Variability in Cold Tolerance of Food and Clinical Listeria monocytogenes Isolates
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Condition
2.2. Growth Rate Determination by Optical Density (OD) Measurement
2.3. Estimation of Growth Rates and Fast or Slow Growth Clustering
2.4. Genotyping and Pan-Genome Analysis
2.5. Measurement of Microbial Growth by Plate Counts in Laboratory Media and Food Matrices
2.6. Data Analysis
3. Results
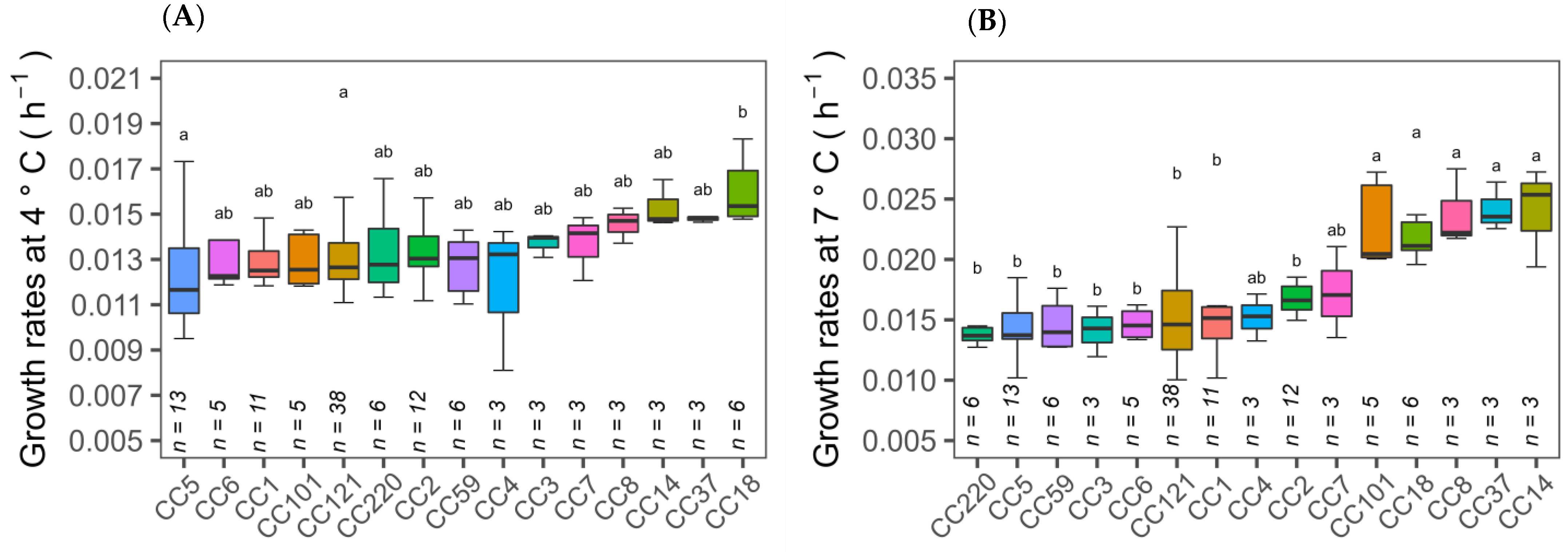
3.1. Cold Tolerance of Isolates
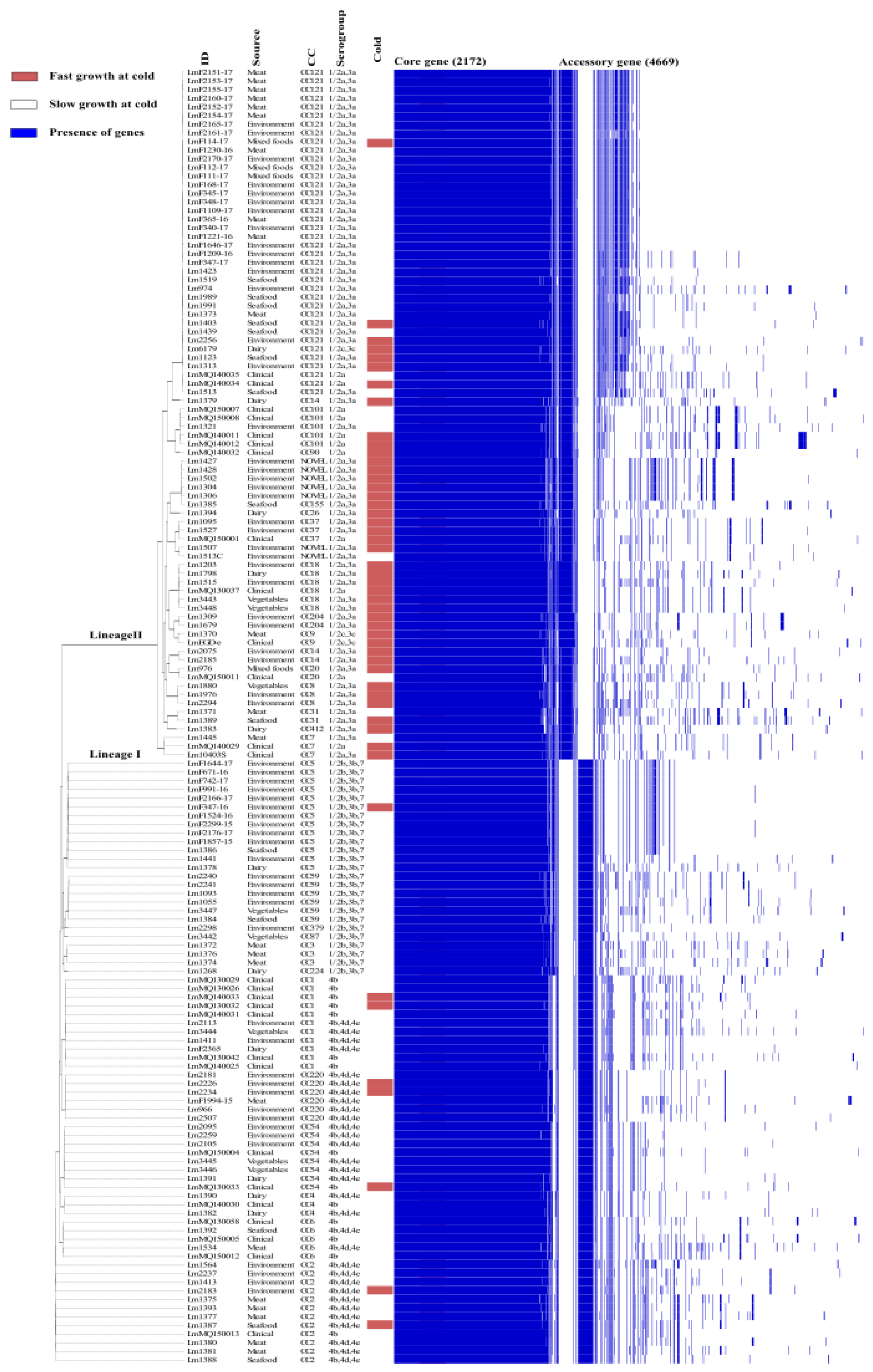
3.2. Pan-Genome Analysis
