Gut Microbiome and Lipidome Signatures in Irritable Bowel Syndrome Patients from a Low-Income, Food-Desert Area: A Pilot Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Sample Collection
2.2. 16S rRNA Gene Sequencing
2.3. Analysis of Lipids
2.4. Microbiome Data Processing and Statistical Analysis
2.5. Procrustes Randomization Test
2.6. Differential Abundance Testing
2.7. Ethics Approval
3. Results
3.1. Patient Demographics
3.2. Gut Microbiome Analysis
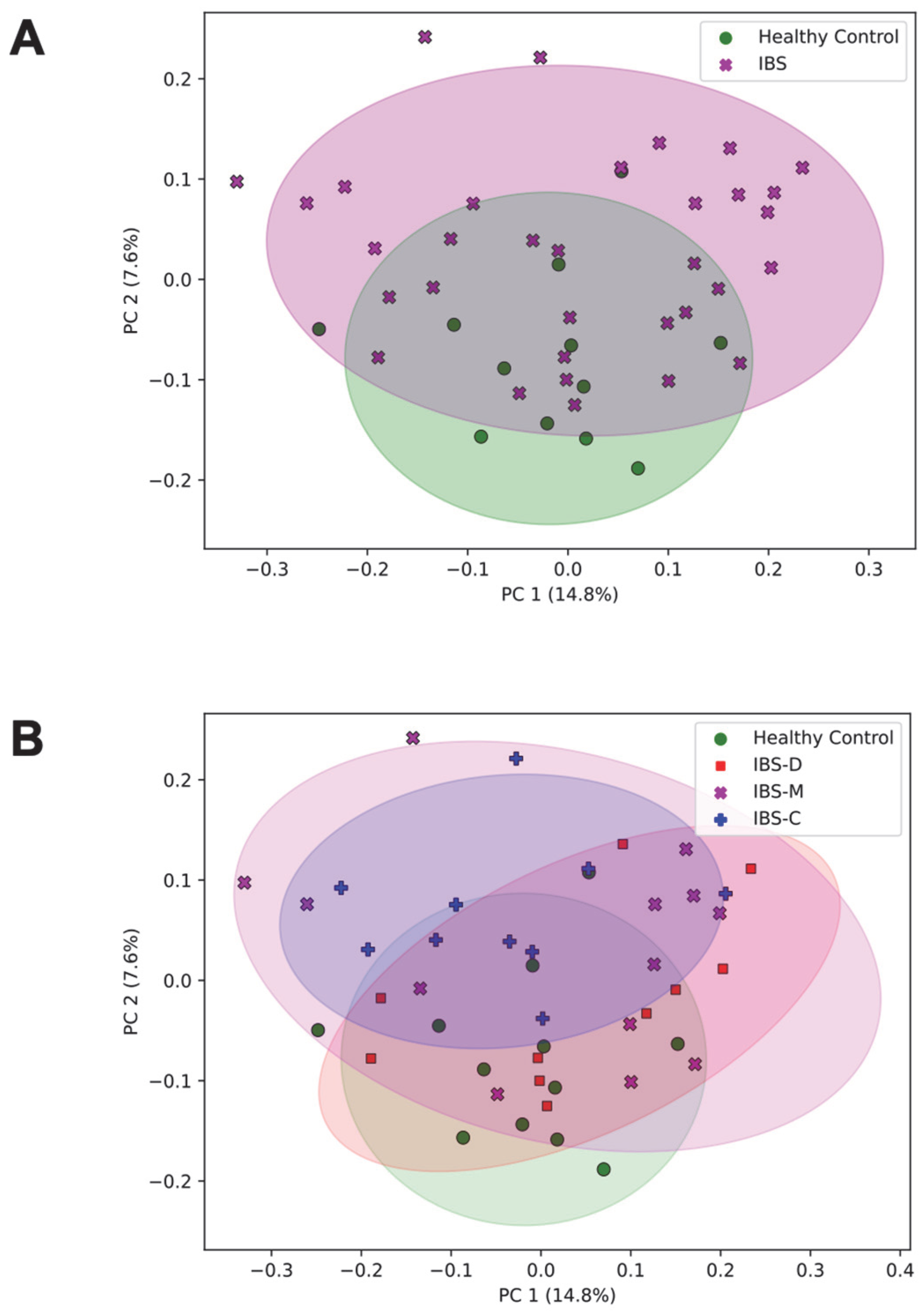
3.2.1. Association of the Community Composition of the Gut Microbiome with IBS Group
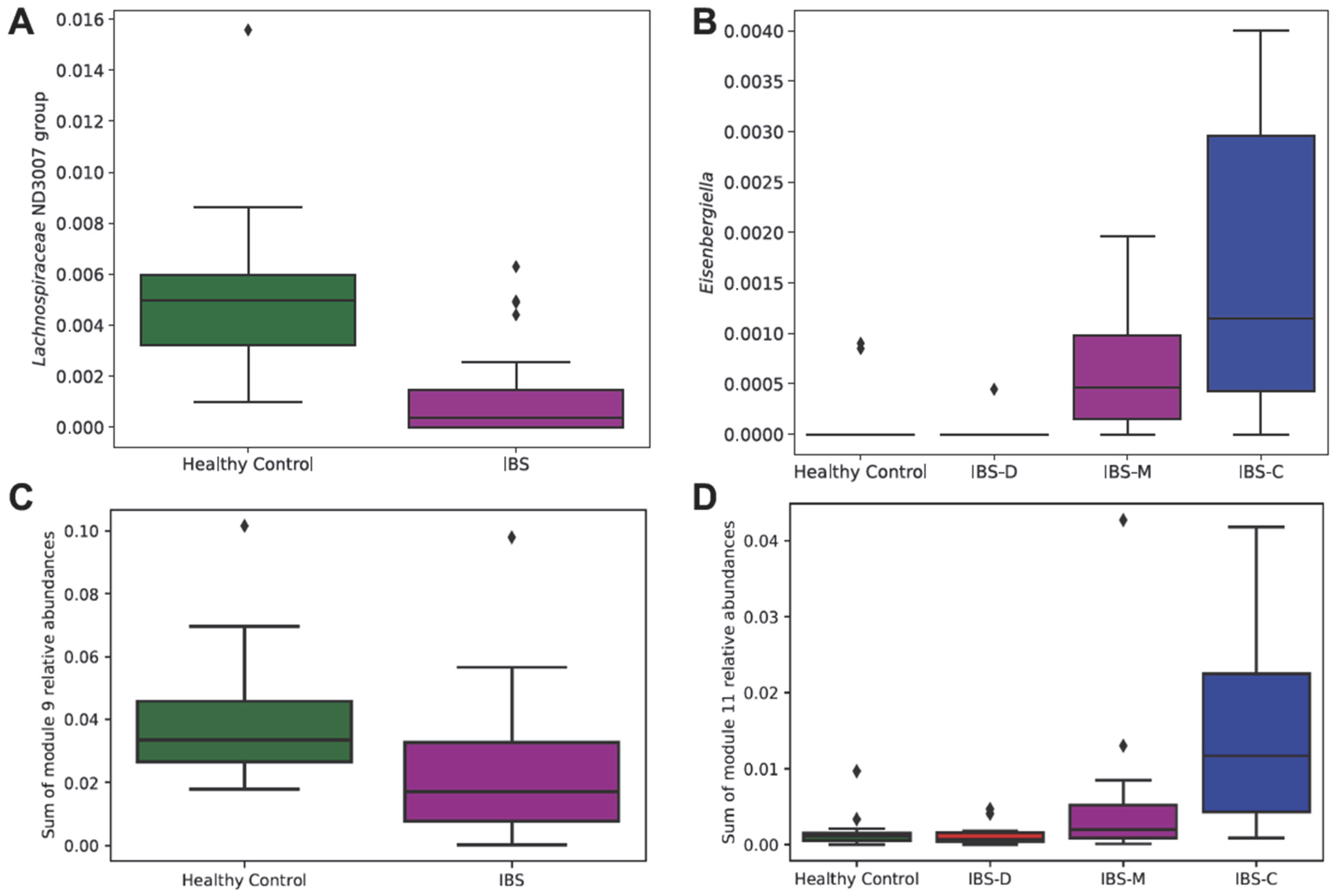
3.2.2. Association of Differentially Abundant Gut Microbiome Taxa with IBS Groups
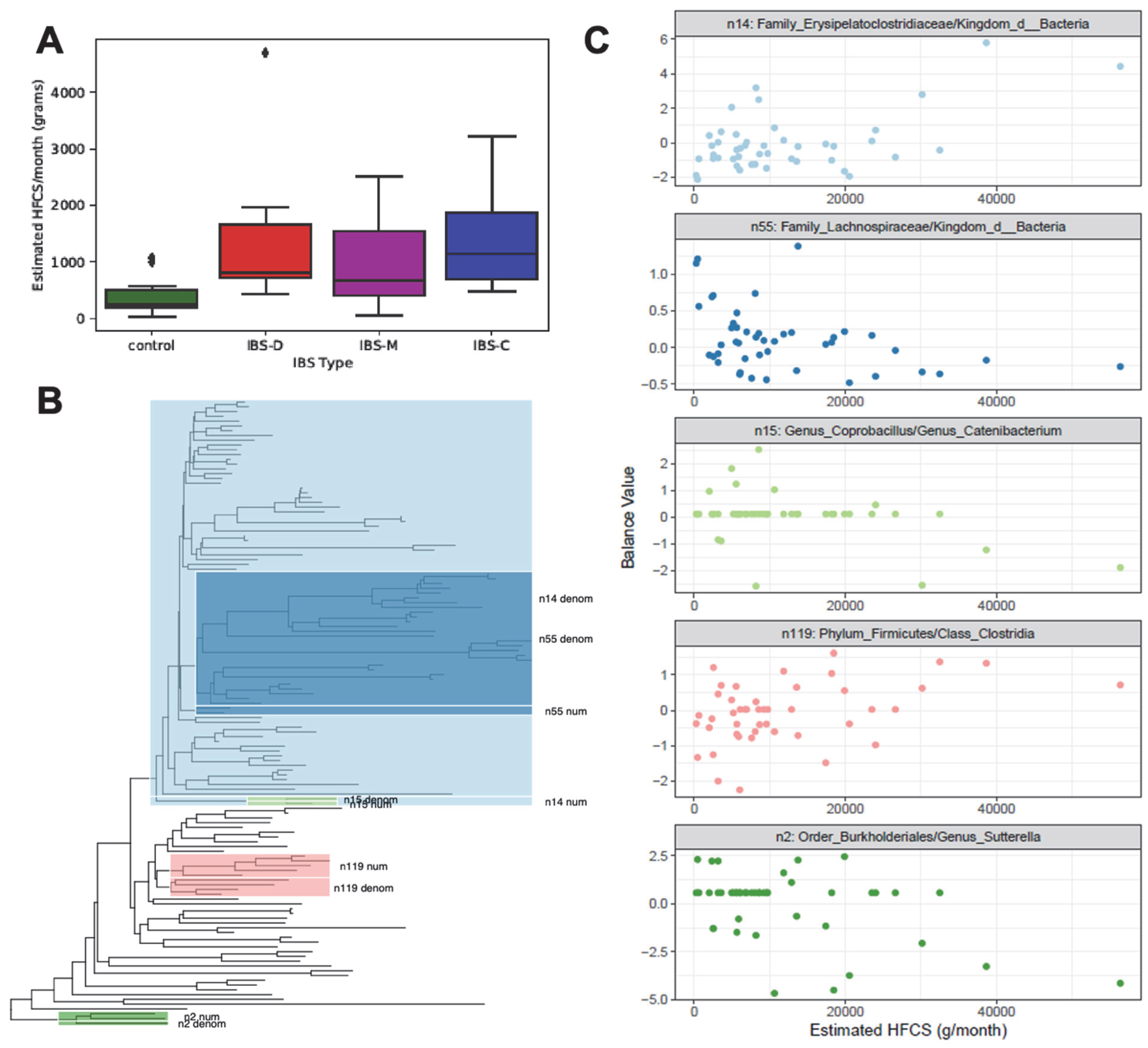
3.3. Monthly HFCS Consumption with Respect to IBS Groups
3.4. Phylogenetic Groups of Taxa in the Gut Microbiome with Respect to Monthly HFCS Consumption
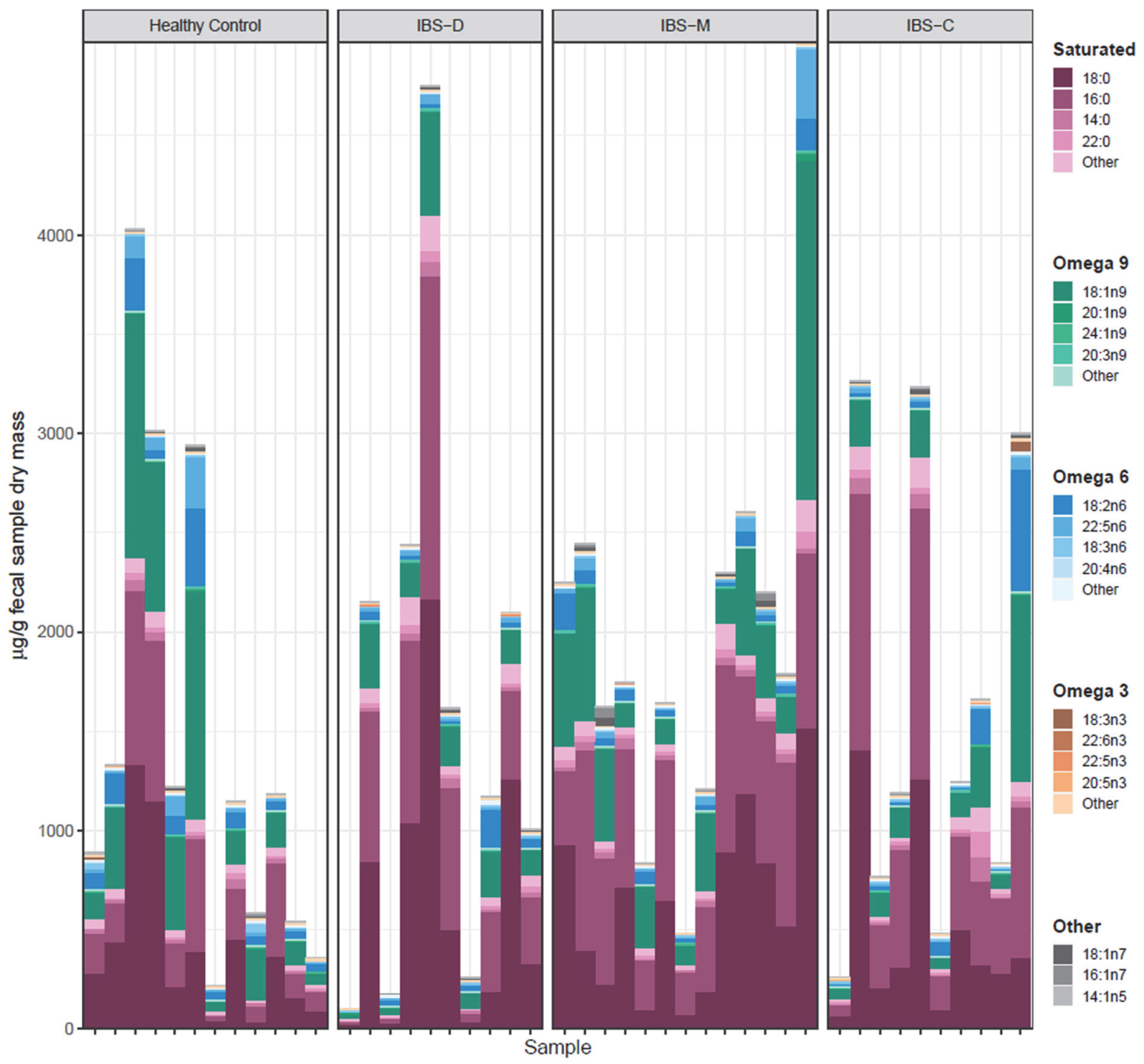
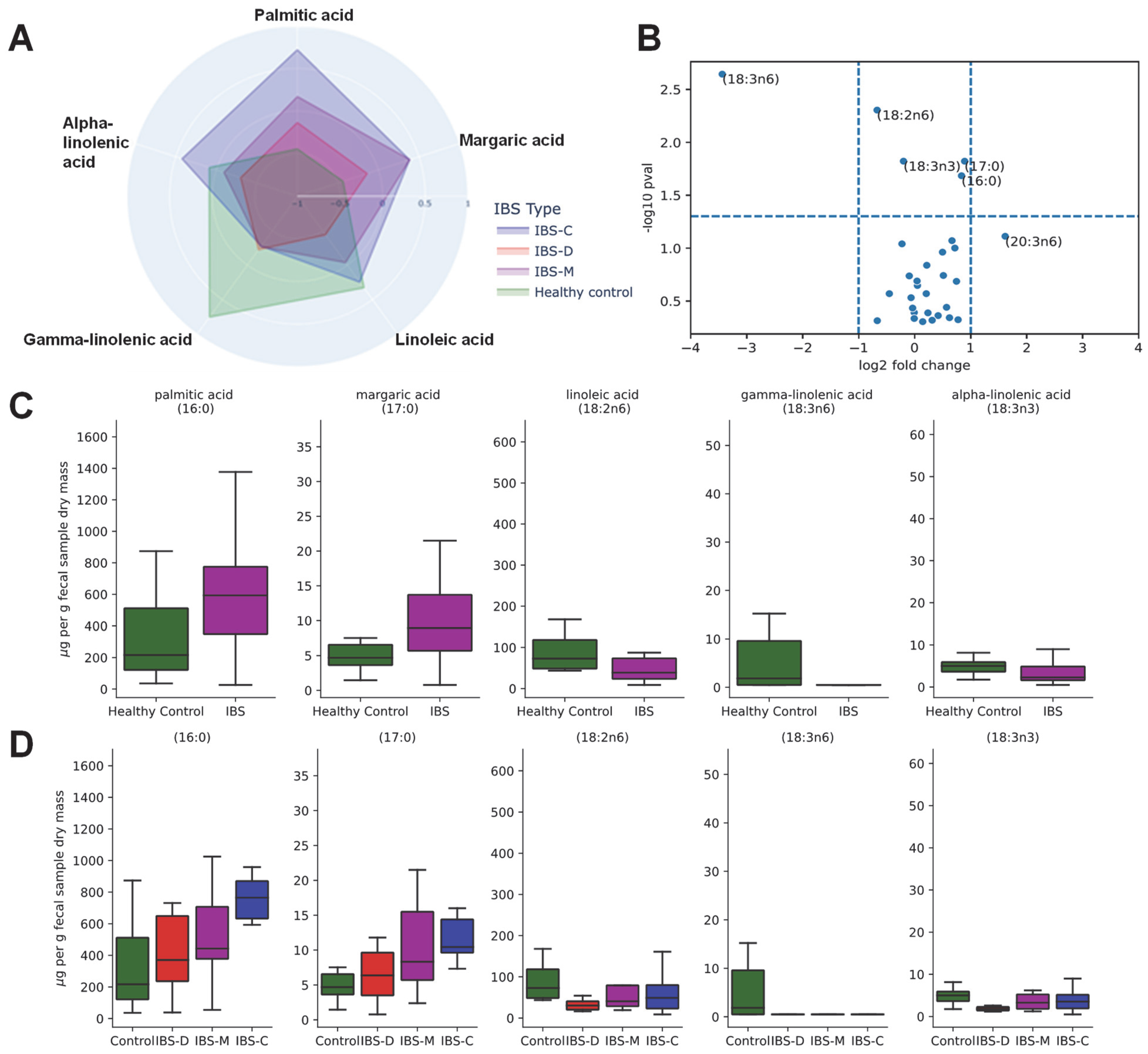
3.5. Association of Fecal Lipids with IBS Groups
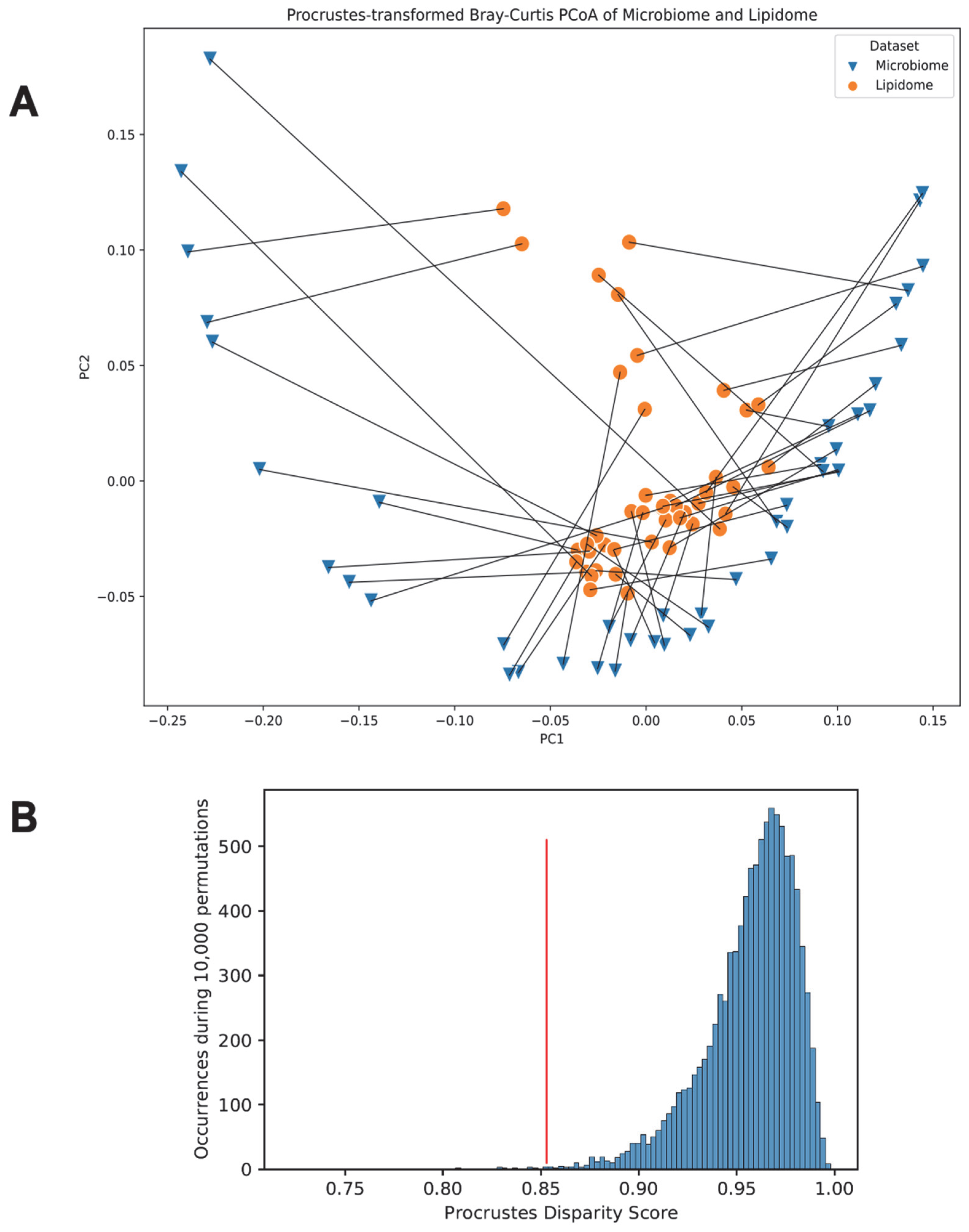
3.6. Link between the Gut Microbiome and the Fecal Lipidome Compositions via Procrustes Randomization Test
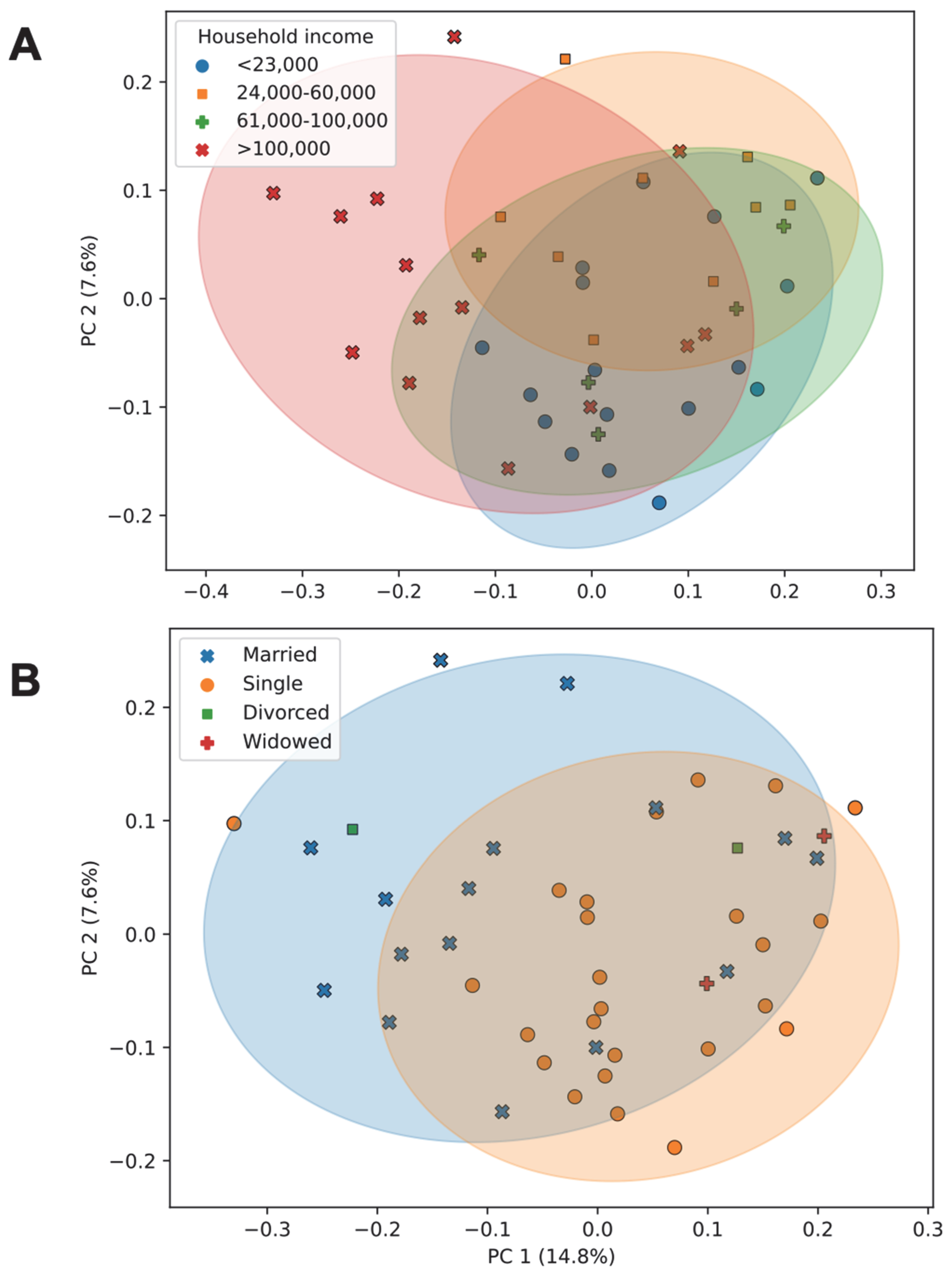
3.7. Association of Socioeconomic Factors with Diversity and Community Composition of the Gut Microbiome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef] [PubMed]
- Ford, A.C.; Lacy, B.E.; Talley, N.J. Irritable Bowel Syndrome. N. Engl. J. Med. 2017, 376, 2566–2578. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Ma, X.; Tian, W.; Zhang, J.; Wei, Y.; Zhang, B.; Wang, F.; Tang, X. Global Research Trends in Irritable Bowel Syndrome: A Bibliometric and Visualized Study. Front. Med. 2022, 9, 922063. [Google Scholar] [CrossRef]
- Maxion-Bergemann, S.; Thielecke, F.; Abel, F.; Bergemann, R. Costs of irritable bowel syndrome in the UK and US. PharmacoEconomics 2006, 24, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Drossman, D.A.; Hasler, W.L. Rome IV-Functional GI Disorders: Disorders of Gut-Brain Interaction. Gastroenterology 2016, 150, 1257–1261. [Google Scholar] [CrossRef]
- Camilleri, M.; Halawi, H.; Oduyebo, I. Biomarkers as a diagnostic tool for irritable bowel syndrome: Where are we? Expert Rev. Gastroenterol. Hepatol. 2017, 11, 303–316. [Google Scholar] [CrossRef]
- Oshima, T.; Siah, K.T.H.; Yoshimoto, T.; Miura, K.; Tomita, T.; Fukui, H.; Miwa, H. Impacts of the COVID-19 pandemic on functional dyspepsia and irritable bowel syndrome: A population-based survey. J. Gastroenterol. Hepatol. 2021, 36, 1820–1827. [Google Scholar] [CrossRef]
- Alcendor, D.J. Racial Disparities-Associated COVID-19 Mortality among Minority Populations in the US. J. Clin. Med. 2020, 9, 2442. [Google Scholar] [CrossRef]
