Improvement of Carotenoids’ Production by Increasing the Activity of Beta-Carotene Ketolase with Different Strategies
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain, Culture Medium, and Culture Conditions
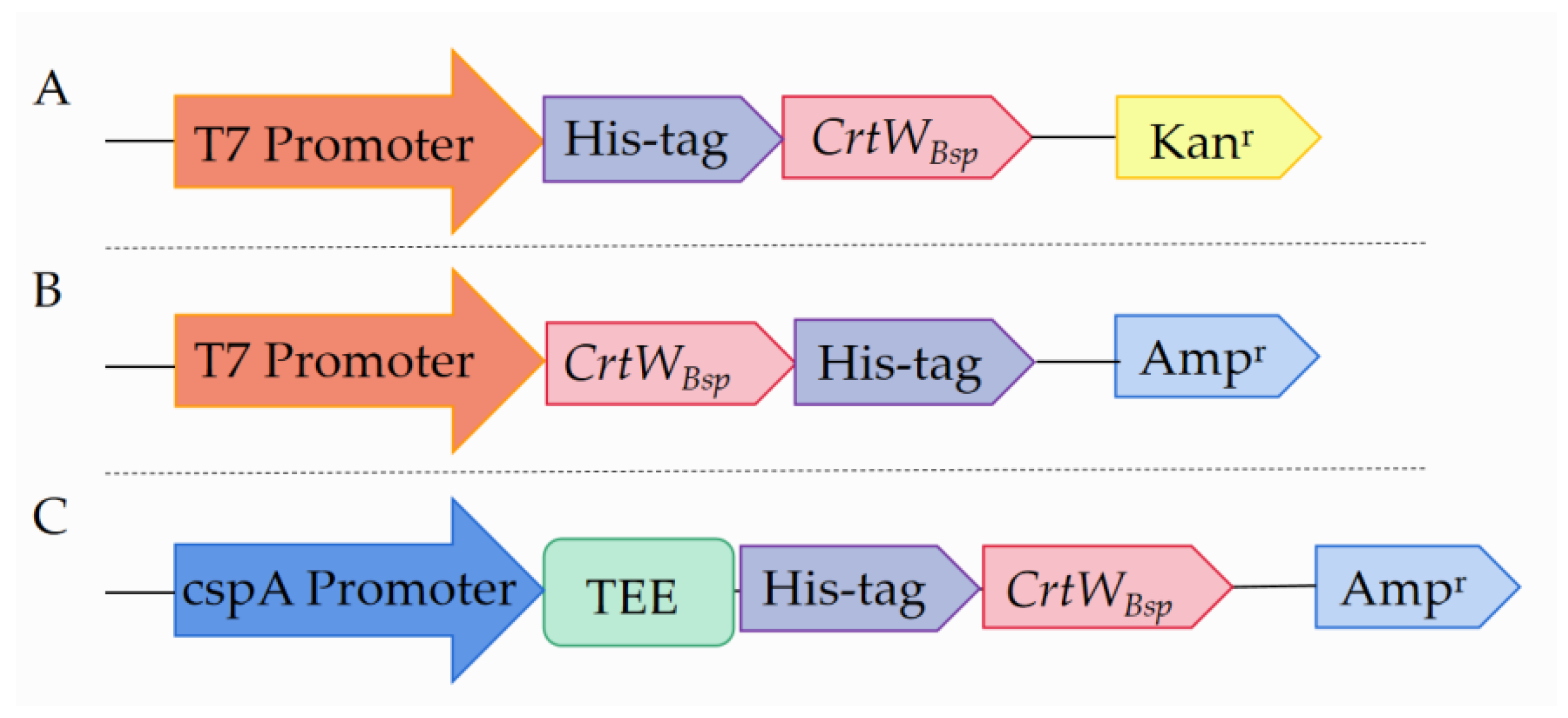
2.2. Construction of Plasmids
2.3. Induced Production of Recombinant Protein
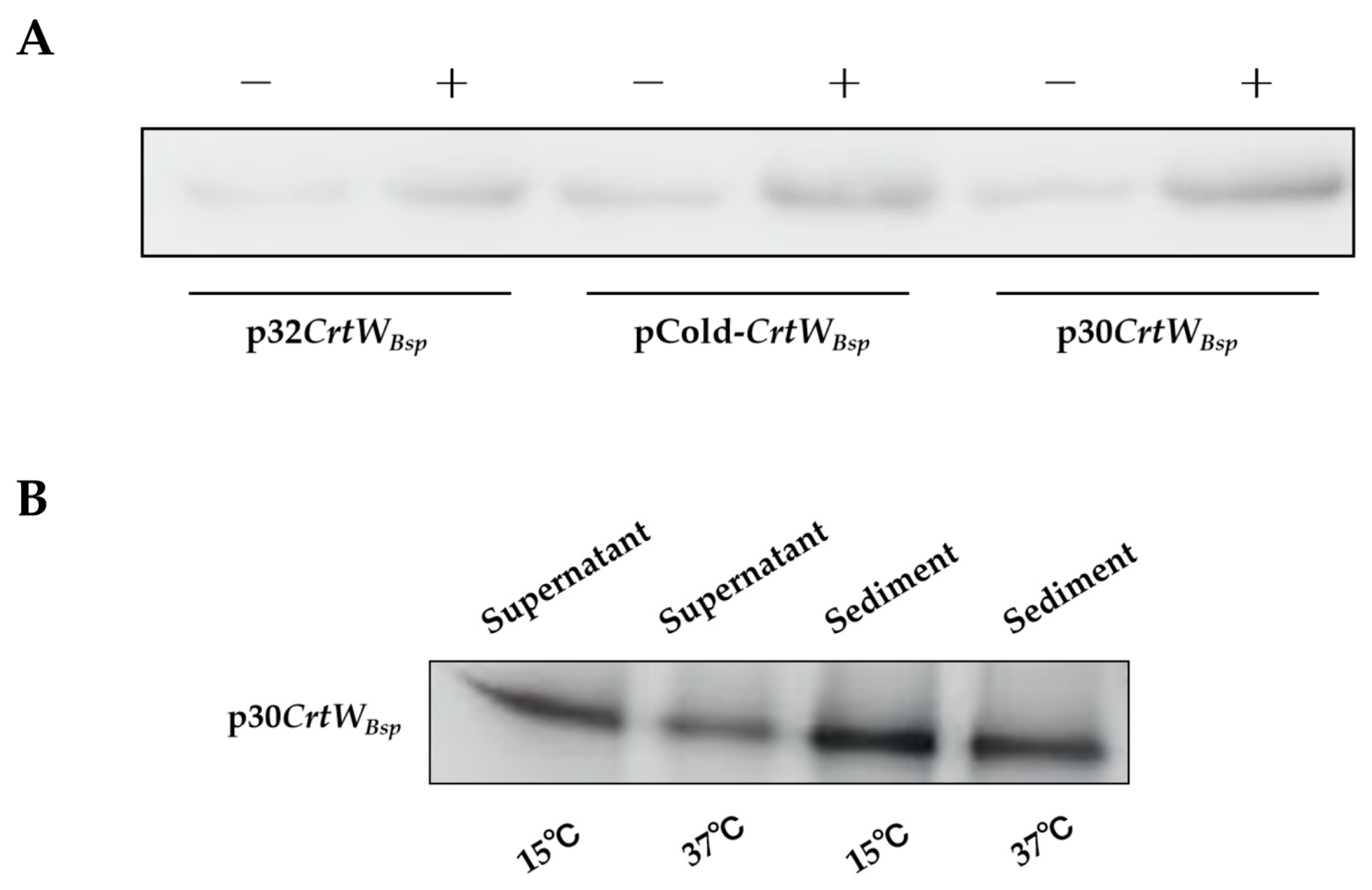
2.4. Western Blot Analysis
2.5. Enzymatic Reaction In Vitro
2.6. Carotenoids Production in E. coli
2.7. Preparation of Enzyme Reaction Solution
2.8. HPLC Analysis
2.9. Data Analysis and Statistics
3. Results
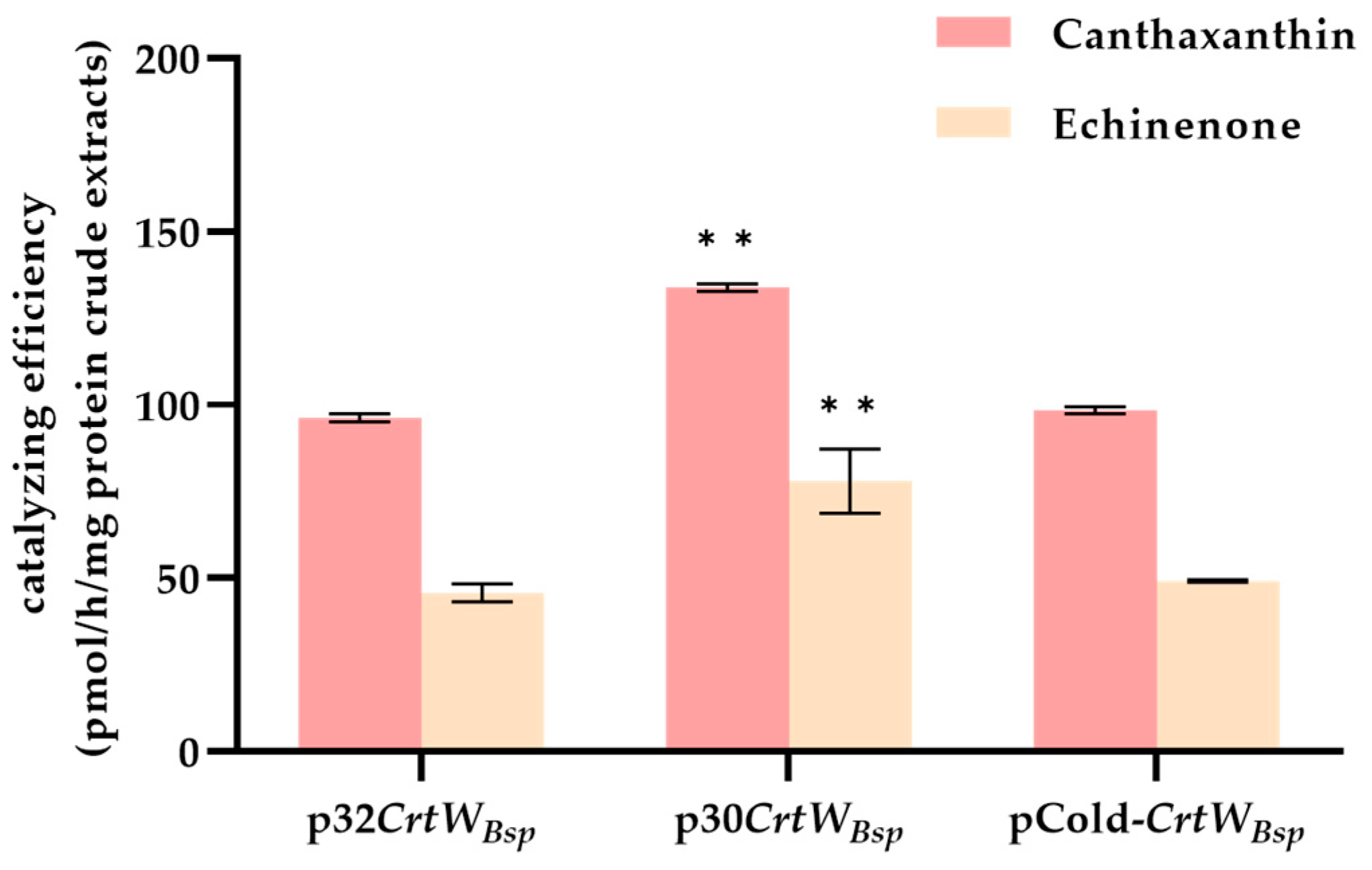
3.1. Soluble Production and Activity of β-Carotene Ketolase Using Different Vectors
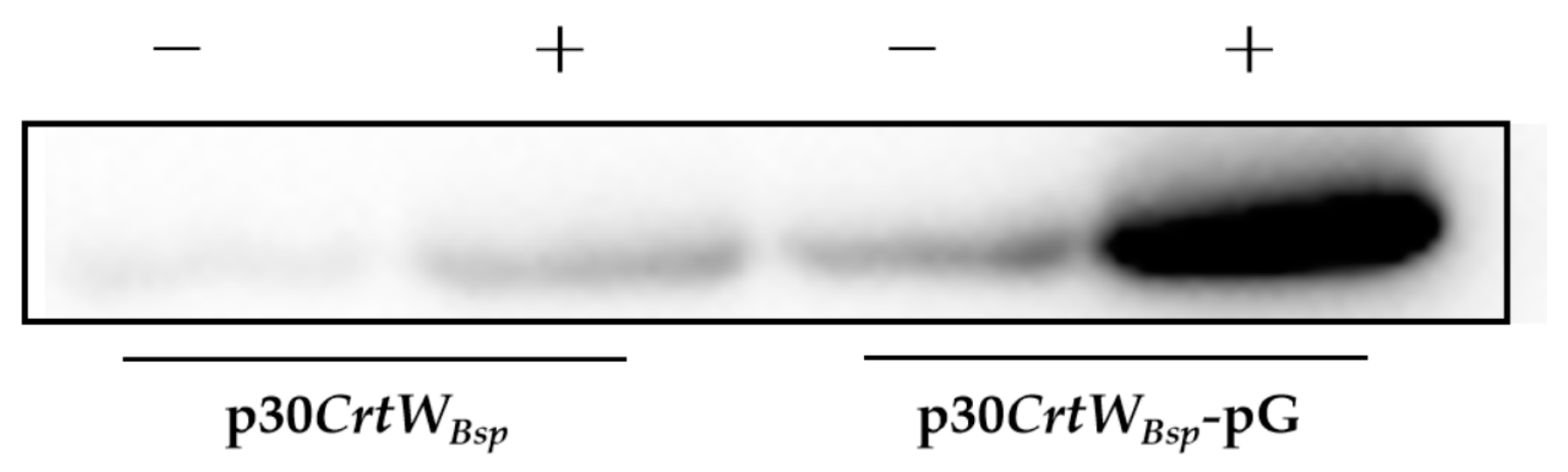
3.2. Effect of Molecular Chaperone on the Soluble Production and Activity of β-Carotene Ketolase
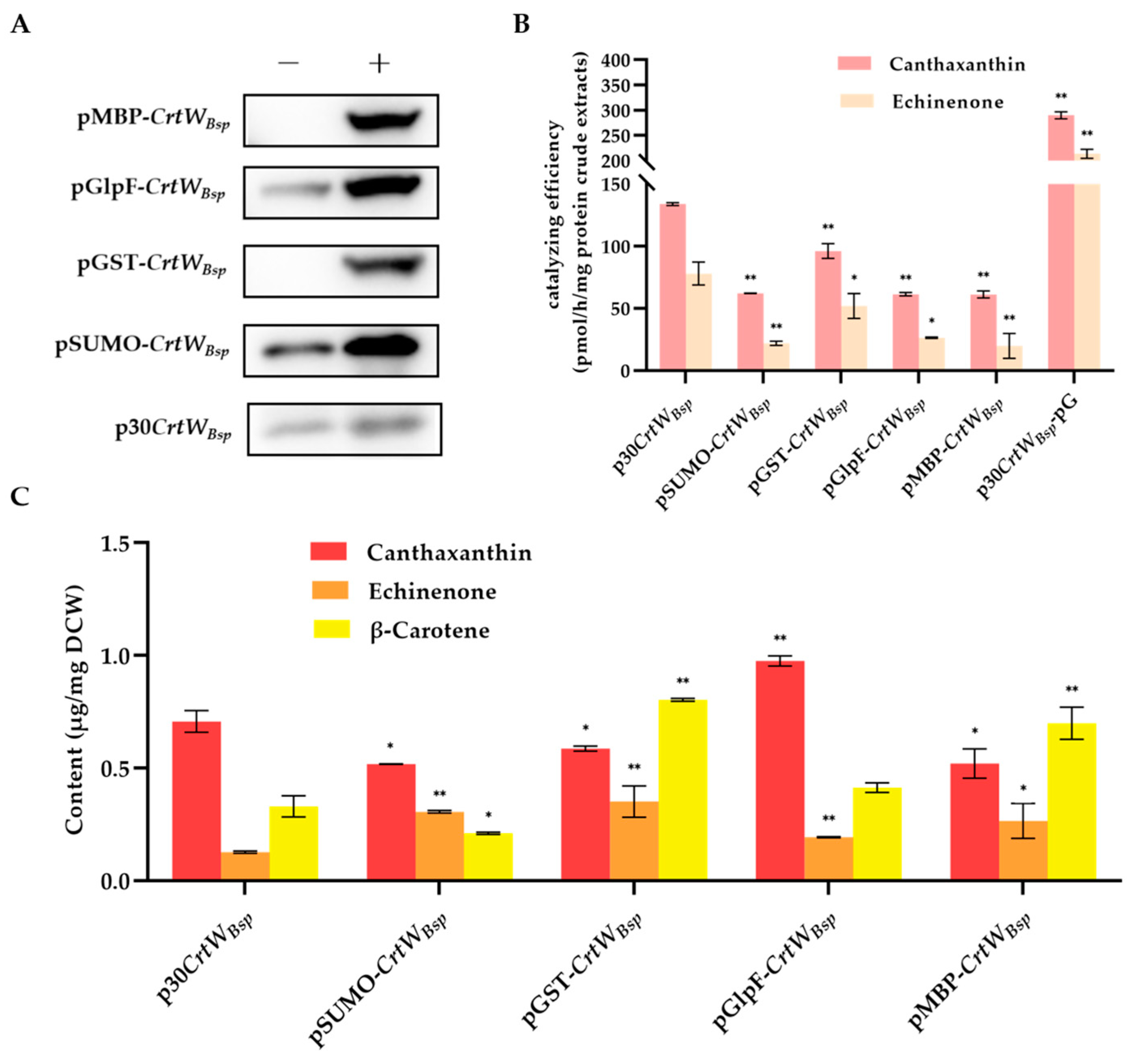
3.3. Effect of Fusion Tags on Soluble Production and Activity of β-Carotene Ketolase
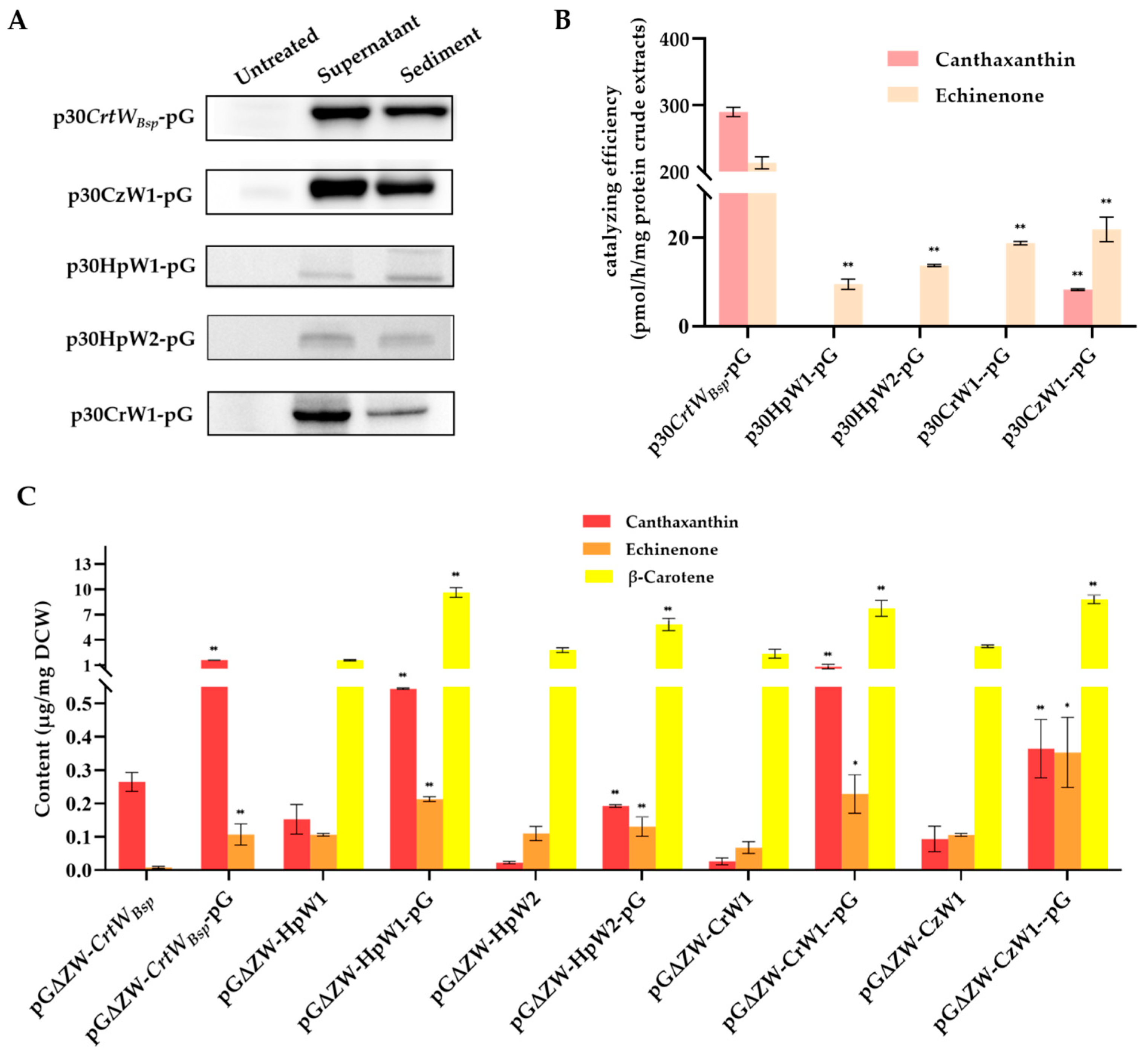
3.4. Effect of Molecular Chaperone on the Soluble Production and Activity of Other β-Carotene Ketolases Derived from Microalgae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hua-Bin, L.; Fan, K.W.; Chen, F. Isolation and purification of canthaxanthin from the microalga Chlorella zofingiensis by high-speed counter-current chromatography. J. Sep. Sci. 2006, 29, 699–703. