Whole Genome Sequencing of the Novel Probiotic Strain Lactiplantibacillus plantarum FCa3L
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Lactiplantibacillus Strains
2.2. Species Identification
2.3. Genomic DNA Extraction and Sequencing
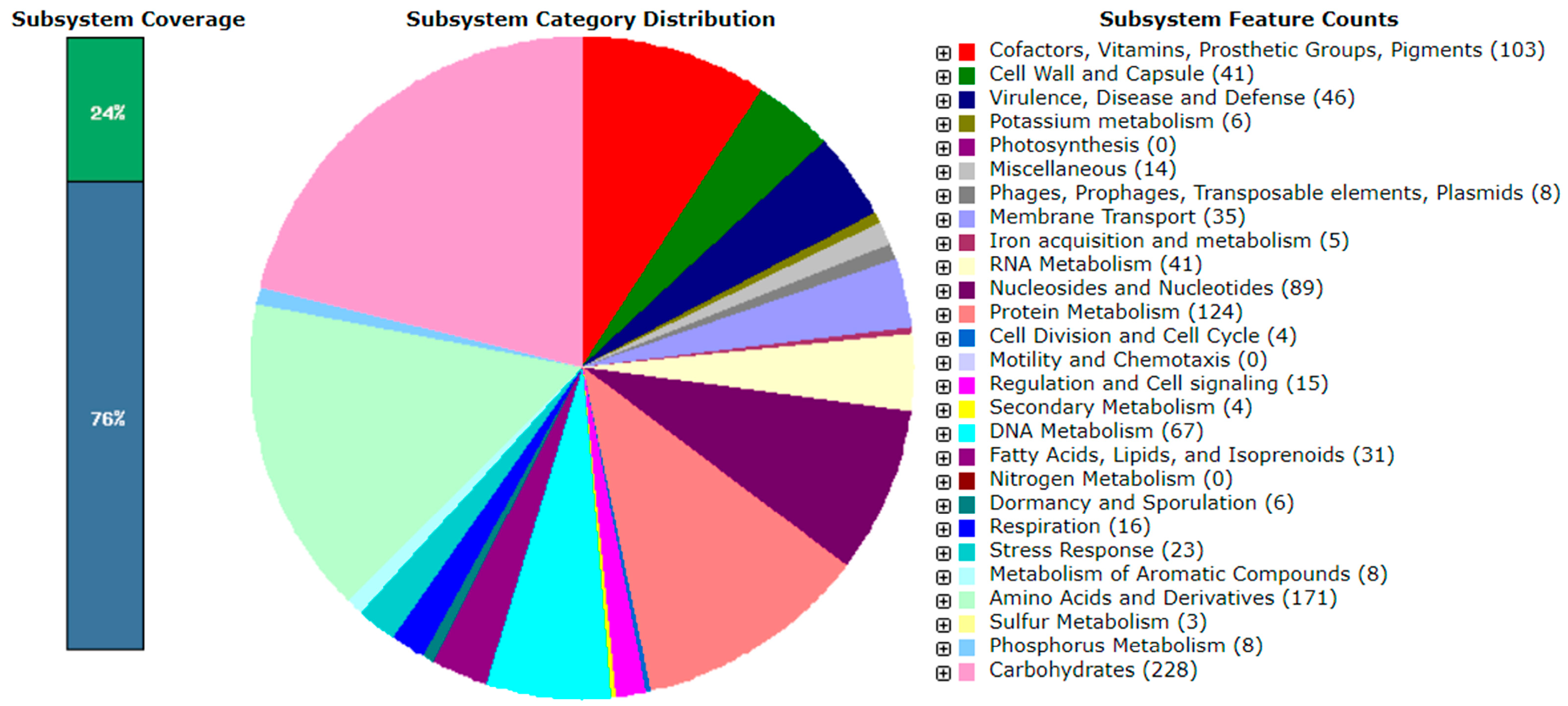
2.4. Genome Assembly, Annotation, Phylogenetic and Functional Analysis
2.5. Atomic Force Microscopy
2.6. Assessment of Probiotic Properties of L. Plantarum FCa3L
2.6.1. Acid and Bile Tolerance
2.6.2. Antagonistic Activity
2.6.3. Acidification Rate
2.6.4. H2O2 Determination
2.6.5. MATS Method
2.6.6. Autoaggregation Assay
2.6.7. Antibiotic Resistance
2.6.8. Antioxidant Activity
2.7. Statistical Analysis
3. Results and Discussion
3.1. L. plantarum FCa3L Identification and Morphology
3.2. The Genome of L. plantarum FCa3L
3.3. Probiotic Properties of L. plantarum FCa3L
3.3.1. Tolerance of L. plantarum FCa3L to Acid and Bile In Vitro
3.3.2. The Antibacterial, Acidifying, and Antioxidant Activities of L. plantarum FCa3L
3.3.3. Cell Surface Properties and Adhesiveness
3.3.4. Antibiotic Susceptibility
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration. GRAS Notices. 2022. Available online: https://www.cfsanappsexternal.fda.gov/scripts/fdcc/index.cfm?set=GRASNotices (accessed on 12 April 2023).