- Ikechi, R.; Fischer, B.D.; DeSipio, J.; Phadtare, S. Irritable Bowel Syndrome: Clinical Manifestations, Dietary Influences, and Management. Healthcare 2017, 5, 21. [Google Scholar] [CrossRef]
- Drossman, D.A.; Camilleri, M.; Mayer, E.A.; Whitehead, W.E. AGA technical review on irritable bowel syndrome. Gastroenterology 2002, 123, 2108–2131. [Google Scholar] [CrossRef]
- DiNicolantonio, J.J.; Lucan, S.C. Is fructose malabsorption a cause of irritable bowel syndrome? Med. Hypotheses 2015, 85, 295–297. [Google Scholar] [CrossRef] [PubMed]
- Peery, A.F.; Dellon, E.S.; Lund, J.; Crockett, S.D.; McGowan, C.E.; Bulsiewicz, W.J.; Gangarosa, L.M.; Thiny, M.T.; Stizenberg, K.; Morgan, D.R.; et al. Burden of gastrointestinal disease in the United States: 2012 update. Gastroenterology 2012, 143, 1179–1187.e3. [Google Scholar] [CrossRef] [PubMed]
- Paripati, N.; Dugan, C.; Nesi, L.; Mone, A.; Patel, S.; Gaughan, J.; DeSipio, J.; Phadtare, S. Relationship between irritable bowel syndrome, psychological comorbidities and the consumption of high-fructose corn syrup in a low-income community in a food desert area. Clin. Nutr. Open Sci. 2022, 46, 1–12. [Google Scholar] [CrossRef]
- Unites States Sensus Data; Data USA: Camden City, N.J.Q.U.S.C.B. 2019. Available online: https://gistbok.ucgis.org/bok-topics/united-states-census-data (accessed on 1 January 2023).
- Chrisinger, B.W.; Ramos, A.; Shaykis, F.; Martinez, T.; Banchoff, A.W.; Winter, S.J.; King, A.C. Leveraging Citizen Science for Healthier Food Environments: A Pilot Study to Evaluate Corner Stores in Camden, New Jersey. Front. Public Health 2018, 6, 89. [Google Scholar] [CrossRef] [PubMed]
- Chong, P.P.; Chin, V.K.; Looi, C.Y.; Wong, W.F.; Madhavan, P.; Yong, V.C. The Microbiome and Irritable Bowel Syndrome—A Review on the Pathophysiology, Current Research and Future Therapy. Front. Microbiol. 2019, 10, 1136. [Google Scholar] [CrossRef] [PubMed]
- Tap, J.; Derrien, M.; Tornblom, H.; Brazeilles, R.; Cools-Portier, S.; Dore, J.; Storsrud, S.; Le Neve, B.; Ohman, L.; Simren, M. Identification of an Intestinal Microbiota Signature Associated With Severity of Irritable Bowel Syndrome. Gastroenterology 2017, 152, 111–123.e8. [Google Scholar] [CrossRef]
- Singh, P.; Lembo, A. Emerging Role of the Gut Microbiome in Irritable Bowel Syndrome. Gastroenterol. Clin. N. Am. 2021, 50, 523–545. [Google Scholar] [CrossRef]
- Kim, G.H.; Lee, K.; Shim, J.O. Gut Bacterial Dysbiosis in Irritable Bowel Syndrome: A Case-Control Study and a Cross-Cohort Analysis Using Publicly Available Data Sets. Microbiol. Spectr. 2023, 11, e0212522. [Google Scholar] [CrossRef]