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Concepcion, M.; Avalos, J.; Bonet, M.L.; Boronat, A.; Gomez-Gomez, L.; Hornero-Mendez, D.; Limon, M.C.; Meléndez-Martínez, A.J.; Olmedilla-Alonso, B.; Palou, A.; et al. A global perspective on carotenoids: Metabolism, biotechnology, and benefits for nutrition and health. Prog. Lipid Res. 2018, 70, 62–93. [Google Scholar] [CrossRef] [PubMed]
- Ashokkumar, V.; Flora, G.; Sevanan, M.; Sripriya, R.; Chen, W.H.; Park, J.-H.; Rajesh banu, J.; Kumar, G. Technological advances in the production of carotenoids and their applications—A critical review. Bioresour. Technol. 2023, 367, 128215. [Google Scholar] [CrossRef] [PubMed]
- Bernier, S.C.; Cantin, L.; Salesse, C. Systematic analysis of the expression, solubility and purification of a passenger protein in fusion with different tags. Protein Expr. Purif. 2018, 152, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Joshi, K.; Kumar, P.; Kataria, R. Microbial carotenoid production and their potential applications as antioxidants: A current update. Process Biochem. 2023, 128, 190–205. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Duan, C.; Yi, S.; Gao, Z.; Xiao, C.; Agathos, S.N.; Wang, G.; Li, J. Biotechnological production of astaxanthin from the microalga Haematococcus pluvialis. Biotechnol. Adv. 2020, 43, 107602. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Meng, C.; Sun, F.; Wei, Z.; Chen, L.; Chen, W.; Tong, S.; Du, H.; Gao, J.; Ren, J.; et al. Sustainable production of astaxanthin in microorganisms: The past, present, and future. Crit. Rev. Food Sci. Nutr. 2023, 63, 10239–10255. [Google Scholar] [CrossRef]