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Hilbert, F.; et al. Update of the list of QPS-recommended microbiological agents intentionally added to food or feed as notified to EFSA 16: Suitability of taxonomic units notified to EFSA until March 2022. EFSA J. 2022, 20, e07408. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, B.; Bangar, S.P.; Echegaray, N.; Suri, S.; Tomasevic, I.; Lorenzo, J.M.; Ozogul, F. The impacts of Lactiplantibacillus plantarum on the functional properties of fermented foods: A review of current knowledge. Microorganisms 2022, 10, 826. [Google Scholar] [CrossRef] [PubMed]
- Ladha, G.; Jeevaratnam, K. A novel antibacterial compound produced by Lactobacillus plantarum LJR13 isolated from rumen liquor of goat effectively controls multi-drug resistant human pathogens. Microbiol. Res. 2020, 241, 126563. [Google Scholar] [CrossRef]
- Algboory, H.L.; Muhialdin, B.J. Novel peptides contribute to the antimicrobial activity of camel milk fermented with Lactobacillus plantarum IS10. Food Control. 2021, 126, 108057. [Google Scholar] [CrossRef]
- Tenea, G.N.; Ascanta, P. Bioprospecting of ribosomally synthesized and post-translationally modified peptides through genome characterization of a novel probiotic Lactiplantibacillus plantarum UTNGt21A strain: A promising natural antimicrobials factory. Front. Microbiol. 2022, 13, 868025. [Google Scholar] [CrossRef]
- Fidanza, M.; Panigrahi, P.; Kollmann, T.R. Lactiplantibacillus plantarum–Nomad and ideal probiotic. Front. Microbiol. 2021, 12, 712236. [Google Scholar] [CrossRef]
- Garcia-Gonzalez, N.; Battista, N.; Prete, R.; Corsetti, A. Health-promoting role of Lactiplantibacillus plantarum isolated from fermented foods. Microorganisms 2021, 9, 349. [Google Scholar] [CrossRef]
- Echegaray, N.; Yilmaz, B.; Sharma, H.; Kumar, M.; Pateiro, M.; Ozogul, F.; Lorenzo, J.M. A novel approach to Lactiplantibacillus plantarum: From probiotic properties to the omics insights. Microbiol. Res. 2023, 268, 127289. [Google Scholar] [CrossRef]
- Report of a Joint FAO/WHO Expert Consultation. Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria. 2001. Available online: http://www.fao.org/tempref/docrep/fao/meeting/009/y6398e.pdf (accessed on 12 April 2023).
