pH Drives Differences in Bacterial Community β-Diversity in Hydrologically Connected Lake Sediments
Abstract
:1. Introduction
2. Materials and Methods
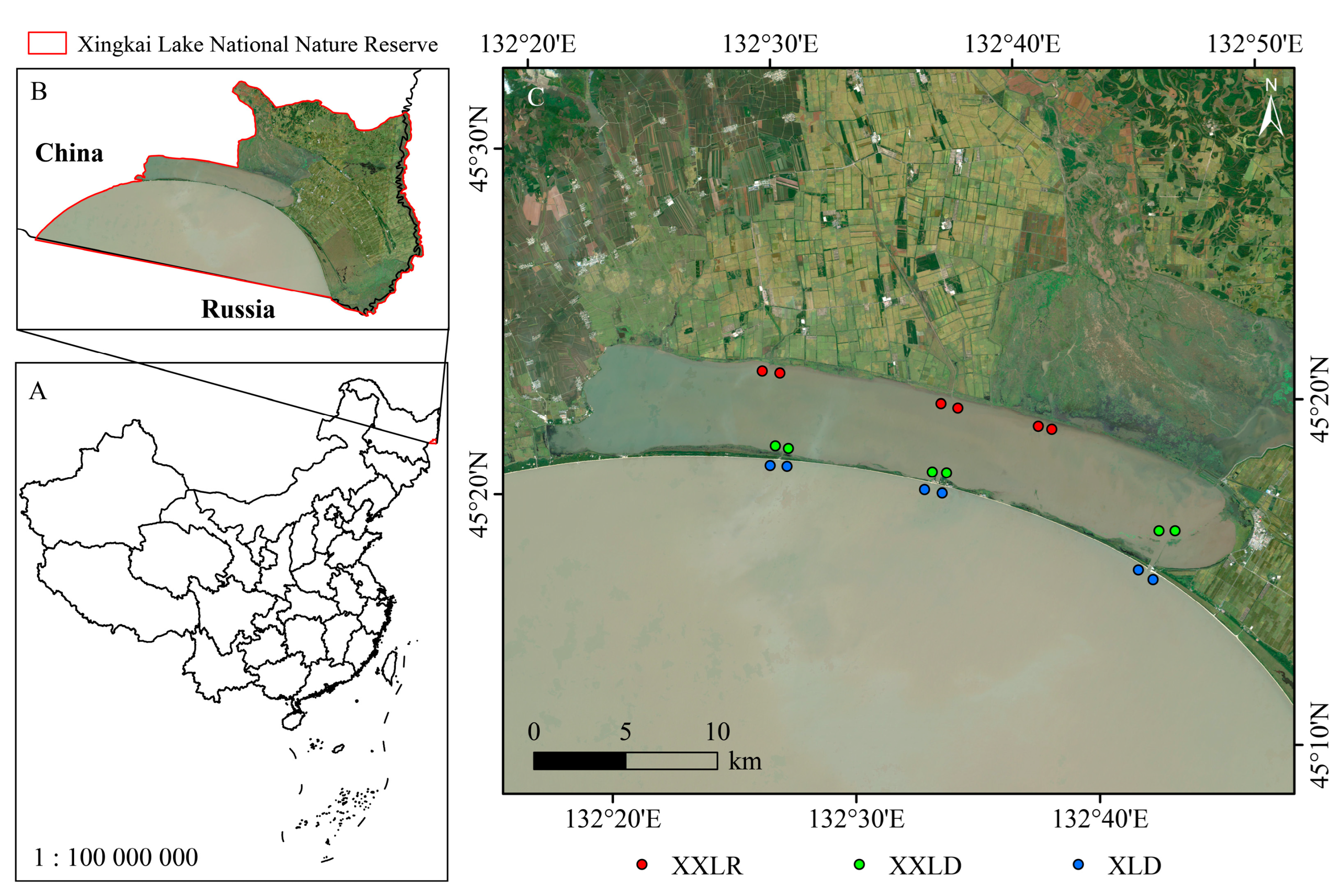
2.1. Sample Sites and Sample Collections
2.2. Sediment Property Analysis
2.3. DNA Extraction and Sequence Processing
2.4. Community Assembly Processes
2.5. Co-Occurrence Network Construction
2.6. Statistical Analysis
3. Results
3.1. Sediment Chemical Properties
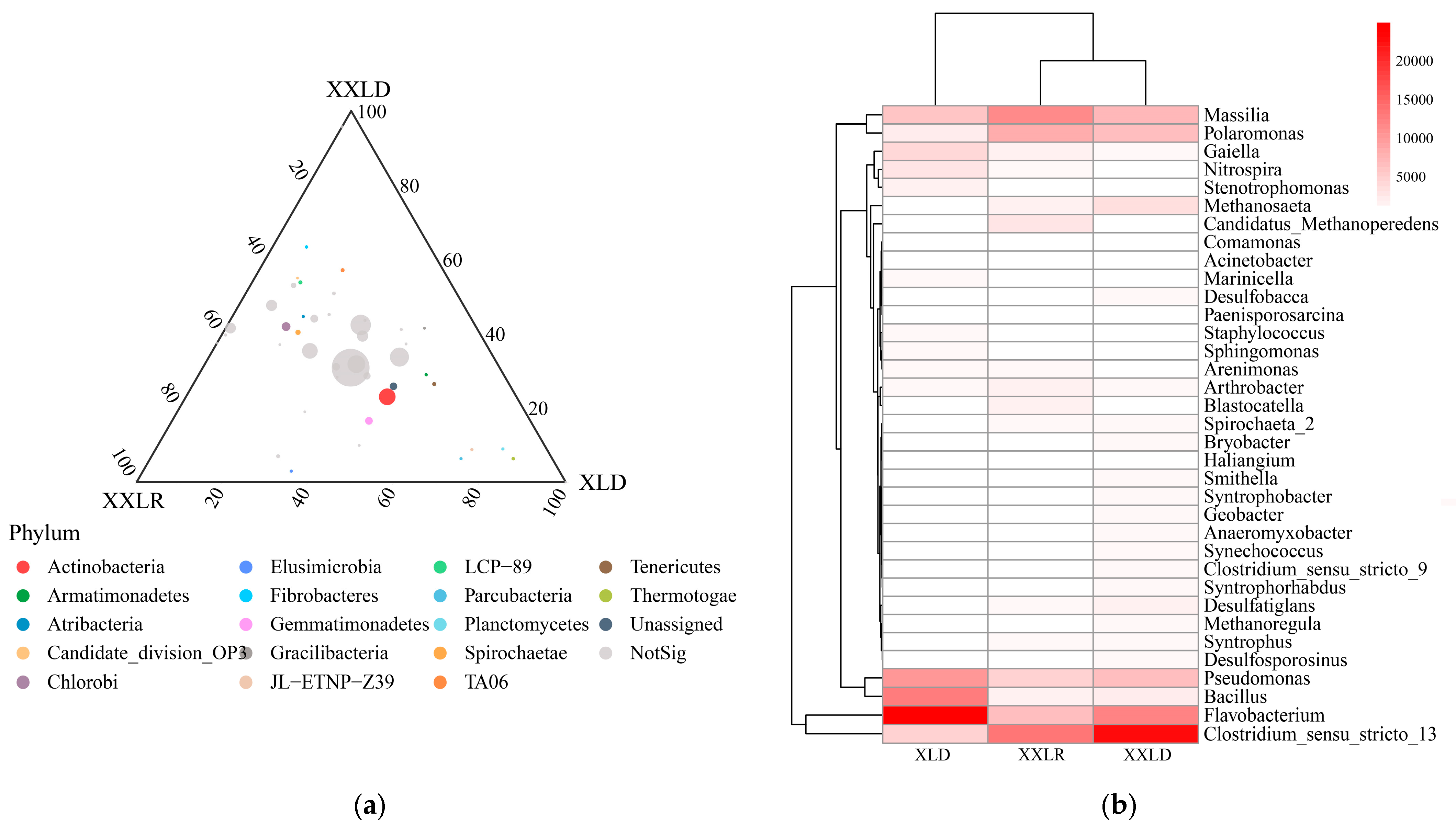
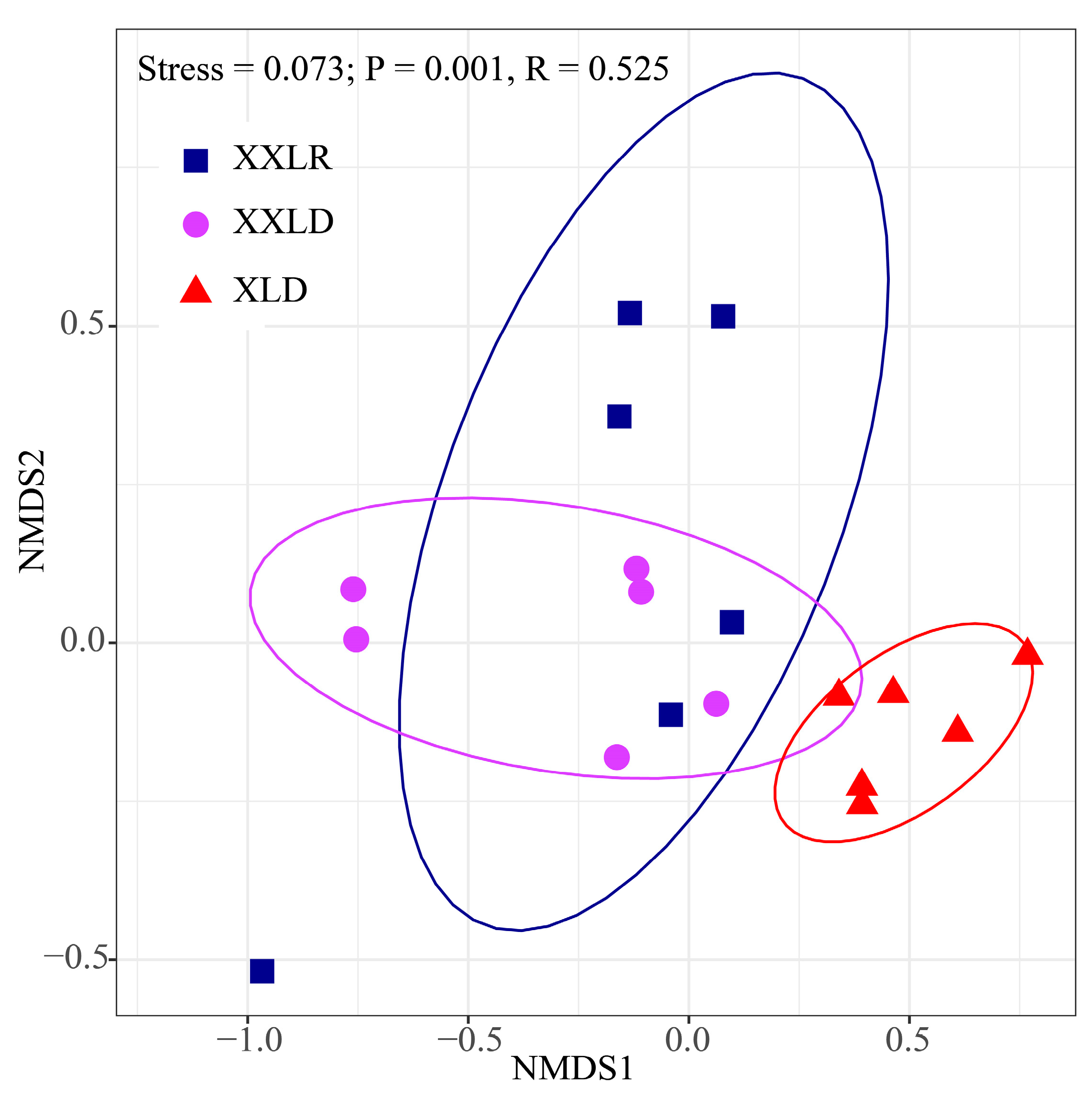
3.2. Bacterial Community Composition and Diversity
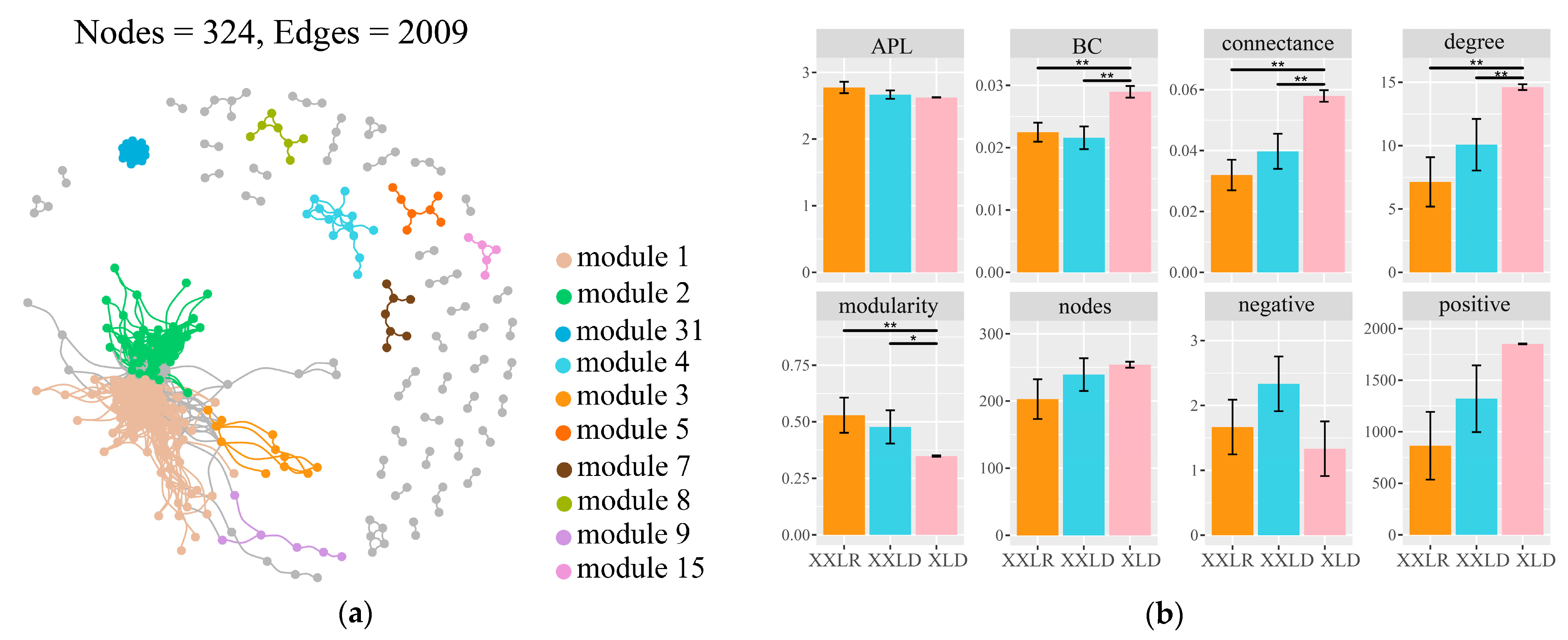
3.3. Bacterial Community Assembly Processes and Co-Occurrence Network
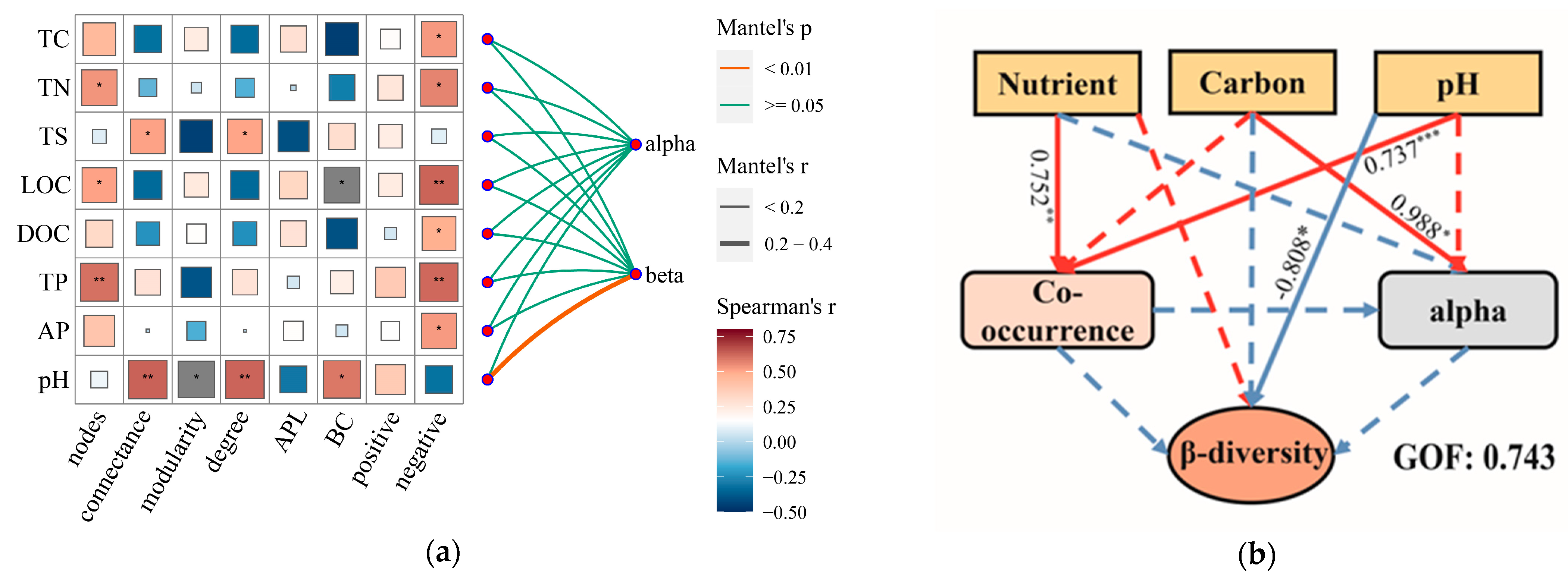
3.4. Factors Shaping the Community Diversity in Sediment
4. Discussion
4.1. Variation Characteristics of Sediment Properties
4.2. Community Assembly Process and Microbial Interactions
4.3. Structure and Diversity of Sediment Bacterial Communities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dodds, W.K.; Perkin, J.S.; Gerken, J.E. Human Impact on Freshwater Ecosystem Services: A Global Perspective. Environ. Sci. Technol. 2013, 47, 9061–9068. [Google Scholar] [CrossRef] [PubMed]
- Loeks, B.M.; Cotner, J.B. Upper Midwest Lakes Are Supersaturated with N2. Proc. Natl. Acad. Sci. USA 2020, 117, 17063–17067. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Jiang, H.; Dong, H.; Liu, Y. A Comprehensive Census of Lake Microbial Diversity on a Global Scale. Sci. China Life Sci. 2019, 62, 1320–1331. [Google Scholar] [CrossRef] [PubMed]
- Verpoorter, C.; Kutser, T.; Seekell, D.A.; Tranvik, L.J. A Global Inventory of Lakes Based on High-Resolution Satellite Imagery. Geophys. Res. Lett. 2014, 41, 6396–6402. [Google Scholar] [CrossRef]
- von Wachenfeldt, E.; Sobek, S.; Bastviken, D.; Tranvik, L.J. Linking Allochthonous Dissolved Organic Matter and Boreal Lake Sediment Carbon Sequestration: The Role of Light-Mediated Flocculation. Limnol. Oceanogr. 2008, 53, 2416–2426. [Google Scholar] [CrossRef]
- Von Wachenfeldt, E.; Tranvik, L.J. Sedimentation in Boreal Lakes—the Role of Flocculation of Allochthonous Dissolved Organic Matter in the Water Column. Ecosystems 2008, 11, 803–814. [Google Scholar] [CrossRef]
- Wang, W.; Yang, Y.; Zhou, Y.; Zhang, S.; Wang, X.; Yang, Z. Impact of Anthropogenic Activities on the Sediment Microbial Communities of Baiyangdian Shallow Lake. Int. J. Sediment Res. 2020, 35, 180–192. [Google Scholar] [CrossRef]
- Yi, Y.; Lin, C.; Wang, W.; Song, J. Habitat and Seasonal Variations in Bacterial Community Structure and Diversity in Sediments of a Shallow Lake. Ecol. Indic. 2021, 120, 106959. [Google Scholar] [CrossRef]
- Chu, S.; Cai, M.; Xu, X. Soil–Plant Transfer of Polychlorinated Biphenyls in Paddy Fields. Sci. Total Environ. 1999, 234, 119–126. [Google Scholar] [CrossRef]