- Choi, S.K.; Nishida, Y.; Matsuda, S.; Adachi, K.; Kasai, H.; Peng, X.; Komemushi, S.; Miki, W.; Misawa, N. Characterization of beta-carotene ketolases, CrtW, from marine bacteria by complementation analysis in Escherichia coli. Mar. Biotechnol. 2005, 7, 515–522. [Google Scholar] [CrossRef]
- Cordwell, S.J. Technologies for bacterial surface proteomics. Curr. Opin. Microbiol. 2006, 9, 320–329. [Google Scholar] [CrossRef]
- Costa, S.; Almeida, A.; Castro, A.; Domingues, L. Fusion tags for protein solubility, purification and immunogenicity in Escherichia coli: The novel Fh8 system. Front. Microbiol. 2014, 5, 63. [Google Scholar] [CrossRef]
- Esposito, D.; Chatterjee, D.K. Enhancement of soluble protein expression through the use of fusion tags. Curr. Opin. Biotechnol. 2006, 17, 353–358. [Google Scholar] [CrossRef]
- Du, F.; Liu, Y.Q.; Xu, Y.S.; Li, Z.J.; Wang, Y.Z.; Zhang, Z.X.; Sun, X.M. Regulating the T7 RNA polymerase expression in E. coli BL21 (DE3) to provide more host options for recombinant protein production. Microb. Cell Fact. 2021, 20, 189. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.O. Guiding tail-anchored membrane proteins to the endoplasmic reticulum in a chaperone cascade. J. Biol. Chem. 2019, 294, 16577–16586. [Google Scholar] [CrossRef] [PubMed]
- Rabouille, C. Pathways of Unconventional Protein Secretion. Trends Cell Biol. 2017, 27, 230–240. [Google Scholar] [CrossRef]