- Pittayanon, R.; Lau, J.T.; Yuan, Y.; Leontiadis, G.I.; Tse, F.; Surette, M.; Moayyedi, P. Gut Microbiota in Patients With Irritable Bowel Syndrome-A Systematic Review. Gastroenterology 2019, 157, 97–108. [Google Scholar] [CrossRef]
- Wang, L.; Alammar, N.; Singh, R.; Nanavati, J.; Song, Y.; Chaudhary, R.; Mullin, G.E. Gut Microbial Dysbiosis in the Irritable Bowel Syndrome: A Systematic Review and Meta-Analysis of Case-Control Studies. J. Acad. Nutr. Diet. 2020, 120, 565–586. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Gu, X.; Li, L.X.; Li, M.; Li, B.; Cui, X.; Zuo, X.L. Microbial and metabolomic profiles in correlation with depression and anxiety co-morbidities in diarrhoea-predominant IBS patients. BMC Microbiol. 2020, 20, 168. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zhao, L.; Zhou, Y.; Yang, C.; Xiong, T.; Lu, L.; Deng, Y.; Luo, W.; Chen, Y.; Qiu, Q.; et al. Altered metabolome and microbiome features provide clues in understanding irritable bowel syndrome and depression comorbidity. ISME J. 2022, 16, 983–996. [Google Scholar] [CrossRef] [PubMed]
- Mujagic, Z.; Kasapi, M.; Jonkers, D.M.; Garcia-Perez, I.; Vork, L.; Weerts, Z.; Serrano-Contreras, J.I.; Zhernakova, A.; Kurilshikov, A.; Scotcher, J.; et al. Integrated fecal microbiome-metabolome signatures reflect stress and serotonin metabolism in irritable bowel syndrome. Gut Microbes 2022, 14, 2063016. [Google Scholar] [CrossRef] [PubMed]
- Vervier, K.; Moss, S.; Kumar, N.; Adoum, A.; Barne, M.; Browne, H.; Kaser, A.; Kiely, C.J.; Neville, B.A.; Powell, N.; et al. Two microbiota subtypes identified in irritable bowel syndrome with distinct responses to the low FODMAP diet. Gut 2022, 71, 1821–1830. [Google Scholar] [CrossRef]
- Vandeputte, D.; Joossens, M. Effects of Low and High FODMAP Diets on Human Gastrointestinal Microbiota Composition in Adults with Intestinal Diseases: A Systematic Review. Microorganisms 2020, 8, 1638. [Google Scholar] [CrossRef]
- van Lanen, A.S.; de Bree, A.; Greyling, A. Efficacy of a low-FODMAP diet in adult irritable bowel syndrome: A systematic review and meta-analysis. Eur. J. Nutr. 2021, 60, 3505–3522. [Google Scholar] [CrossRef]
- Schmulson, M.J.; Drossman, D.A. What Is New in Rome IV. J. Neurogastroenterol. Motil. 2017, 23, 151–163. [Google Scholar] [CrossRef]
- Mearin, F.; Lacy, B.E.; Chang, L.; Chey, W.D.; Lembo, A.J.; Simren, M.; Spiller, R. Bowel Disorders. Gastroenterology 2016, 150, 1393–1407.E5. [Google Scholar] [CrossRef]
- White, B.; Sterrett, J.D.; Grigoryan, Z.; Lally, L.; Heinze, J.D.; Alikhan, H.; Lowry, C.A.; Perez, L.J.; DeSipio, J.; Phadtare, S. Characterization of gut microbiome and metabolome in Helicobacter pylori patients in an underprivileged community in the United States. World J. Gastroenterol. 2021, 27, 5575–5594. [Google Scholar] [CrossRef]