- Ducrotte, P.; Sawant, P.; Jayanthi, V. Clinical trial: Lactobacillus plantarum 299v (DSM 9843) improves symptoms of irritable bowel syndrome. World J. Gastroenterol. 2012, 18, 4012–4018. [Google Scholar] [CrossRef]
- Yang, B.; Yue, Y.; Chen, Y.; Ding, M.; Li, B.; Wang, L.; Wang, Q.; Stanton, C.; Ross, R.P.; Zhao, J.; et al. Lactobacillus plantarum CCFM1143 alleviates chronic diarrhea via inflammation regulation and gut microbiota modulation: A double-blind, randomized, placebo-controlled study. Front. Immunol. 2021, 12, 746585. [Google Scholar] [CrossRef] [PubMed]
- Prakoeswa, C.R.S.; Bonita, L.; Karim, A.; Herwanto, N.; Umborowati, M.A.; Setyaningrum, T.; Hidayati, A.N.; Surono, I.S. Beneficial effect of Lactobacillus plantarum IS-10506 supplementation in adults with atopic dermatitis: A randomized controlled trial. J. Dermatolog. Treat. 2022, 33, 1491–1498. [Google Scholar] [CrossRef] [PubMed]
- Cappello, C.; Acin-Albiac, M.; Pinto, D.; Polo, A.; Filannino, P.; Rinaldi, F.; Gobbetti, M.; Di Cagno, R. Do nomadic lactobacilli fit as potential vaginal probiotics? The answer lies in a successful selective multi-step and scoring approach. Microb. Cell Fact. 2023, 22, 27. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, J.; Tian, Z.; Si, Y.; Chen, H.; Gan, J. The mechanisms of the potential probiotic Lactiplantibacillus plantarum against cardiovascular disease and the recent developments in its fermented foods. Foods 2022, 11, 2549. [Google Scholar] [CrossRef]
- Park, J.; Kwon, M.; Lee, J.; Park, S.; Seo, J.; Roh, S. Anti-cancer effects of Lactobacillus plantarum L-14 cell-free extract on human malignant melanoma A375 cells. Molecules 2020, 25, 3895. [Google Scholar] [CrossRef]
- Botta, C.; Spyridopoulou, K.; Bertolino, M.; Rantsiou, K.; Chlichlia, K.; Cocolin, L. Lactiplantibacillus plantarum inhibits colon cancer cell proliferation as function of its butyrogenic capability. Biomed. Pharmacother. 2022, 149, 112755. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, S.; Jiang, Y.; Zhao, Z.; Shen, Y.; Zhang, J.; Zhao, L. Neuroprotective effect of Lactobacillus plantarum DP189 on MPTP-induced Parkinson’s disease model mice. J. Funct. Foods 2021, 85, 104635. [Google Scholar] [CrossRef]
- Lee, Y.Z.; Cheng, S.-H.; Chang, M.-Y.; Lin, Y.-F.; Wu, C.-C.; Tsai, Y.-C. Neuroprotective effects of Lactobacillus plantarum PS128 in a mouse model of Parkinson’s disease: The role of gut microbiota and microRNAs. Int. J. Mol. Sci. 2023, 24, 6794. [Google Scholar] [CrossRef]
- Kleerebezem, M.; Boekhorst, J.; van Kranenburg, R.; Molenaar, D.; Kuipers, O.P.; Leer, R.; Tarchini, R.; Peters, S.A.; Sandbrink, H.M.; Fiers, M.W.; et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 2003, 100, 1990–1995. [Google Scholar] [CrossRef]
- van den Nieuwboer, M.; van Hemert, S.; Claassen, E.; de Vos, W.M. Lactobacillus plantarum WCFS1 and its host interaction: A dozen years after the genome. Microb. Biotech. 2016, 9, 452–465. [Google Scholar] [CrossRef]
- Ventura, M.; Turroni, F.; van Sinderen, D. Probiogenomics as a tool to obtain genetic insights into adaptation of probiotic bacteria to the human gut. Bioeng. Bugs 2012, 3, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Evanovich, E.; de Souza Mendonça Mattos, P.J.; Guerreiro, J.F. Comparative genomic analysis of Lactobacillus plantarum: An overview. Int. J. Genom. 2019, 2019, 4973214. [Google Scholar] [CrossRef]