- Frink, C.R. Nutrient Budget: Rational Analysis of Eutrophication in a Connecticut Lake. Environ. Sci. Technol. 1967, 1, 425–428. [Google Scholar] [CrossRef]
- Oni, O.E.; Schmidt, F.; Miyatake, T.; Kasten, S.; Witt, M.; Hinrichs, K.-U.; Friedrich, M.W. Microbial Communities and Organic Matter Composition in Surface and Subsurface Sediments of the Helgoland Mud Area, North Sea. Front. Microbiol. 2015, 6, 1290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.; Jiang, X. Profiling of Sediment Microbial Community in Dongting Lake before and after Impoundment of the Three Gorges Dam. Int. J. Environ. Res. Public. Health 2016, 13, 617. [Google Scholar] [CrossRef] [PubMed]
- Gerbersdorf, S.U.; Hollert, H.; Brinkmann, M.; Wieprecht, S.; Schüttrumpf, H.; Manz, W. Anthropogenic Pollutants Affect Ecosystem Services of Freshwater Sediments: The Need for a “Triad plus x” Approach. J. Soils Sediments 2011, 11, 1099–1114. [Google Scholar] [CrossRef]
- Obi, C.C.; Adebusoye, S.A.; Ugoji, E.O.; Ilori, M.O.; Amund, O.O.; Hickey, W.J. Microbial Communities in Sediments of Lagos Lagoon, Nigeria: Elucidation of Community Structure and Potential Impacts of Contamination by Municipal and Industrial Wastes. Front. Microbiol. 2016, 7, 1213. [Google Scholar] [CrossRef] [Green Version]
- Cui, H.; Ou, Y.; Wang, L.; Liang, A.; Yan, B.; Li, Y. Dynamic Changes in Microbial Communities and Nutrient Stoichiometry Associated with Soil Aggregate Structure in Restored Wetlands. Catena 2021, 197, 104984. [Google Scholar] [CrossRef]
- Li, M.; Fang, A.; Yu, X.; Zhang, K.; He, Z.; Wang, C.; Peng, Y.; Xiao, F.; Yang, T.; Zhang, W. Microbially-Driven Sulfur Cycling Microbial Communities in Different Mangrove Sediments. Chemosphere 2021, 273, 128597. [Google Scholar] [CrossRef]
- Song, K.; Wang, Z.; Li, L.; Tedesco, L.; Li, F.; Jin, C.; Du, J. Wetlands Shrinkage, Fragmentation and Their Links to Agriculture in the Muleng–Xingkai Plain, China. J. Environ. Manag. 2012, 111, 120–132. [Google Scholar] [CrossRef]
- Yuan, Y.; Jiang, M.; Liu, X.; Yu, H.; Otte, M.L.; Ma, C.; Her, Y.G. Environmental Variables Influencing Phytoplankton Communities in Hydrologically Connected Aquatic Habitats in the Lake Xingkai Basin. Ecol. Indic. 2018, 91, 1–12. [Google Scholar] [CrossRef]
- Jiang, M.; Wang, Q.; Tian, X.; Zhu, X.; Dong, X.; Wu, Z.; Yuan, Y. Spatiotemporal Variation and Ecological Risk Assessment of Sediment Heavy Metals in Two Hydrologically Connected Lakes. Front. Ecol. Evol. 2022, 10, 1005194. [Google Scholar] [CrossRef]
- Sun, W.; Zhang, E.; Chen, R.; Shen, J. Lacustrine Carbon Cycling since the Last Interglaciation in Northeast China: Evidence from n-Alkanes in the Sediments of Lake Xingkai. Quat. Int. 2019, 523, 101–108. [Google Scholar] [CrossRef]