- O’Neill, L.; Pandya, V.; Grigoryan, Z.; Patel, R.; DeSipio, J.; Judge, T.; Phadtare, S. Effects of Milkfat on the Gut Microbiome of Patients After Bariatric Surgery, a Pilot Study. Obes. Surg. 2022, 32, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Awan, I.; Schultz, E.; Sterrett, J.D.; Dawud, L.M.; Kessler, L.R.; Schoch, D.; Lowry, C.A.; Feldman-Winter, L.; Phadtare, S. A Pilot Study Exploring Temporal Development of Gut Microbiome/Metabolome in Breastfed Neonates during the First Week of Life. Pediatr. Gastroenterol. Hepatol. Nutr. 2023, 26, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Janssen, S.; McDonald, D.; Gonzalez, A.; Navas-Molina, J.A.; Jiang, L.; Xu, Z.Z.; Winker, K.; Kado, D.M.; Orwoll, E.; Manary, M.; et al. Phylogenetic Placement of Exact Amplicon Sequences Improves Associations with Clinical Information. mSystems 2018, 3, e00021-18. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 27. [Google Scholar] [CrossRef]
- Dahl, E.M.; Neer, E.; Bowie, K.R.; Leung, E.T.; Karstens, L. microshades: An R Package for Improving Color Accessibility and Organization of Microbiome Data. Microbiol. Resour. Announc. 2022, 11, e0079522. [Google Scholar] [CrossRef]
- Jackson, D.A. PROTEST: A PROcrustean Randomization TEST of community environment concordance. Écoscience 1995, 2, 297–303. [Google Scholar] [CrossRef]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbo, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef]
- Friedman, J.; Alm, E.J. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef]
- Blondel, V.D.; Guillaume, J.-L.; Lambiotte, R.; Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. 2008, 10008, 1–12. [Google Scholar] [CrossRef]
- Silverman, J.D.; Washburne, A.D.; Mukherjee, S.; David, L.A. A phylogenetic transform enhances analysis of compositional microbiota data. eLife 2017, 6, e21887. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. Using ggtree to Visualize Data on Tree-like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef] [PubMed]
- Lovell, R.M.; Ford, A.C. Effect of gender on prevalence of irritable bowel syndrome in the community: Systematic review and meta-analysis. Am. J. Gastroenterol. 2012, 107, 991–1000. [Google Scholar] [CrossRef]
- Wigington, W.C.; Johnson, W.D.; Minocha, A. Epidemiology of irritable bowel syndrome among African Americans as compared with whites: A population-based study. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2005, 3, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Sperber, A.D.; Bangdiwala, S.I.; Drossman, D.A.; Ghoshal, U.C.; Simren, M.; Tack, J.; Whitehead, W.E.; Dumitrascu, D.L.; Fang, X.; Fukudo, S.; et al. Worldwide Prevalence and Burden of Functional Gastrointestinal Disorders, Results of Rome Foundation Global Study. Gastroenterology 2021, 160, 99–114.e3. [Google Scholar] [CrossRef]