- Martino, M.E.; Bayjanov, J.R.; Caffrey, B.E.; Wels, M.; Joncour, P.; Hughes, S.; Gillet, B.; Kleerebezem, M.; van Hijum, S.A.; Leulier, F. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Envirol. Microbiol. 2016, 18, 4974–4989. [Google Scholar] [CrossRef] [PubMed]
- Filannino, P.; De Angelis, M.; Di Cagno, R.; Gozzi, G.; Riciputi, Y.; Gobbetti, M. How Lactobacillus plantarum shapes its transcriptome in response to contrasting habitats. Envirol. Microbiol. 2018, 20, 3700–3716. [Google Scholar] [CrossRef]
- Ramos, C.L.; Thorsen, L.; Schwan, R.F.; Jespersen, L. Strain-specific probiotics properties of Lactobacillus fermentum, Lactobacillus plantarum and Lactobacillus brevis isolates from Brazilian food products. Food Microbiol. 2013, 36, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, E.A.; Yarullina, D.R.; Ilinskaya, O.N. Antagonistic activity of lactobacilli isolated from natural ecotopes. Microbiology 2017, 86, 708–713. [Google Scholar] [CrossRef]
- Tsapieva, A.; Duplik, N.; Suvorov, A. Structure of plantaricin locus of Lactobacillus plantarum 8P-A3. Benef. Microbes 2011, 2, 255–261. [Google Scholar] [CrossRef]
- Anisimova, E.A.; Yarullina, D.R. Antibiotic resistance of Lactobacillus strains. Curr. Microbiol. 2019, 76, 1407–1416. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 12 April 2023).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Overbeek, R.; Begley, T.; Butler, R.M.; Choudhuri, J.V.; Chuang, H.Y.; Cohoon, M.; de Crécy-Lagard, V.; Diaz, N.; Disz, T.; Edwards, R.; et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005, 33, 5691–5702. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Stern, N.J.; Svetoch, E.A.; Eruslanov, B.V.; Perelygin, V.V.; Mitsevich, E.V.; Mitsevich, I.P.; Pokhilenko, V.D.; Levchuk, V.P.; Svetoch, O.E.; Seal, B.S. Isolation of a Lactobacillus salivarius strain and purification of its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrob. Agents Chemother. 2006, 50, 3111–3116. [Google Scholar] [CrossRef] [PubMed]
- Minnullina, L.; Pudova, D.; Shagimardanova, E.; Shigapova, L.; Sharipova, M.; Mardanova, A. Comparative genome analysis of uropathogenic Morganella morganii strains. Front. Cell Infect. Microbiol. 2019, 9, 167. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.H.; Guan, J.J.; Wang, J.S.; Yin, H.C.; Qiao, F.D.; Jia, F. Physicochemical and sensory characterization of ginger-juice yogurt during fermentation. Food Sci. Biotechnol. 2012, 21, 1541–1548. [Google Scholar] [CrossRef]
- Juarez Tomas, M.S.; Claudia Otero, M.; Ocana, V.; NaderMacias, M.E. Production of antimicrobial substances by lactic acid bacteria I: Determination of hydrogen peroxide. Methods Mol. Biol. 2004, 268, 337–346. [Google Scholar] [CrossRef]
- Rosenberg, M.; Gutnick, D.; Rosenberg, E. Adherence of bacteria to hydrocarbons: A simple method for measuring cell-surface hydrophobicity. FEMS Microbiol. Lett. 1980, 9, 29–33. [Google Scholar] [CrossRef]