- Wang, L.; Yu, X.; Xue, Z.; Huo, L.; Jiang, M.; Lu, X.; Zou, Y. Distribution Characteristics of Iron, Carbon, Nitrogen and Phosphorus in the Surface Soils of Different Land Use Types near Xingkai Lake. J. Soils Sediments 2019, 19, 275–285. [Google Scholar] [CrossRef]
- Xing, M.; Wang, Q.; Li, X.; Li, Y.; Zhou, X. Selection of Keystone Species Based on Stable Carbon and Nitrogen Isotopes to Construct a Typical Food Web on the Shore of Xingkai Lake, China. Ecol. Indic. 2021, 132, 108263. [Google Scholar] [CrossRef]
- Hong, S.; Piao, S.; Chen, A.; Liu, Y.; Liu, L.; Peng, S.; Sardans, J.; Sun, Y.; Peñuelas, J.; Zeng, H. Afforestation Neutralizes Soil PH. Nat. Commun. 2018, 9, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, X.; Yu, X.; Li, Z.; Wang, Q.; Liu, Z.; Zou, Y. Iron-Bound Organic Carbon Is Conserved in the Rhizosphere Soil of Freshwater Wetlands. Soil Biol. Biochem. 2020, 149, 107949. [Google Scholar] [CrossRef]
- Lefroy, R.D.B.; Blair, G.J.; Strong, W.M. Changes in Soil Organic Matter with Cropping as Measured by Organic Carbon Fractions and 13C Natural Isotope Abundance. Plant Soil 1993, 155–156, 399–402. [Google Scholar] [CrossRef]
- Loginow, W.; Wisniewski, W.; Gonet, S.S.; Ciescinska, B. Fractionation of Organic Carbon Based on Susceptibility to Oxidation. Pol. J. Soil Sci. 1987, 20, 47–52. [Google Scholar]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Author Correction: Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Konopka, A.E.; Fredrickson, J.K. Stochastic and Deterministic Assembly Processes in Subsurface Microbial Communities. ISME J. 2012, 6, 1653–1664. [Google Scholar] [CrossRef] [Green Version]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L.; Konopka, A. Quantifying Community Assembly Processes and Identifying Features That Impose Them. ISME J. 2013, 7, 2069–2079. [Google Scholar] [CrossRef]
- Kang, L.; Chen, L.; Zhang, D.; Peng, Y.; Song, Y.; Kou, D.; Deng, Y.; Yang, Y. Stochastic Processes Regulate Belowground Community Assembly in Alpine Grasslands on the Tibetan Plateau. Environ. Microbiol. 2022, 24, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R Tools for Integrating Phylogenies and Ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Cheng, K.; Li, K.; Jin, Y.; He, X. Deciphering the Diversity Patterns and Community Assembly of Rare and Abundant Bacterial Communities in a Wetland System. Sci. Total Environ. 2022, 838, 156334. [Google Scholar] [CrossRef] [PubMed]