- Nishiwaki, H.; Hamaguchi, T.; Ito, M.; Ishida, T.; Maeda, T.; Kashihara, K.; Tsuboi, Y.; Ueyama, J.; Shimamura, T.; Mori, H.; et al. Short-Chain Fatty Acid-Producing Gut Microbiota Is Decreased in Parkinson’s Disease but Not in Rapid-Eye-Movement Sleep Behavior Disorder. mSystems 2020, 5, e00797-20. [Google Scholar] [CrossRef]
- Zhang, Q.; Xiao, X.; Li, M.; Yu, M.; Ping, F.; Zheng, J.; Wang, T.; Wang, X. Vildagliptin increases butyrate-producing bacteria in the gut of diabetic rats. PLoS ONE 2017, 12, e0184735. [Google Scholar] [CrossRef]
- Pozuelo, M.; Panda, S.; Santiago, A.; Mendez, S.; Accarino, A.; Santos, J.; Guarner, F.; Azpiroz, F.; Manichanh, C. Reduction of butyrate- and methane-producing microorganisms in patients with Irritable Bowel Syndrome. Sci. Rep. 2015, 5, 12693. [Google Scholar] [CrossRef]
- Eribo, O.A.; du Plessis, N.; Ozturk, M.; Guler, R.; Walzl, G.; Chegou, N.N. The gut microbiome in tuberculosis susceptibility and treatment response: Guilty or not guilty? Cell. Mol. Life Sci. 2020, 77, 1497–1509. [Google Scholar] [CrossRef] [PubMed]
- Bellono, N.W.; Bayrer, J.R.; Leitch, D.B.; Castro, J.; Zhang, C.; O’Donnell, T.A.; Brierley, S.M.; Ingraham, H.A.; Julius, D. Enterochromaffin Cells Are Gut Chemosensors that Couple to Sensory Neural Pathways. Cell 2017, 170, 185–198.e6. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, M. Serotonin in the gastrointestinal tract. Curr. Opin. Endocrinol. Diabetes Obes. 2009, 16, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Lobionda, S.; Sittipo, P.; Kwon, H.Y.; Lee, Y.K. The Role of Gut Microbiota in Intestinal Inflammation with Respect to Diet and Extrinsic Stressors. Microorganisms 2019, 7, 271. [Google Scholar] [CrossRef]
- Ma, E.; Maskarinec, G.; Lim, U.; Boushey, C.; Wilkens, L.; Setiawan, V.; Le Marchand, L.; Randolph, T.W.; Jenkins, I.C.; Curtis, K.R.; et al. Long-term association between diet quality and characteristics of the gut microbiome in the multiethnic cohort study. Br. J. Nutr. 2022, 128, 93–102. [Google Scholar] [CrossRef]
- Kleigrewe, K.; Haack, M.; Baudin, M.; Menabreaz, T.; Crovadore, J.; Masri, M.; Beyrer, M.; Andlauer, W.; Lefort, F.; Dawid, C.; et al. Dietary Modulation of the Human Gut Microbiota and Metabolome with Flaxseed Preparations. Int. J. Mol. Sci. 2022, 23, 10473. [Google Scholar] [CrossRef]