- Kirillova, A.V.; Danilushkina, A.A.; Irisov, D.S.; Bruslik, N.L.; Fakhrullin, R.F.; Zakharov, Y.A.; Bukhmin, V.S.; Yarullina, D.R. Assessment of resistance and bioremediation ability of Lactobacillus strains to lead and cadmium. Int. J. Microbiol. 2017, 2017, 9869145. [Google Scholar] [CrossRef]
- Kos, B.; Susković, J.; Vuković, S.; Simpraga, M.; Frece, J.; Matosić, S. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 2003, 94, 981–987. [Google Scholar] [CrossRef]
- Charteris, W.P.; Kelly, P.M.; Morelli, L.; Collins, J.K. Antibiotic susceptibility of potentially probiotic Lactobacillus species. J. Food Prot. 1998, 61, 1636–1643. [Google Scholar] [CrossRef]
- Melo, T.A.; Dos Santos, T.F.; Pereira, L.R.; Passos, H.M.; Rezende, R.P.; Romano, C.C. Functional profile evaluation of Lactobacillus fermentum TCUESC01: A new potential probiotic strain isolated during cocoa fermentation. Biomed Res. Int. 2017, 2017, 5165916. [Google Scholar] [CrossRef]
- Blois, M.S. Antioxidant determinations by the use of a stable free radical. Nature 1958, 181, 1199–1200. [Google Scholar] [CrossRef]
- Hwang, I.C.; Kim, S.H.; Kang, D.K. Complete genome sequence of Lactobacillus plantarum SK156, a candidate vehicle for mucosal vaccine delivery. J. Anim. Sci. Technol. 2020, 62, 956–958. [Google Scholar] [CrossRef] [PubMed]
- Arena, M.P.; Fiocco, D.; Massa, S.; Capozzi, V.; Russo, P.; Spano, G. Lactobacillus plantarum as a strategy for an in situ production of vitamin B2. J. Food Nutr. Disor. 2014, S1-004. [Google Scholar] [CrossRef]
- Sanger, F.; Air, G.M.; Barrell, B.G.; Brown, N.L.; Coulson, A.R.; Fiddes, C.A.; Hutchison, C.A.; Slocombe, P.M.; Smith, M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature 1977, 265, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Kiani, A.K.; Anpilogov, K.; Dautaj, A.; Marceddu, G.; Sonna, W.N.; Percio, M.; Dundar, M.; Beccari, T.; Bertelli, M. Bacteriophages in food supplements obtained from natural sources. Acta Biomed. 2020, 91, e2020025. [Google Scholar] [CrossRef]
- Ermolenko, E.I.; Isakov, V.A.; Zhdan-Pushkina, S.K.; Tets, V.V. Kolichestvennaia otsenka antagonisticheskoĭ aktivnosti lactobatsill [Quantitative characterization of antagonistic activity of lactobacilli]. Zh. Mikrobiol. Epidemiol. Immunobiol. 2004, 5, 94–98. [Google Scholar]
- Van Loosdrecht, M.; Norde, W.; Zehnder, A. Physical chemical description of bacterial adhesion. J. Biomater. Appl. 1990, 5, 91–106. [Google Scholar] [CrossRef]
- de Wouters, T.; Jans, C.; Niederberger, T.; Fischer, P.; Rühs, P.A. Adhesion potential of intestinal microbes predicted by physico-chemical characterization methods. PLoS ONE 2015, 10, e0136437. [Google Scholar] [CrossRef]
- Li, J.; McLandsborough, L.A. The effects of the surface charge and hydrophobicity of Escherichia coli on its adhesion to beef muscle. Int. J. Food. Microbiol. 1999, 53, 185–193. [Google Scholar] [CrossRef]
- Pelletier, C.; Bouley, C.; Cayuela, C.; Bouttier, S.; Bourlioux, P.; Bellon-Fontaine, M.N. Cell surface characteristics of Lactobacillus casei subsp. casei, Lactobacillus paracasei subsp. paracasei, and Lactobacillus rhamnosus strains. Appl. Environ. Microbiol. 1997, 63, 1725–1731. [Google Scholar] [CrossRef]