- Chase, J.M.; Myers, J.A. Disentangling the Importance of Ecological Niches from Stochastic Processes across Scales. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 2351–2363. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Ning, D. Stochastic Community Assembly: Does It Matter in Microbial Ecology? Microbiol. Mol. Biol. Rev. 2017, 81, e00002-17. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Bai, J.; Wang, J.; Zhang, G.; Wang, W.; Wang, X.; Zhang, L.; Wang, Y.; Liu, X.; Cui, B. Different Stochastic Processes Regulate Bacterial and Fungal Community Assembly in Estuarine Wetland Soils. Soil Biol. Biochem. 2022, 167, 108586. [Google Scholar] [CrossRef]
- Yuan, M.M.; Guo, X.; Wu, L.; Zhang, Y.A.; Xiao, N.; Ning, D.; Shi, Z.; Zhou, X.; Wu, L.; Yang, Y. Climate Warming Enhances Microbial Network Complexity and Stability. Nat. Clim. Chang. 2021, 11, 343–348. [Google Scholar] [CrossRef]
- Anderson, M.J. Distance-Based Tests for Homogeneity of Multivariate Dispersions. Biometrics 2006, 62, 245–253. [Google Scholar] [CrossRef]
- Levens, R. Evolution in Changing Environments, Some Theoretical Explorations; Princeton University Press: Princeton, NJ, USA, 1968. [Google Scholar]
- Wu, W.; Lu, H.-P.; Sastri, A.; Yeh, Y.-C.; Gong, G.-C.; Chou, W.-C.; Hsieh, C.-H. Contrasting the Relative Importance of Species Sorting and Dispersal Limitation in Shaping Marine Bacterial versus Protist Communities. ISME J. 2018, 12, 485–494. [Google Scholar] [CrossRef] [Green Version]
- Csiki, S.J.; Rhoads, B.L. Influence of Four Run-of-River Dams on Channel Morphology and Sediment Characteristics in Illinois, USA. Geomorphology 2014, 206, 215–229. [Google Scholar] [CrossRef]
- Li, D.; Yao, P.; Bianchi, T.S.; Zhang, T.; Zhao, B.; Pan, H.; Wang, J.; Yu, Z. Organic Carbon Cycling in Sediments of the Changjiang Estuary and Adjacent Shelf: Implication for the Influence of Three Gorges Dam. J. Mar. Syst. 2014, 139, 409–419. [Google Scholar] [CrossRef]
- Luo, X.; Xiang, X.; Huang, G.; Song, X.; Wang, P.; Fu, K. Bacterial Abundance and Physicochemical Characteristics of Water and Sediment Associated with Hydroelectric Dam on the Lancang River China. Int. J. Environ. Res. Public. Health 2019, 16, 2031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Jiang, L.; Huang, X.; Cai, Z. Identifying Nitrogen Source and Transport Characteristics of the Urban Estuaries and Gate-Controlled Rivers in Northern Taihu Lake, China. Ecol. Indic. 2021, 130, 108035. [Google Scholar] [CrossRef]
- Shoja, H.; Rahimi, G.; Fallah, M.; Ebrahimi, E. Investigation of Phosphorus Fractions and Isotherm Equation on the Lake Sediments in Ekbatan Dam (Iran). Environ. Earth Sci. 2017, 76, 1–15. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Y.; Cai, Z.; Zhang, M.; Ye, C. Response of the Nitrogen Load and Its Driving Forces in Estuarine Water to Dam Construction in Taihu Lake, China. Environ. Sci. Pollut. Res. 2020, 27, 31458–31467. [Google Scholar] [CrossRef]