- Chopra, A.K.; Brasier, A.R.; Das, M.; Xu, X.J.; Peterson, J.W. Improved synthesis of Salmonella typhimurium enterotoxin using gene fusion expression systems. Gene 1994, 144, 81–85. [Google Scholar] [CrossRef]
- Kaur, J.; Kumar, A.; Kaur, J. Strategies for optimization of heterologous protein expression in E. coli: Roadblocks and reinforcements. Int. J. Biol. Macromol. 2018, 106, 803–822. [Google Scholar] [CrossRef]
- Chen, J.-P.; Gong, J.-S.; Su, C.; Li, H.; Xu, Z.-H.; Shi, J.-S. Improving the soluble expression of difficult-to-express proteins in prokaryotic expression system via protein engineering and synthetic biology strategies. Metab. Eng. 2023, 78, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Adnan, M.; Ma, X.; Olsson, S.; Wang, J.; Liu, G. Promoter regulation and genetic engineering strategies for enhanced cellulase expression in Trichoderma reesei. Microbiol. Res. 2022, 259, 127011. [Google Scholar] [CrossRef]
- Fraser, P.D.; Shimada, H.; Misawa, N. Enzymic confirmation of reactions involved in routes to astaxanthin formation, elucidated using a direct substrate in vitro assay. Eur. J. Biochem. 1998, 252, 229–236. [Google Scholar] [CrossRef]
- Huang, D.; Liu, W.; Li, A.; Wang, C.; Hu, Z. Discovery of Geranylgeranyl Pyrophosphate Synthase (GGPPS) Paralogs from Haematococcus pluvialis Based on Iso-Seq Analysis and Their Function on Astaxanthin Biosynthesis. Mar. Drugs 2019, 17, 696. [Google Scholar] [CrossRef]
- Zhu, C.; Naqvi, S.; Capell, T.; Christou, P. Metabolic engineering of ketocarotenoid biosynthesis in higher plants. Arch. Biochem. Biophys. 2009, 483, 182–190. [Google Scholar] [CrossRef]