- Monteagudo-Mera, A.; Rastall, R.A.; Gibson, G.R.; Charalampopoulos, D.; Chatzifragkou, A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 2019, 103, 6463–6472. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Bawazir, M.; Dhall, A.; Kim, H.E.; He, L.; Heo, J.; Hwang, G. Implication of surface properties, bacterial motility, and hydrodynamic conditions on bacterial surface sensing and their initial adhesion. Front. Bioeng. Biotechnol. 2021, 9, 643722. [Google Scholar] [CrossRef] [PubMed]
- Karagulyan, M.; Goebel, M.O.; Diehl, D.; Abu Quba, A.A.; Kästner, M.; Bachmann, J.; Wick, L.Y.; Schaumann, G.E.; Miltner, A. Water stress-driven changes in bacterial cell surface properties. Appl. Environ. Microbiol. 2022, 88, e0073222. [Google Scholar] [CrossRef] [PubMed]
- Kemgang, T.S.; Kapila, S.; Shanmugam, V.P.; Kapila, R. Cross-talk between probiotic lactobacilli and host immune system. J. Appl. Microbiol. 2014, 117, 303–319. [Google Scholar] [CrossRef] [PubMed]
- Tuo, Y.; Yu, H.; Ai, L.; Wu, Z.; Guo, B.; Chen, W. Aggregation and adhesion properties of 22 Lactobacillus strains. J. Dairy Sci. 2013, 96, 4252–4257. [Google Scholar] [CrossRef]
- Bouchard, D.S.; Seridan, B.; Saraoui, T.; Rault, L.; Germon, P.; Gonzalez-Moreno, C.; Nader-Macias, F.M.; Baud, D.; François, P.; Chuat, V.; et al. Lactic acid bacteria isolated from bovine mammary microbiota: Potential allies against bovine mastitis. PLoS ONE 2015, 10, e0144831. [Google Scholar] [CrossRef]
- Kaushik, J.K.; Kumar, A.; Duary, R.K.; Mohanty, A.K.; Grover, S.; Batish, V.K. Functional and probiotic attributes of an indigenous isolate of Lactobacillus plantarum. PLoS ONE 2009, 4, e8099. [Google Scholar] [CrossRef]
- Remus, D.M.; Bongers, R.S.; Meijerink, M.; Fusetti, F.; Poolman, B.; de Vos, P.; Wells, J.M.; Kleerebezem, M.; Bron, P.A. Impact of Lactobacillus plantarum sortase on target protein sorting, gastrointestinal persistence, and host immune response modulation. J. Bacteriol. 2013, 195, 502–509. [Google Scholar] [CrossRef]
- Walter, J.; Schwab, C.; Loach, D.M.; Gänzle, M.G.; Tannock, G.W. Glucosyltransferase A (GtfA) and inulosucrase (Inu) of Lactobacillus reuteri TMW1.106 contribute to cell aggregation, in vitro biofilm formation, and colonization of the mouse gastrointestinal tract. Microbiology 2008, 154, 72–80. [Google Scholar] [CrossRef]
- Anisimova, E.; Yarullina, D. Characterization of erythromycin and tetracycline resistance in Lactobacillus fermentum strains. Int. J. Microbiol. 2018, 2018, 3912326. [Google Scholar] [CrossRef]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Clarke, S.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O’Toole, P.W. Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2018, 85, e01738-18. [Google Scholar] [CrossRef] [PubMed]



| Probiotic Properties | FCa3L | 8PA3 |
|---|---|---|
| Survival in GI tract, % 1 | ||
| Ox gall 2% | ||
| 2 h | 103.8 | 113.7 |
| 4 h | 65.0 | 52.9 |
| HCl, pH = 2 | ||
| 2 h | 12.8 | 15.2 |
| 4 h | 11.3 | 10.0 |
| Adhesion | ||
| % of adhesion ± SD to: | ||
| Hexadecane | 21.9 ± 1.6 | 24.9 ± 8.9 |
| Ethyl acetate | 25.9 ± 3.7 | 22.9 ± 3.2 |
| Chloroform | 92.8 ± 3.2 | 87.9 ± 2.9 |
| AFM analysis of the surface structure and nonspecific adhesion of bacterial cells | ||
| Surface roughness (500 × 500 nm), Sq, nm | 4.5 ± 1.6 * | 1.9 ± 0.6 |
| Surface roughness (500 × 500 nm), Sa, nm | 3.4 ± 1.3 | 1.5 ± 0.5 |