- Wang, J.; Shen, J.I.; Wu, Y.; Tu, C.; Soininen, J.; Stegen, J.C.; He, J.; Liu, X.; Zhang, L.; Zhang, E. Phylogenetic Beta Diversity in Bacterial Assemblages across Ecosystems: Deterministic versus Stochastic Processes. ISME J. 2013, 7, 1310–1321. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Jiang, H.; Sun, X.; Chen, J.; Xie, Z.; Dong, H. Minerals Play Key Roles in Driving Prokaryotic and Fungal Communities in the Surface Sediments of the Qinghai-Tibetan Lakes. FEMS Microbiol. Ecol. 2020, 96, fiaa035. [Google Scholar] [CrossRef]
- Qin, M.; Xu, H.; Zeng, J.; Zhao, D.; Yu, Z.; Wu, Q.L. Composition and Assembly of Bacterial Communities in Surface and Deeper Sediments from Aquaculture-Influenced Sites in Eastern Lake Taihu, China. Aquat. Sci. 2020, 82, 1–13. [Google Scholar] [CrossRef]
- He, Q.; Wang, S.; Hou, W.; Feng, K.; Li, F.; Hai, W.; Zhang, Y.; Sun, Y.; Deng, Y. Temperature and Microbial Interactions Drive the Deterministic Assembly Processes in Sediments of Hot Springs. Sci. Total Environ. 2021, 772, 145465. [Google Scholar] [CrossRef]
- Kuang, B.; Xiao, R.; Wang, C.; Zhang, L.; Wei, Z.; Bai, J.; Zhang, K.; Campos, M.; Jorquera, M.A. Bacterial Community Assembly in Surface Sediments of a Eutrophic Shallow Lake in Northern China. Ecohydrol. Hydrobiol. 2022. [Google Scholar] [CrossRef]
- Li, Y.; Gao, Y.; Zhang, W.; Wang, C.; Wang, P.; Niu, L.; Wu, H. Homogeneous Selection Dominates the Microbial Community Assembly in the Sediment of the Three Gorges Reservoir. Sci. Total Environ. 2019, 690, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; Zhou, R.; Zeng, S.; Wei, D.; Deng, X.; Xing, C.; Weng, S.; He, J.; Huang, Z. Stochastic Processes Shape the Bacterial Community Assembly in Shrimp Cultural Pond Sediments. Appl. Microbiol. Biotechnol. 2021, 105, 5013–5022. [Google Scholar] [CrossRef] [PubMed]
- Chase, J.M. Spatial Scale Resolves the Niche versus Neutral Theory Debate. J. Veg. Sci. 2014, 25, 319–322. [Google Scholar] [CrossRef]
- Shi, Y.; Li, Y.; Xiang, X.; Sun, R.; Yang, T.; He, D.; Zhang, K.; Ni, Y.; Zhu, Y.-G.; Adams, J.M. Spatial Scale Affects the Relative Role of Stochasticity versus Determinism in Soil Bacterial Communities in Wheat Fields across the North China Plain. Microbiome 2018, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P. Patterns and Processes of Microbial Community Assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, B.M.; Stegen, J.C.; Kim, M.; Dong, K.; Adams, J.M.; Lee, Y.K. Soil PH Mediates the Balance between Stochastic and Deterministic Assembly of Bacteria. ISME J. 2018, 12, 1072–1083. [Google Scholar] [CrossRef]
- Feng, Y.; Chen, R.; Stegen, J.C.; Guo, Z.; Zhang, J.; Li, Z.; Lin, X. Two Key Features Influencing Community Assembly Processes at Regional Scale: Initial State and Degree of Change in Environmental Conditions. Mol. Ecol. 2018, 27, 5238–5251. [Google Scholar] [CrossRef]