- Taladrid, D.; de Celis, M.; Belda, I.; Bartolome, B.; Moreno-Arribas, M.V. Hypertension- and glycaemia-lowering effects of a grape-pomace-derived seasoning in high-cardiovascular risk and healthy subjects. Interplay with the gut microbiome. Food Funct. 2022, 13, 2068–2082. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, L.; Li, X.; Hao, R.; Li, J. Effects of high fructose corn syrup on intestinal microbiota structure and obesity in mice. NPJ Sci. Food 2022, 6, 17. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef]
- Beaumont, M.; Goodrich, J.K.; Jackson, M.A.; Yet, I.; Davenport, E.R.; Vieira-Silva, S.; Debelius, J.; Pallister, T.; Mangino, M.; Raes, J.; et al. Heritable components of the human fecal microbiome are associated with visceral fat. Genome Biol. 2016, 17, 189. [Google Scholar] [CrossRef]
- Bhat, S.F.; Pinney, S.E.; Kennedy, K.M.; McCourt, C.R.; Mundy, M.A.; Surette, M.G.; Sloboda, D.M.; Simmons, R.A. Exposure to high fructose corn syrup during adolescence in the mouse alters hepatic metabolism and the microbiome in a sex-specific manner. J. Physiol. 2021, 599, 1487–1511. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, X.; Yu, E.; Wang, N.; Cai, Q.; Shuai, Q.; Yan, F.; Jiang, L.; Wang, H.; Liu, J.; et al. Changes in gut microbiota and plasma inflammatory factors across the stages of colorectal tumorigenesis: A case-control study. BMC Microbiol. 2018, 18, 92. [Google Scholar] [CrossRef] [PubMed]
- Baranzini, S.; iMSMS Consortium. Gut microbiome of multiple sclerosis patients and paired household healthy controls reveal associations with disease risk and course. Cell 2022, 185, 3467–3486. [Google Scholar]
- Bernard, K.; Burdz, T.; Wiebe, D.; Balcewich, B.M.; Zimmerman, T.; Lagace-Wiens, P.; Hoang, L.M.N.; Bernier, A.M. Characterization of isolates of Eisenbergiella tayi, a strictly anaerobic Gram-stain variable bacillus recovered from human clinical materials in Canada. Anaerobe 2017, 44, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, F.; Mao, L.; Feng, T.; Wang, K.; Xu, M.; Lv, B.; Wang, X. Bifico relieves irritable bowel syndrome by regulating gut microbiota dysbiosis and inflammatory cytokines. Eur. J. Nutr. 2023, 62, 139–155. [Google Scholar] [CrossRef]
- Bailen, M.; Bressa, C.; Martinez-Lopez, S.; Gonzalez-Soltero, R.; Montalvo Lominchar, M.G.; San Juan, C.; Larrosa, M. Microbiota Features Associated with a High-Fat/Low-Fiber Diet in Healthy Adults. Front. Nutr. 2020, 7, 583608. [Google Scholar] [CrossRef]
- Jacobs, J.P.; Lagishetty, V.; Hauer, M.C.; Labus, J.S.; Dong, T.S.; Toma, R.; Vuyisich, M.; Naliboff, B.D.; Lackner, J.M.; Gupta, A.; et al. Multi-omics profiles of the intestinal microbiome in irritable bowel syndrome and its bowel habit subtypes. Microbiome 2023, 11, 5. [Google Scholar] [CrossRef]
- Van Hoorn, R.; Kapoor, R.; Kamphuis, J. A short review on sources and health benefits of GLA, The GOOD omega-6. Oléagineux Corps Gras Lipides 2008, 15, 262–264. [Google Scholar] [CrossRef]