- Ye, L.; Zhu, X.; Wu, T.; Wang, W.; Zhao, D.; Bi, C.; Zhang, X. Optimizing the localization of astaxanthin enzymes for improved productivity. Biotechnol. Biofuels 2018, 11, 278. [Google Scholar] [CrossRef]
- Ye, R.W.; Stead, K.J.; Yao, H.; He, H. Mutational and functional analysis of the β-carotene ketolase involved in the production of canthaxanthin and astaxanthin. Appl. Environ. Microbiol. 2006, 72, 5829–5837. [Google Scholar] [CrossRef]
- Otzen, D.E.; Andersen, K.K. Folding of outer membrane proteins. Arch. Biochem. Biophys. 2013, 531, 34–43. [Google Scholar] [CrossRef]
- Tastan, O.; Dutta, A.; Booth, P.; Klein-Seetharaman, J. Retinal proteins as model systems for membrane protein folding. Biochim. Biophys. Acta (BBA)-Bioenerg. 2014, 1837, 656–663. [Google Scholar] [CrossRef]
- Mital, S.; Christie, G.; Dikicioglu, D. Recombinant expression of insoluble enzymes in Escherichia coli: A systematic review of experimental design and its manufacturing implications. Microb. Cell. Fact. 2021, 20, 208. [Google Scholar] [CrossRef]
- Gupta, S.K.; Shukla, P. Advanced technologies for improved expression of recombinant proteins in bacteria: Perspectives and applications. Crit. Rev. Biotechnol. 2016, 36, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Bartolo-Aguilar, Y.; Chávez-Cabrera, C.; Flores-Cotera, L.B.; Badillo-Corona, J.A.; Oliver-Salvador, C.; Marsch, R. The potential of cold-shock promoters for the expression of recombinant proteins in microbes and mammalian cells. J. Genet. Eng. Biotechnol. 2022, 20, 173. [Google Scholar] [CrossRef] [PubMed]