3.3. Association of Cold Growth Phenotypes with Isolate Subtypes and Stress Genes
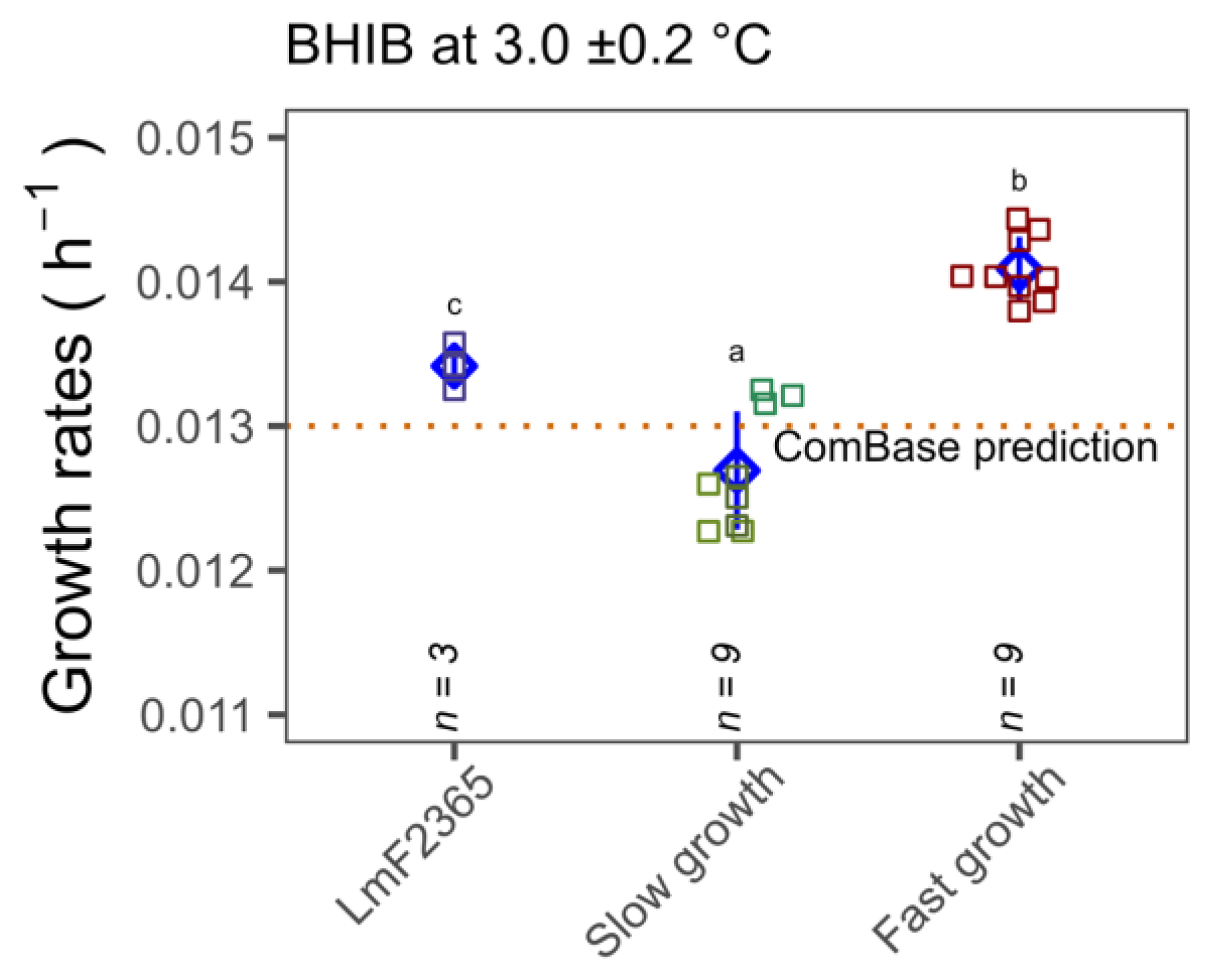
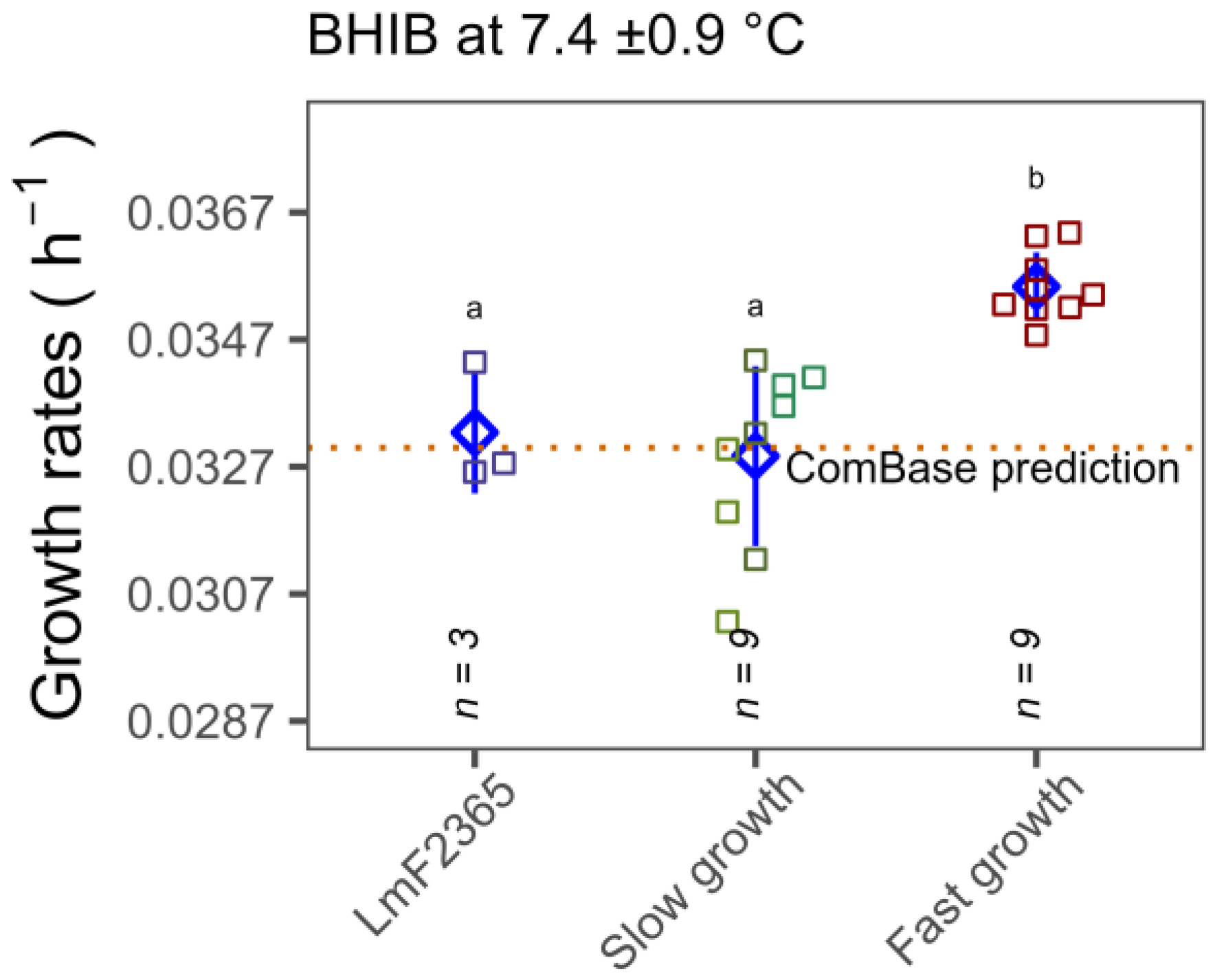
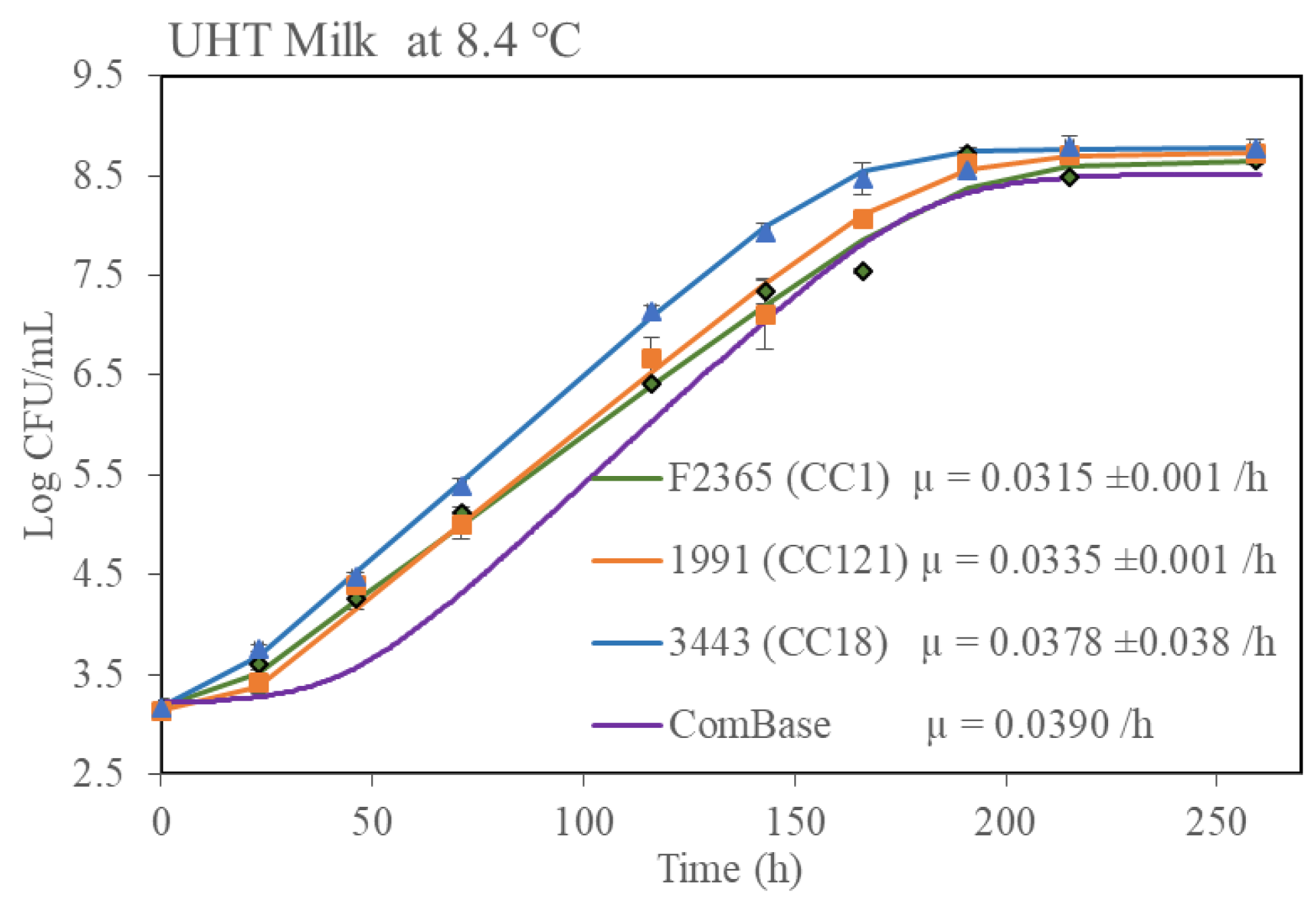
3.4. Growth Rates Determined by Standard Plate Count in Media and Food Matrices
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, e06971. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Table 1. Annual Reported Cases of Notifiable Diseases and Rates per 100,000, United States, Excluding U.S. Territories and Non-U.S. Residents. 2019. Available online: https://wonder.cdc.gov/nndss/static/2019/annual/2019-table1.html (accessed on 3 June 2022).
