Abstract
Bacillus subtilis, a probiotic bacterium with engineering potential, is widely used for the expression of exogenous proteins. In this study, we utilized the integrative plasmid pDG364 to integrate the hemagglutinin–neuraminidase (HN) gene from Newcastle disease virus (NDV) into the genome of the B. subtilis 168 model strain. We successfully constructed a recombinant B. subtilis strain (designated B. subtilis RH) that displays a truncated HN antigen fragment on the surface of its spores and further evaluated its immunogenic effects in mice. Using ELISA, we quantified the levels of IgG in serum and secretory IgA (sIgA) in intestinal contents. The results revealed that the recombinant B. subtilis RH elicited robust specific mucosal and humoral immune responses in mice. Furthermore, B. subtilis RH demonstrated potential mucosal immune adjuvant properties by fostering the development of immune organs and augmenting the number of lymphocytes in the small intestinal villi. Additionally, the strain significantly upregulated the relative expression of inflammatory cytokines such as IL-1β, IL-6, IL-10, TNF-α, and IFN-γ in the small intestinal mucosa. In conclusion, the B. subtilis RH strain developed in this study exhibits promising mucosal immunogenic effects. It holds potential as a candidate for an anti-NDV mucosal subunit vaccine and offers a novel preventive strategy for the poultry industry against this disease.
1. Introduction
Bacillus subtilis is a probiotic bacterium that is widely used for engineering purposes to express exogenous proteins [1,2]. Under conditions of extreme stress or nutrient scarcity, B. subtilis generates a distinct cellular form known as the spore. The spore’s core and cortex are crucial for its formation and integrity. Encompassing the cortex is the spore’s coat layer, composed of approximately 80 unique coat proteins, which collectively define the spore surface [3]. This robust architecture endows B. subtilis spores with high resistance, enabling them to withstand acid and bile salt challenges within the intestine tract [4,5]. Beyond its capacity to elicit both mucosal and humoral immune responses within the host, B. subtilis also functions as an effective carrier or adjuvant for mucosal vaccine delivery [6]. Leveraging spore surface display technology enables the stable presentation of antigenic proteins on the spore surface. Research efforts have yielded the development of protective antigens through the utilization of diverse coat proteins, such as cotB, cotC, cotG, etc., as anchoring agents. This approach facilitates the expression of antigenic proteins on the spore surface and culminates in the creation of recombinant probiotics that are rich in immunogenic properties [7,8,9]. Recombinant strains of B. subtilis are characterized by their ease of storage and transport, which simplifies the immunization process and reduces stress in animals. These strains exhibit a broad spectrum of applications in the field of animal immunization. Recent advances have led to the successful display of various antigenic proteins on the spore surface, including tetanus toxoid fragment C, ovalbumin [10,11], cholera toxin [12], the transmissible gastroenteritis virus spike protein [13], the highly pathogenic avian influenza H5N1 hemagglutinin protein [14], and the Vibrio OmpK protein [15]. The induction of specific immune responses by these recombinant probiotics underscores their exceptional immune attributes and probiotic functionalities.
Newcastle disease virus (NDV) is the causative agent of Newcastle disease (ND), a significant poultry disease. Characterized by high contagiousness, ND is categorized by the World Organization for Animal Health (WOAH) as a List A disease in animals, necessitating legal reporting [16]. In China, the Ministry of Agriculture ranks it as a Class Ⅱ animal epidemic disease. Challenges in eradicating NDV stem from suboptimal vaccination, antigenic drift, limited duration of immunity, and immunosuppressive conditions [17,18,19]. The hemagglutinin–neuraminidase (HN) protein, a surface glycoprotein of NDV, is renowned for its ability to elicit the production of neutralizing antibodies within poultry, thereby serving as a pivotal protective antigen. The globular head domain of the HN protein houses the essential functional regions, including the active site and all antigenic sites [20,21]. This domain is instrumental in the virus’s infectivity and pathogenicity, positioning the HN protein as an optimal target for the genetic engineering of vaccines aimed at combating ND [22,23,24]. Currently, the main types of genetically engineered vaccines for ND include nucleic acid vaccines, subunit vaccines, and live-vector vaccines [25,26,27]. The attachment and entry of NDV into host cells are facilitated by its binding to two major salic acid (SA) receptors present on the membrane of target cells. Specifically, SAα2,3-Gal receptors are prevalent in chicken small intestinal epithelial cells, while SAα2,6-Gal receptors are more common in avian trachea ciliated epithelial cells [28,29]. Consequently, NDV exhibits a specific mucosal tropism and can readily infect birds through the respiratory and digestive tracts. Mucosal vaccines have the capacity to elicit the production of sIgA, establishing mucosal immunity through various immunization routes such as intranasal, ocular, drinking water, and aerosol administration. These methods significantly enhance local immunity within the respiratory and digestive tracts [30,31]. In summary, mucosal immunization plays a crucial role in managing ND, and the development of recombinant probiotic-based mucosal vaccines against NDV is currently a paramount strategy in this field.