- Camilloni, C.; Bonetti, D.; Morrone, A.; Giri, R.; Dobson, C.M.; Brunori, M.; Gianni, S.; Vendruscolo, M. Towards a structural biology of the hydrophobic effect in protein folding. Sci. Rep. 2016, 6, 28285. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, C.; Liu, H. Enhanced Recombinant Protein Production Under Special Environmental Stress. Front. Microbiol. 2021, 12, 630814. [Google Scholar] [CrossRef]
- Vera, A.; González-Montalbán, N.; Arís, A.; Villaverde, A. The conformational quality of insoluble recombinant proteins is enhanced at low growth temperatures. Biotechnol. Bioeng. 2007, 96, 1101–1106. [Google Scholar] [CrossRef]
- Hewitt, S.N.; Choi, R.; Kelley, A.; Crowther, G.J.; Napuli, A.J.; Van Voorhis, W.C. Expression of proteins in Escherichia coli as fusions with maltose-binding protein to rescue non-expressed targets in a high-throughput protein-expression and purification pipeline. Acta Crystallogr. Sect. F-Struct. Biol. Commun. 2011, 67, 1006–1009. [Google Scholar] [CrossRef]
- Köppl, C.; Lingg, N.; Fischer, A.; Kröß, C.; Loibl, J.; Buchinger, W.; Schneider, R.; Jungbauer, A.; Striedner, G.; Cserjan-Puschmann, M. Fusion Tag Design Influences Soluble Recombinant Protein Production in Escherichia coli. Int. J. Mol. Sci. 2022, 23, 7678. [Google Scholar] [CrossRef] [PubMed]