- Zhang, K.; Shi, Y.; Cui, X.; Yue, P.; Li, K.; Liu, X.; Tripathi, B.M.; Chu, H. Salinity Is a Key Determinant for Soil Microbial Communities in a Desert Ecosystem. Msystems 2019, 4, e00225-18. [Google Scholar] [CrossRef] [Green Version]
- Jiao, S.; Lu, Y. Abundant Fungi Adapt to Broader Environmental Gradients than Rare Fungi in Agricultural Fields. Glob. Chang. Biol. 2020, 26, 4506–4520. [Google Scholar] [CrossRef]
- Wu, L.; Yang, P.; Zhang, L.; Luo, L.; Hong, Y.; Zhu, W.; Zheng, L.; Zhao, G.; Tong, C.; Peñuelas, J. Sediment Sulfate Content Determines Assembly Processes and Network Stability of Bacteria Communities of Coastal Land-Based Shrimp Aquaculture Ponds. Aquaculture 2023, 563, 738953. [Google Scholar] [CrossRef]
- Broman, E.; Sachpazidou, V.; Pinhassi, J.; Dopson, M. Oxygenation of Hypoxic Coastal Baltic Sea Sediments Impacts on Chemistry, Microbial Community Composition, and Metabolism. Front. Microbiol. 2017, 8, 2453. [Google Scholar] [CrossRef] [PubMed]
- Choi, A.; Cho, H.; Kim, B.; Kim, H.C.; Jung, R.-H.; Lee, W.-C.; Hyun, J.-H. Effects of Finfish Aquaculture on Biogeochemistry and Bacterial Communities Associated with Sulfur Cycles in Highly Sulfidic Sediments. Aquac. Environ. Interact. 2018, 10, 413–427. [Google Scholar] [CrossRef] [Green Version]
- Carter, M.R. Microbial Biomass and Mineralizable Nitrogen in Solonetzic Soils: Influence of Gypsum and Lime Amendments. Soil Biol. Biochem. 1986, 18, 531–537. [Google Scholar] [CrossRef]
- Chase, J.M. Stochastic Community Assembly Causes Higher Biodiversity in More Productive Environments. Science 2010, 328, 1388–1391. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Deng, Y.E.; Zhang, P.; Xue, K.; Liang, Y.; Van Nostrand, J.D.; Yang, Y.; He, Z.; Wu, L.; Stahl, D.A. Stochasticity, Succession, and Environmental Perturbations in a Fluidic Ecosystem. Proc. Natl. Acad. Sci. USA 2014, 111, E836–E845. [Google Scholar] [CrossRef] [Green Version]
- Jiao, S.; Yang, Y.; Xu, Y.; Zhang, J.; Lu, Y. Balance between Community Assembly Processes Mediates Species Coexistence in Agricultural Soil Microbiomes across Eastern China. ISME J. 2020, 14, 202–216. [Google Scholar] [CrossRef]
- Pandit, S.N.; Kolasa, J.; Cottenie, K. Contrasts between Habitat Generalists and Specialists: An Empirical Extension to the Basic Metacommunity Framework. Ecology 2009, 90, 2253–2262. [Google Scholar] [CrossRef] [Green Version]
- Fan, K.; Weisenhorn, P.; Gilbert, J.A.; Shi, Y.; Bai, Y.; Chu, H. Soil PH Correlates with the Co-Occurrence and Assemblage Process of Diazotrophic Communities in Rhizosphere and Bulk Soils of Wheat Fields. Soil Biol. Biochem. 2018, 121, 185–192. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, J.; Liu, Y.; Shi, P.; Wei, G. Co-Occurrence Patterns of Soybean Rhizosphere Microbiome at a Continental Scale. Soil Biol. Biochem. 2018, 118, 178–186. [Google Scholar] [CrossRef]