- Noguchi, M.; Shimizu, M.; Lu, P.; Takahashi, Y.; Yamauchi, Y.; Sato, S.; Kiyono, H.; Kishino, S.; Ogawa, J.; Nagata, K.; et al. Lactic acid bacteria-derived gamma-linolenic acid metabolites are PPARdelta ligands that reduce lipid accumulation in human intestinal organoids. J. Biol. Chem. 2022, 298, 102534. [Google Scholar] [CrossRef]
- Janczy, A.; Szymanski, M.; Stankiewicz, M.; Kaska, L.; Waleron, K.; Stelmanska, E.; Sledzinski, T.; Mika, A. Increased Amount of Polyunsaturated Fatty Acids in the Intestinal Contents of Patients with Morbid Obesity. Obes. Surg. 2023, 33, 1228–1236. [Google Scholar] [CrossRef]
- Kosnicki, K.L.; Penprase, J.C.; Cintora, P.; Torres, P.J.; Harris, G.L.; Brasser, S.M.; Kelley, S.T. Effects of moderate, voluntary ethanol consumption on the rat and human gut microbiome. Addict. Biol. 2019, 24, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Manor, O.; Dai, C.L.; Kornilov, S.A.; Smith, B.; Price, N.D.; Lovejoy, J.C.; Gibbons, S.M.; Magis, A.T. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 2020, 11, 5206. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.; Dinis, L.; Barreiros Mota, I.; Morais, J.; Ismael, S.; Pereira-Leal, J.B.; Cardoso, J.; Ribeiro, P.; Beato, H.; Resende, M.; et al. Impact of Beer and Nonalcoholic Beer Consumption on the Gut Microbiota: A Randomized, Double-Blind, Controlled Trial. J. Agric. Food Chem. 2022, 70, 13062–13070. [Google Scholar] [CrossRef] [PubMed]
- Silvernale, C.; Kuo, B.; Staller, K. Lower socioeconomic status is associated with an increased prevalence of comorbid anxiety and depression among patients with irritable bowel syndrome: Results from a multicenter cohort. Scand. J. Gastroenterol. 2019, 54, 1070–1074. [Google Scholar] [CrossRef] [PubMed]
- Johansen, S.G.; Ness-Jensen, E. The changes in prevalence and risk of irritable bowel syndrome over time in a population-based cohort, the HUNT study, Norway. Scand. J. Gastroenterol. 2022, 1–7. [Google Scholar] [CrossRef]
- Dill-McFarland, K.A.; Tang, Z.Z.; Kemis, J.H.; Kerby, R.L.; Chen, G.; Palloni, A.; Sorenson, T.; Rey, F.E.; Herd, P. Close social relationships correlate with human gut microbiota composition. Sci. Rep. 2019, 9, 703. [Google Scholar] [CrossRef]
- Wilmes, L.; Collins, J.M.; O’Riordan, K.J.; O’Mahony, S.M.; Cryan, J.F.; Clarke, G. Of bowels, brain and behavior: A role for the gut microbiota in psychiatric comorbidities in irritable bowel syndrome. Neurogastroenterol. Motil. Off. J. Eur. Gastrointest. Motil. Soc. 2021, 33, e14095. [Google Scholar] [CrossRef]



| Characteristics | Number of IBS Patients | Number of Healthy Control Participants |
|---|---|---|
| n = 33(%) | n = 12(%) | |