- Yadav, D.K.; Yadav, N.; Yadav, S.; Haque, S.; Tuteja, N. An insight into fusion technology aiding efficient recombinant protein production for functional proteomics. Arch. Biochem. Biophys. 2016, 612, 57–77. [Google Scholar] [CrossRef] [PubMed]
- Mamipour, M.; Yousefi, M.; Hasanzadeh, M. An overview on molecular chaperones enhancing solubility of expressed recombinant proteins with correct folding. Int. J. Biol. Macromol. 2017, 102, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Fan, J.; Han, R.; Xiao, J.; Li, Q.; Xu, G.; Dong, J.; Ni, Y. Enhancing soluble expression of sucrose phosphorylase in Escherichia coli by molecular chaperones. Protein Expr. Purif. 2020, 169, 105571. [Google Scholar] [CrossRef] [PubMed]
- Jhamb, K.; Sahoo, D.K. Production of soluble recombinant proteins in Escherichia coli: Effects of process conditions and chaperone co-expression on cell growth and production of xylanase. Bioresour. Technol. 2012, 123, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-P.; Perrett, S.; Zhou, J.-M. Dimeric Trigger Factor Stably Binds Folding-competent Intermediates and Cooperates with the DnaK-DnaJ-GrpE Chaperone System to Allow Refolding. J. Biol. Chem. 2005, 280, 13315–13320. [Google Scholar] [CrossRef]
- Jang, J.-H.; Choi, K.-Y. Whole Cell Biotransformation of 1-dodecanol by Escherichia coli by Soluble Expression of ADH Enzyme from Yarrowia lipolytica. Biotechnol. Bioprocess. Eng. 2021, 26, 247–255. [Google Scholar] [CrossRef]