In this study, we engineered probiotics using the B. subtilis model strain 168. By integrating a fusion gene into the genome of B. subtilis 168 via the integration plasmid pDG364, we successfully constructed the recombinant B. subtilis RH. This strain displays the truncated HN (HNJD) protein of Newcastle disease virus on its spore surface. We then proceeded to assess the specific immune response and mucosal immune adjuvant effect elicited by B. subtilis RH in mice. The novel oral vaccine developed herein offers an innovative approach for the prevention of NDV-related diseases, presenting a fresh alternative in disease management strategies.
2. Materials and Methods
2.1. Strains, Vaccines, Cell, Plasmids, Primers Sequences, and Experimental Ethics
The bacterial strains (Escherichia coli BL21/DH5α and Bacillus subtilis 168), vaccines (NDV vaccine strain LaSota/CS2), chick embryo fibroblasts (CEF), plasmids (pUCm-T, pET-32a, and pDG364-cotB), and primer sequences utilized in this study are itemized in Supplementary Table S1. All methodologies and animal experimentation were carried out in accordance with the “Guide for the Care and Use of Laboratory Animals”, receiving approval from the Institutional Animal Care and Use Committee of Sichuan Agricultural University (approval number: SYXK Chuan 2019-187).
2.2. Cloning of Truncated HN Gene
The E.Z.N.A® Total RNA Kit II (Omega, Honolulu, HI, USA) was used to extract the total viral RNA from the NDV LaSota vaccine strain (GenBank accession No. AY845400.2). Subsequently, the extracted RNA was reverse-transcribed to synthesize cDNA. The hemagglutinin–neuraminidase-truncated (HNJD) gene was PCR-amplified using the cDNA as a template and HNJD-F1/R1 primers [32]. The amplicon was then T-A-cloned into a pUCm-T vector to generate the recombinant plasmid pUCm-T-HNJD, which was sequenced and subjected to site-directed mutagenesis to eliminate potential restriction sites that might interfere with the subsequent steps (performed by Tsingke Biotechnology Co., Ltd., Beijing, China).
2.3. Preparation of Hyperimmune Serum and Prokaryotic Expression of HNJD
Female New Zealand rabbits weighing 2.0 ± 0.2 kg were used to produce anti-NDV hyperimmune serum. Each rabbit received a subcutaneous injection of 2 mL of the live vaccine (LaSota strain). This was followed by weekly booster immunizations, and serum collection occurred subsequent to three consecutive vaccinations. Hemagglutination inhibition (HI) assays were performed to determine the HI antibody titers against NDV, and optimal dilutions were ascertained and preserved for further use.
The pUCm-T-HNJD plasmid and the pET-32a vector were digested with BamH I and EcoR I enzymes (Takara Bio, Dalian, China). The resulting target fragments were ligated to form the expression plasmid pET-32a-HNJD1, which was then transformed into E. coli BL21. The successfully expressing strain was designated as E. coli BL21/pET-32a-HNJD. The expression of the recombinant protein was induced with isopropyl-β-D-thiogalactoside (IPTG) added to a Luria–Bertani (LB) broth at a final concentration of 1 mM. After incubation at 37 °C with shaking at 160 rpm for 6 h, 1 mL of bacterial culture was collected, while the pET-32a vector alone served as a negative control. The bacterial cells were lysed by ultrasonication and centrifuged at 10,000× g for 1 min. The pellet was collected and subjected to SDS-PAGE, followed by transfer onto a PVDF membrane (Solarbio, Beijing, China). The membrane was blocked with 5% skim milk at 37 °C for 2 h and then incubated overnight at 4 °C with rabbit-derived NDV hyperimmune serum (diluted 1:200 in TBST). HRP-conjugated goat anti-rabbit IgG (diluted 1:2000 in TBST, BOSTER, Wuhan, China) was used as the secondary antibody. After a 2 h incubation at room temperature and washing with TBST, color development was performed using the DAB Color Development Kit (BOSTER, Wuhan, China) according to the manufacturer’s instructions. If the HNJD protein specifically binds to anti-NDV hyperimmune serum, a brown band should be visible on the membrane after immunoblotting.