| Gender | ||
| F | 26(78.8) | 6(50.0) |
| M | 7(21.2) | 6(50.0) |
| Age | ||
| 18–39 | 12(36.4) | 6(50.0) |
| 40–80 | 21(63.6) | 6(50.0) |
| Race | ||
| African American | 2(6.1) | 1(8.3) |
| Hispanic | 1(3) | 0(0) |
| Caucasian | 28(84.8) | 9(75%) |
| Asian | 2(6.1) | 2(16.7) |
| Marital Status | ||
| Widowed | 2(6.1) | 0(0) |
| Single | 15(45.5) | 10(83.3) |
| Separated/Divorced | 2(6.1) | 0(0) |
| Married | 14(42.4) | 2(16.7) |
| IBS Subtype | ||
| IBS-Diarrhea | 10(30.3) | None |
| IBS-Constipation | 10(30.3) | None |
| IBS-Mixed | 13(39.4) | None |
| Hypertension | ||
| Yes | 10(30.3) | 1(8.33) |
| No | 23(69.7) | 11(91.7) |
| Diabetes | ||
| Yes | 3(9.1) | 0(0) |
| No | 30(90.1) | 12(100) |
| High cholesterol | ||
| Yes | 6(18.2) | 0(0) |
| No | 27(81.8) | 12(100) |
| Average BMI | 27.5 | 23.14 |
| Depression | ||
| Yes | 12(36.4) | 0(0) |
| No | 21(63.6) | 12(100) |
| Anxiety | ||
| Yes | 13(39.4) | 0(0) |
| No | 20(60.6) | 12(100) |
| PTSD | ||
| Yes | 2(6.1) | 0(0) |
| No | 31(93.9) | 12(100) |
| Bipolar | ||
| Yes | 2(6.1) | 0(0) |
| No | 31(93.9) | 12(100) |
| Psychosis | ||
| Yes | 0(0) | 0(0) |
| No | 31(93.9) | 12(100) |
| Smoking | ||
| Yes | 1(3) | 0(0) |
| No | 32(97) | 12(100) |
| Alcohol | ||
| Yes | 18(54.5) | 12(100) |
| No | 15(45.5) | 0(0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paripati, N.; Nesi, L.; Sterrett, J.D.; Dawud, L.M.; Kessler, L.R.; Lowry, C.A.; Perez, L.J.; DeSipio, J.; Phadtare, S. Gut Microbiome and Lipidome Signatures in Irritable Bowel Syndrome Patients from a Low-Income, Food-Desert Area: A Pilot Study. Microorganisms 2023, 11, 2503. https://doi.org/10.3390/microorganisms11102503
Paripati N, Nesi L, Sterrett JD, Dawud LM, Kessler LR, Lowry CA, Perez LJ, DeSipio J, Phadtare S. Gut Microbiome and Lipidome Signatures in Irritable Bowel Syndrome Patients from a Low-Income, Food-Desert Area: A Pilot Study. Microorganisms. 2023; 11(10):2503. https://doi.org/10.3390/microorganisms11102503
Chicago/Turabian StyleParipati, Nikita, Lauren Nesi, John D. Sterrett, Lamya’a M. Dawud, Lyanna R. Kessler, Christopher A. Lowry, Lark J. Perez, Joshua DeSipio, and Sangita Phadtare. 2023. "Gut Microbiome and Lipidome Signatures in Irritable Bowel Syndrome Patients from a Low-Income, Food-Desert Area: A Pilot Study" Microorganisms 11, no. 10: 2503. https://doi.org/10.3390/microorganisms11102503
APA StyleParipati, N., Nesi, L., Sterrett, J. D., Dawud, L. M., Kessler, L. R., Lowry, C. A., Perez, L. J., DeSipio, J., & Phadtare, S. (2023). Gut Microbiome and Lipidome Signatures in Irritable Bowel Syndrome Patients from a Low-Income, Food-Desert Area: A Pilot Study. Microorganisms, 11(10), 2503. https://doi.org/10.3390/microorganisms11102503