- Wiedmann, M. Subtyping of Bacterial Foodborne Pathogens. Nutr. Rev. 2002, 60, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; Le Monnier, A.; Brisse, S. A New Perspective on Listeria monocytogenes Evolution. PLoS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef] [PubMed]
- Wareth, G.; Linde, J.; Hammer, P.; Splettstoesser, W.; Pletz, M.; Neubauer, H.; Sprague, L. Molecular Characterization of German Acinetobacter baumannii Isolates and Multilocus Sequence Typing (MLST) Analysis Based on WGS Reveals Novel STs. Pathogens 2021, 10, 690. [Google Scholar] [CrossRef] [PubMed]
- Myintzaw, P.; Pennone, V.; McAuliffe, O.; Begley, M.; Callanan, M. Correlation of organic acid tolerance and genotypic characteristics of Listeria monocytogenes food and clinical isolates. Food Microbiol. 2022, 104, 104004. [Google Scholar] [CrossRef]
- Hingston, P.; Chen, J.; Dhillon, B.K.; Laing, C.; Bertelli, C.; Gannon, V.; Tasara, T.; Allen, K.; Brinkman, F.S.L.; Hansen, L.T.; et al. Genotypes Associated with Listeria monocytogenes Isolates Displaying Impaired or Enhanced Tolerances to Cold, Salt, Acid, or Desiccation Stress. Front. Microbiol. 2017, 8, 369. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Brynildsrud, O.; Bohlin, J.; Scheffer, L.; Eldholm, V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 2016, 17, 238. [Google Scholar] [CrossRef]
- European Commission (EC). Commission Regulation (EC) No 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs. Off. J. Eur. Union 2005, 50, 1–26. [Google Scholar]
- Codex Alimentarius Commission. Guidelines on the Application of General Principles of Food Hygiene to the Control of Listeria Monocytogenes in Ready-to-Eat Foods. Available online: http://www.fao.org/fao-who%02codexalimentarius/ (accessed on 3 June 2022).