- Fang, Y.; Fu, X.; Xie, W.; Li, L.; Liu, Z.; Zhu, C.; Mou, H. Expression, purification and characterisation of chondroitinase AC II with glyceraldehyde-3-phosphate dehydrogenase tag and chaperone (GroEs-GroEL) from Arthrobacter sp. CS01. Int. J. Biol. Macromol. 2019, 129, 471–476. [Google Scholar] [CrossRef]
- Gong, Z.; Wang, H.; Tang, J.; Bi, C.; Li, Q.; Zhang, X. Coordinated Expression of Astaxanthin Biosynthesis Genes for Improved Astaxanthin Production in Escherichia coli. J. Agric. Food Chem. 2020, 68, 14917–14927. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Q.; Huang, D.; Yang, H.; Hong, Z.; Wang, C. Improvement of Carotenoids’ Production by Increasing the Activity of Beta-Carotene Ketolase with Different Strategies. Microorganisms 2024, 12, 377. https://doi.org/10.3390/microorganisms12020377
Zhou Q, Huang D, Yang H, Hong Z, Wang C. Improvement of Carotenoids’ Production by Increasing the Activity of Beta-Carotene Ketolase with Different Strategies. Microorganisms. 2024; 12(2):377. https://doi.org/10.3390/microorganisms12020377
Chicago/Turabian StyleZhou, Qiaomian, Danqiong Huang, Haihong Yang, Zeyu Hong, and Chaogang Wang. 2024. "Improvement of Carotenoids’ Production by Increasing the Activity of Beta-Carotene Ketolase with Different Strategies" Microorganisms 12, no. 2: 377. https://doi.org/10.3390/microorganisms12020377
APA StyleZhou, Q., Huang, D., Yang, H., Hong, Z., & Wang, C. (2024). Improvement of Carotenoids’ Production by Increasing the Activity of Beta-Carotene Ketolase with Different Strategies. Microorganisms, 12(2), 377. https://doi.org/10.3390/microorganisms12020377