| Nonspecific adhesion of the biofilm surface (20 × 20 µm), nN | 3.1 ± 1.1 | 6.8 ± 2.7 |
| Nonspecific adhesion of the cell surface (500 × 500 nm), nN | 2.0 ± 0.1 * | 11.4 ± 2.8 |
| Auto-aggregation, % | ||
| 4 h | 58.7 ± 7.9 * | 23.1 ± 4.2 |
| 24 h | 67.2 ± 3.7 | 72.7 ± 8.4 |
| Total titratable acidity (TTA), mmol/mL | 1.46 ± 0.15 | 1.37 ± 0.23 |
| H2O2 production | ||
| TMB assay | Present | Present |
| XO assay, µM | 53.3 ± 2.3 | 58.0 ± 5.0 |
| Antioxidant activity, % | 9.1 ± 4.7 | 20.5 ± 5.9 |
| Indicator Microorganisms | FCa3L | 8PA3 |
|---|---|---|
| Gram-positive | ||
| Bacillus cereus (Clinical isolate) | 8.60 ± 1.14 * | 2.75 ± 0.50 |
| Enterococcus faecalis (Clinical isolate) | 3.40 ± 0.89 | 1.25 ± 0.96 |
| Micrococcus luteus (Clinical isolate) | 1.20 ± 0.84 | 0.75 ± 0.50 |
| Staphylococcus aureus ATCC 29213 | 7.20 ± 0.45 * | 1.75 ± 0.95 |
| Gram-negative | ||
| Escherichia coli K-12 | 5.20 ± 0.84 * | 3.00 ± 0.82 |
| Pseudomonas aeruginosa ATCC 27853 | 5.00 ± 0.71 * | 1.25 ± 0.50 |
| Klebsiella pneumonia (Clinical isolate) | 2.80 ± 0.45 | 2.00 ± 1.41 |
| Morganella morganii MM190 (Clinical isolate) | 8.50 ± 0.00 * | 5.25 ± 0.50 |
| Serratia marcescens (Clinical isolate) | 1.80 ± 0.45 | 1.50 ± 0.57 |
| Antibiotics | Amount per disc, μg | LAB Strains | |
|---|---|---|---|
| FCa3L | 8PA3 | ||
| Ampicillin | 10 | S (25.0 ± 3.5) | S (43.0 ± 2.8) |
| Amikacin | 30 | R (5.0 ± 0.0) | R (7.0 ± 2.8) |
| Chloramphenicol | 30 | S (28.0 ± 2.8) | S (19.0 ± 3.0) |
| Ciprofloxacin | 5 | R (6.0 ± 0.0) | R (7.0 ± 2.0) |
| Clindamycin | 2 | S (12.5 ± 3.5) | MS (11.5 ± 2.1) |
| Erythromycin | 15 | S (20.5 ± 1.4) | MS (14.0 ± 3.5) |
| Gentamicin | 10 | R (5.5 ± 0.7) | R (5.0 ± 0.0) |
| Kanamycin | 30 | R (6.0 ± 0.0) | R (6.0 ± 0.0) |
| Rifampicin | 5 | S (21.0 ± 0.0) | S (20.5 ± 0.7) |
| Streptomycin | 30 | R (5.5 ± 0.7) | R (7.0 ± 2.8) |
| Tetracycline | 30 | S (19.0 ± 1.4) | S (26.0 ± 4.0) |
| Vancomycin | 30 | R (5.0 ± 0.0) | S (21.0 ± 4.2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karaseva, O.; Ozhegov, G.; Khusnutdinova, D.; Siniagina, M.; Anisimova, E.; Akhatova, F.; Fakhrullin, R.; Yarullina, D. Whole Genome Sequencing of the Novel Probiotic Strain Lactiplantibacillus plantarum FCa3L. Microorganisms 2023, 11, 1234. https://doi.org/10.3390/microorganisms11051234
Karaseva O, Ozhegov G, Khusnutdinova D, Siniagina M, Anisimova E, Akhatova F, Fakhrullin R, Yarullina D. Whole Genome Sequencing of the Novel Probiotic Strain Lactiplantibacillus plantarum FCa3L. Microorganisms. 2023; 11(5):1234. https://doi.org/10.3390/microorganisms11051234
Chicago/Turabian StyleKaraseva, Olga, Georgii Ozhegov, Dilyara Khusnutdinova, Maria Siniagina, Elizaveta Anisimova, Farida Akhatova, Rawil Fakhrullin, and Dina Yarullina. 2023. "Whole Genome Sequencing of the Novel Probiotic Strain Lactiplantibacillus plantarum FCa3L" Microorganisms 11, no. 5: 1234. https://doi.org/10.3390/microorganisms11051234
APA StyleKaraseva, O., Ozhegov, G., Khusnutdinova, D., Siniagina, M., Anisimova, E., Akhatova, F., Fakhrullin, R., & Yarullina, D. (2023). Whole Genome Sequencing of the Novel Probiotic Strain Lactiplantibacillus plantarum FCa3L. Microorganisms, 11(5), 1234. https://doi.org/10.3390/microorganisms11051234