- Laguerre, O.; Derens, E.; Palagos, B. Study of domestic refrigerator temperature and analysis of factors affecting temperature: A French survey Etude sur les réfrigérateurs domestiques et analyse des facteurs influençant la température: Enquête française. Int. J. Refrig. 2002, 25, 653–659. [Google Scholar] [CrossRef]
- Walker, S.J.; Archer, P.W.; Banks, J.G. Growth of Listeria monocytogenes at Refrigeration. J. Appl. Bacteriol. 1990, 68, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Tasara, T.; Stephan, R. Cold Stress Tolerance of Listeria monocytogenes: A Review of Molecular Adaptive Mechanisms and Food Safety Implications. J. Food Prot. 2006, 69, 1473–1484. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, L.; Felten, A.; Palma, F.; Mariet, J.-F.; Radomski, N.; Mistou, M.-Y.; Augustin, J.-C.; Guillier, L. Insights from genome-wide approaches to identify variants associated to phenotypes at pan-genome scale: Application to L. monocytogenes’ ability to grow in cold conditions. Int. J. Food Microbiol. 2019, 291, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Bergis, H.; Bonanno, L.; Asséré, A.; Lombard, B. EURL Lm Technical Guidance Document on Challenge Tests and Durability Studies for Assessing Shelf-Life of Ready-to-Eat Foods Related to Listeria monocytogenes. EURL Lm 2021, 4, 60. [Google Scholar]
- Sprouffske, K.; Wagner, A. Growthcurver: An R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinform. 2016, 17, 172. [Google Scholar] [CrossRef] [PubMed]
- Yeh, H.-Y.; Line, J.E.; Hinton, A. Community-Level Physiological Profiling for Microbial Community Function in Broiler Ceca. Curr. Microbiol. 2019, 76, 173–177. [Google Scholar] [CrossRef]
- Cuccuru, G.; Orsini, M.; Pinna, A.; Sbardellati, A.; Soranzo, N.; Travaglione, A.; Uva, P.; Zanetti, G.; Fotia, G. Orione, a web-based framework for NGS analysis in microbiology. Bioinformatics 2014, 30, 1928–1929. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Hadfield, J.; Croucher, N.J.; Goater, R.J.; AbuDahab, K.; Aanensen, D.M.; Harris, S.R. Phandango: An interactive viewer for bacterial population genomics. Bioinformatics 2018, 34, 292–293. [Google Scholar] [CrossRef]
- Gaudy, A.F.; Abu-Niaaj, F.; Gaudy, E.T. Statistical Study of the Spot-Plate Technique for Viable-Cell Counts. Appl. Microbiol. 1963, 11, 305–309. [Google Scholar] [CrossRef]
- Bergholz, T.M.; Shah, M.K.; Burall, L.S.; Rakic-Martinez, M.; Datta, A.R. Genomic and phenotypic diversity of Listeria monocytogenes clonal complexes associated with human listeriosis. Appl. Microbiol. Biotechnol. 2018, 102, 3475–3485. [Google Scholar] [CrossRef] [PubMed]
- Bland, R.N.; Johnson, J.D.; Waite-Cusic, J.G.; Weisberg, A.J.; Riutta, E.R.; Chang, J.H.; Kovacevic, J. Application of Whole Genome Sequencing to Understand Diversity and Presence of Genes Associated with Sanitizer Tolerance in Listeria monocytogenes from Produce Handling Sources. Foods 2021, 10, 2454. [Google Scholar] [CrossRef] [PubMed]
- Kuenne, C.; Billion, A.; Abu Mraheil, M.; Strittmatter, A.; Daniel, R.; Goesmann, A.; Barbuddhe, S.; Hain, T.; Chakraborty, T. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genom. 2013, 14, 47. [Google Scholar] [CrossRef] [PubMed]
- Pennone, V.; Gonzales-Barron, U.; Hunt, K.; Cadavez, V.; McAuliffe, O.; Butler, F. Omnibus Modeling of Listeria monocytogenes Growth Rates at Low Temperatures. Foods 2021, 10, 1099. [Google Scholar] [CrossRef] [PubMed]