- Wang, Y.; Sheng, H.-F.; He, Y.; Wu, J.-Y.; Jiang, Y.-X.; Tam, N.F.-Y.; Zhou, H.-W. Comparison of the Levels of Bacterial Diversity in Freshwater, Intertidal Wetland, and Marine Sediments by Using Millions of Illumina Tags. Appl. Environ. Microbiol. 2012, 78, 8264–8271. [Google Scholar] [CrossRef] [Green Version]
- Hollister, E.B.; Engledow, A.S.; Hammett, A.J.M.; Provin, T.L.; Wilkinson, H.H.; Gentry, T.J. Shifts in Microbial Community Structure along an Ecological Gradient of Hypersaline Soils and Sediments. ISME J. 2010, 4, 829–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newton, R.J.; Jones, S.E.; Eiler, A.; McMahon, K.D.; Bertilsson, S. A Guide to the Natural History of Freshwater Lake Bacteria. Microbiol. Mol. Biol. Rev. 2011, 75, 14–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Ren, H.; Shen, L.; Lou, L.; Tian, G.; Zheng, P.; Hu, B. PH Levels Drive Bacterial Community Structure in Sediments of the Qiantang River as Determined by 454 Pyrosequencing. Front. Microbiol. 2015, 6, 285. [Google Scholar] [CrossRef] [PubMed]
- Andreote, F.D.; Jiménez, D.J.; Chaves, D.; Dias, A.C.F.; Luvizotto, D.M.; Dini-Andreote, F.; Fasanella, C.C.; Lopez, M.V.; Baena, S.; Taketani, R.G. The Microbiome of Brazilian Mangrove Sediments as Revealed by Metagenomics. PLoS ONE 2012, 7, e38600. [Google Scholar] [CrossRef]
- Sauvain, L.; Bueche, M.; Junier, T.; Masson, M.; Wunderlin, T.; Kohler-Milleret, R.; Gascon Diez, E.; Loizeau, J.-L.; Tercier-Waeber, M.-L.; Junier, P. Bacterial Communities in Trace Metal Contaminated Lake Sediments Are Dominated by Endospore-Forming Bacteria. Aquat. Sci. 2014, 76, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Zeng, C.; Li, Y.; Lu, A.; Ding, H.; Wang, X.; Wang, C. Electrochemical Interaction of a Heterotrophic Bacteria Alcaligenes Faecalis with a Graphite Cathode. Geomicrobiol. J. 2012, 29, 244–249. [Google Scholar] [CrossRef]
- Mor, G.; Kwon, J.-Y. Trophoblast-Microbiome Interaction: A New Paradigm on Immune Regulation. Am. J. Obstet. Gynecol. 2015, 213, S131–S137. [Google Scholar] [CrossRef] [Green Version]
- Rajilić-Stojanović, M.; De Vos, W.M. The First 1000 Cultured Species of the Human Gastrointestinal Microbiota. FEMS Microbiol. Rev. 2014, 38, 996–1047. [Google Scholar] [CrossRef]
- Crump, B.C.; Peranteau, C.; Beckingham, B.; Cornwell, J.C. Respiratory Succession and Community Succession of Bacterioplankton in Seasonally Anoxic Estuarine Waters. Appl. Environ. Microbiol. 2007, 73, 6802–6810. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Y.; Zou, Y.; Chen, B.; Grebmeier, J.M.; Li, H.; Yu, Y.; Zheng, T. Phylogenetic Diversity of Sediment Bacteria in the Northern Bering Sea. Polar Biol. 2011, 34, 907–919. [Google Scholar] [CrossRef]
