vélez, Marta, Elisa Ferragud, Cristina Redondo, James H. Graham, and Jaime Cubero. 2023. "Chemotactic Responses of Xanthomonas with Different Host Ranges" Microorganisms 11, no. 1: 43. https://doi.org/10.3390/microorganisms11010043
APA StyleSena-Vélez, M., Ferragud, E., Redondo, C., Graham, J. H., & Cubero, J. (2023). Chemotactic Responses of Xanthomonas with Different Host Ranges. Microorganisms, 11(1), 43. https://doi.org/10.3390/microorganisms11010043