- Verheyen, D.; Van Impe, J.F.M. The Inclusion of the Food Microstructural Influence in Predictive Microbiology: State-of-the-Art. Foods 2021, 10, 2119. [Google Scholar] [CrossRef]
- Muchaamba, F.; Stephan, R.; Tasara, T. β-Phenylethylamine as a Natural Food Additive Shows Antimicrobial Activity against Listeria monocytogenes on Ready-to-Eat Foods. Foods 2020, 9, 1363. [Google Scholar] [CrossRef] [PubMed]
- Tasara, T.; Klumpp, J.; Bille, J.; Stephan, R. Genome Sequences of Listeria monocytogenes Strains Responsible for Cheese- and Cooked Ham Product-Associated Swiss Listeriosis Outbreaks in 2005 and 2011. Genome Announc. 2016, 4, e00106-16. [Google Scholar] [CrossRef] [PubMed]
- Lobb, B.; Tremblay, B.J.-M.; Moreno-Hagelsieb, G.; Doxey, A.C. An assessment of genome annotation coverage across the bacterial tree of life. Microb. Genom. 2020, 6, e000341. [Google Scholar] [CrossRef]
- Nyhan, L.; Begley, M.; Mutel, A.; Qu, Y.; Johnson, N.; Callanan, M. Predicting the combinatorial effects of water activity, pH and organic acids on Listeria growth in media and complex food matrices. Food Microbiol. 2018, 74, 75–85. [Google Scholar] [CrossRef]
- Verheyen, D.; Xu, X.M.; Govaert, M.; Baka, M.; Skåra, T.; Van Impe, J.F. Food Microstructure and Fat Content Affect Growth Morphology, Growth Kinetics, and Preferred Phase for Cell Growth of Listeria monocytogenes in Fish-Based Model Systems. Appl. Environ. Microbiol. 2019, 85, e00707-19. [Google Scholar] [CrossRef]
- Borezee, E.; Pellegrini, E.; Berche, P. OppA of Listeria monocytogenes, an Oligopeptide-Binding Protein Required for Bacterial Growth at Low Temperature and Involved in Intracellular Survival. Infect. Immun. 2000, 68, 7069–7077. [Google Scholar] [CrossRef] [PubMed]
- Angelidis, A.S.; Smith, G.M. Role of the Glycine Betaine and Carnitine Transporters in Adaptation of Listeria monocytogenes to Chill Stress in Defined Medium. Appl. Environ. Microbiol. 2003, 69, 7492–7498. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Graham, J.E.; Bigelow, L.; Morse, P.D.; Wilkinson, B.J. Identification of Listeria monocytogenes Genes Expressed in Response to Growth at Low Temperature. Appl. Environ. Microbiol. 2002, 68, 1697–1705. [Google Scholar] [CrossRef] [PubMed]
- Brøndsted, L.; Kallipolitis, B.H.; Ingmer, H.; Knöchel, S. kdpE and a putative RsbQ homologue contribute to growth of Listeria monocytogenes at high osmolarity and low temperature. FEMS Microbiol. Lett. 2003, 219, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Becker, L.A.; Evans, S.N.; Hutkins, R.W.; Benson, A.K. Role of ςB in Adaptation of Listeria monocytogenes to Growth at Low Temperature. J. Bacteriol. 2000, 182, 7083–7087. [Google Scholar] [CrossRef]
- Christiansen, J.K.; Larsen, M.H.; Ingmer, H.; Søgaard-Andersen, L.; Kallipolitis, B.H. The RNA-Binding Protein Hfq of Listeria monocytogenes: Role in Stress Tolerance and Virulence. J. Bacteriol. 2004, 186, 3355–3362. [Google Scholar] [CrossRef]
- Knudsen, G.M.; Olsen, J.E.; Dons, L. Characterization of DegU, a response regulator in Listeria monocytogenes, involved in regulation of motility and contributes to virulence. FEMS Microbiol. Lett. 2004, 240, 171–179. [Google Scholar] [CrossRef]
- Schmid, B.; Klumpp, J.; Raimann, E.; Loessner, M.J.; Stephan, R.; Tasara, T. Role of Cold Shock Proteins in Growth of Listeria monocytogenes under Cold and Osmotic Stress Conditions. Appl. Environ. Microbiol. 2009, 75, 1621–1627. [Google Scholar] [CrossRef]