- Jones, R.T.; Robeson, M.S.; Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. A Comprehensive Survey of Soil Acidobacterial Diversity Using Pyrosequencing and Clone Library Analyses. ISME J. 2009, 3, 442–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozupone, C.A.; Knight, R. Global Patterns in Bacterial Diversity. Proc. Natl. Acad. Sci. USA 2007, 104, 11436–11440. [Google Scholar] [CrossRef] [Green Version]
- Torsvik, V.; Øvreås, L.; Thingstad, T.F. Prokaryotic Diversity–Magnitude, Dynamics, and Controlling Factors. Science 2002, 296, 1064–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, C.; Zhang, W.; Zhong, Z.; Han, X.; Yang, G.; Feng, Y.; Ren, G. Differential Responses of Soil Microbial Biomass, Diversity, and Compositions to Altitudinal Gradients Depend on Plant and Soil Characteristics. Sci. Total Environ. 2018, 610, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Siles, J.A.; Margesin, R. Abundance and Diversity of Bacterial, Archaeal, and Fungal Communities along an Altitudinal Gradient in Alpine Forest Soils: What Are the Driving Factors? Microb. Ecol. 2016, 72, 207–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, J.; Yin, Y.; Zhu, W.; Zhou, Y. Variations in Soil Bacterial Community Diversity and Structures among Different Revegetation Types in the Baishilazi Nature Reserve. Front. Microbiol. 2018, 9, 2874. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, B.M.; Kim, M.; Singh, D.; Lee-Cruz, L.; Lai-Hoe, A.; Ainuddin, A.N.; Go, R.; Rahim, R.A.; Husni, M.H.A.; Chun, J. Tropical Soil Bacterial Communities in Malaysia: PH Dominates in the Equatorial Tropics Too. Microb. Ecol. 2012, 64, 474–484. [Google Scholar] [CrossRef]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil Bacterial and Fungal Communities across a PH Gradient in an Arable Soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- Han, X.; Schubert, C.J.; Fiskal, A.; Dubois, N.; Lever, M.A. Eutrophication as a Driver of Microbial Community Structure in Lake Sediments. Environ. Microbiol. 2020, 22, 3446–3462. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pu, H.; Yuan, Y.; Qin, L.; Liu, X. pH Drives Differences in Bacterial Community β-Diversity in Hydrologically Connected Lake Sediments. Microorganisms 2023, 11, 676. https://doi.org/10.3390/microorganisms11030676
Pu H, Yuan Y, Qin L, Liu X. pH Drives Differences in Bacterial Community β-Diversity in Hydrologically Connected Lake Sediments. Microorganisms. 2023; 11(3):676. https://doi.org/10.3390/microorganisms11030676
Chicago/Turabian StylePu, Haiguang, Yuxiang Yuan, Lei Qin, and Xiaohui Liu. 2023. "pH Drives Differences in Bacterial Community β-Diversity in Hydrologically Connected Lake Sediments" Microorganisms 11, no. 3: 676. https://doi.org/10.3390/microorganisms11030676