- Zheng, W.; Kathariou, S. Differentiation of epidemic-associated strains of Listeria monocytogenes by restriction fragment length polymorphism in a gene region essential for growth at low temperatures (4 °C). Appl. Environ. Microbiol. 1995, 61, 4310–4314. [Google Scholar] [CrossRef]
- Markkula, A.; Mattila, M.; Lindström, M.; Korkeala, H. Genes encoding putative DEAD-box RNA helicases in Listeria monocytogenes EGD-e are needed for growth and motility at 3 °C. Environ. Microbiol. 2012, 14, 2223–2232. [Google Scholar] [CrossRef]
- Arguedas-Villa, C.; Stephan, R.; Tasara, T. Evaluation of cold growth and related gene transcription responses associated with Listeria monocytogenes strains of different origins. Food Microbiol. 2010, 27, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Pöntinen, A.; Markkula, A.; Lindström, M.; Korkeala, H. Two-Component-System Histidine Kinases Involved in Growth of Listeria monocytogenes EGD-e at Low Temperatures. Appl. Environ. Microbiol. 2015, 81, 3994–4004. [Google Scholar] [CrossRef] [PubMed]
- Mattila, M.; Lindström, M.; Somervuo, P.; Markkula, A.; Korkeala, H. Role of flhA and motA in growth of Listeria monocytogenes at low temperatures. Int. J. Food Microbiol. 2011, 148, 177–183. [Google Scholar] [CrossRef] [PubMed]



| Product | pH | List of Ingredients |
|---|---|---|
| Milk (UHT) | 6.77 | UHT Milk 3.2% fat content |
| Chocolate milk (UHT) | 6.89 | Semi Skimmed Milk (96%), Sugar, Fat Reduced Cocoa Powder, Stabilisers (Guar Gum, Carrageenan), Flavourings, Vitamin D. |
| Fish pie | 6.06 | Organic Whole Milk 35%, Organic Carrots 19%, Organic Potatoes 19%, Organic Salmon (Fish) 10%, Organic Green Beans 7%, Organic Leeks 4%, Organic Onions 2%, Organic Breadcrumbs (Wheat Flour, Yeast) 2%, Organic Unsalted Butter (Milk) 1%, Organic Parsley 0.2%, Organic Peppercorns < 0.1% |
| Isolates | CC | Category |
|---|---|---|
| LmF2365 * | 1 | Reference Isolate |
| Lm1372 | 3 | Slow growth |
| Lm1378 | 5 | Slow growth |
| Lm1991 * | 121 | Slow growth |
| Lm1515 | 18 | Fast Growth |
| Lm3443 * | 18 | Fast Growth |
| Lm3448 | 18 | Fast Growth |
| Gene | Accession | Query (bp) | Identity (%) | Predicted Function |
|---|---|---|---|---|
| group_3291 | WP_003727783.1 | 591 | 100 | phage scaffolding protein |
| group_2594 | WP_003734899.1 | 825 | 100 | phage tail family protein |
| group_2595 | HAC4747847.1 | 405 | 98.51 | hypothetical protein |
| group_4413 | EAG7277673.1 | 1014 | 99.7 | phage tail protein |
| group_4415 | EAF7046744.1 | 444 | 99.32 | hypothetical protein |
| group_2328 | WP_003727788.1 | 408 | 99.26 | minor capsid protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Myintzaw, P.; Pennone, V.; McAuliffe, O.; Begley, M.; Callanan, M. Variability in Cold Tolerance of Food and Clinical Listeria monocytogenes Isolates. Microorganisms 2023, 11, 65. https://doi.org/10.3390/microorganisms11010065
Myintzaw P, Pennone V, McAuliffe O, Begley M, Callanan M. Variability in Cold Tolerance of Food and Clinical Listeria monocytogenes Isolates. Microorganisms. 2023; 11(1):65. https://doi.org/10.3390/microorganisms11010065
Chicago/Turabian StyleMyintzaw, Peter, Vincenzo Pennone, Olivia McAuliffe, Máire Begley, and Michael Callanan. 2023. "Variability in Cold Tolerance of Food and Clinical Listeria monocytogenes Isolates" Microorganisms 11, no. 1: 65. https://doi.org/10.3390/microorganisms11010065
APA StyleMyintzaw, P., Pennone, V., McAuliffe, O., Begley, M., & Callanan, M. (2023). Variability in Cold Tolerance of Food and Clinical Listeria monocytogenes Isolates. Microorganisms, 11(1), 65. https://doi.org/10.3390/microorganisms11010065