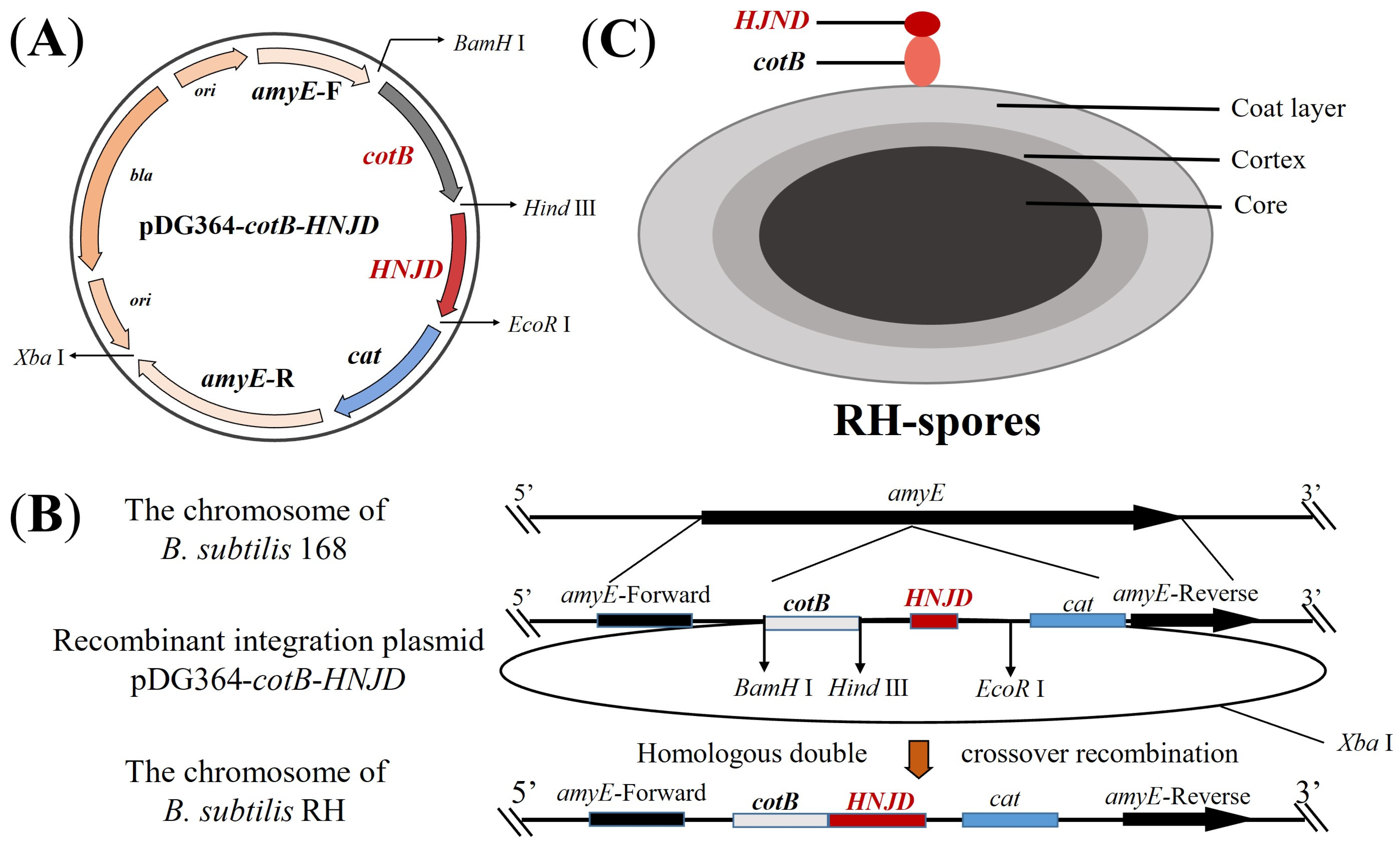
2.4. Construction and Transformation of Recombinant Integration Plasmid
The plasmid pDG364-cotB developed in our previous work [33], was utilized. Employing the methodology detailed in Section 2.3, we constructed the recombinant plasmid pDG364-cotB-HNJD. Competent cells of B. subtilis 168 were prepared following Julkowska et al.’s protocol [34]. The plasmid pDG364-cotB-HNJD was linearized with an Xba I enzyme digestion prior to being introduced into the competent cells. Through homologous double-crossover recombination, the target gene was integrated into the amyE (amylase) locus of B. subtilis (Figure 1B). Positive clones were identified on chloramphenicol (5 μg/mL) resistance plates and further screened on a 1% starch nutrient agar medium. Genomic DNA from the recombinant bacteria was extracted, and a PCR analysis was performed with primer pairs amyE-F/R, amyE-F/HNJD-R2, cotB-F/HNJD-R2, and HNJD-F2/R2. Amplification products were verified by gel electrophoresis. The correctly identified recombinant strain was designated B. subtilis RH.
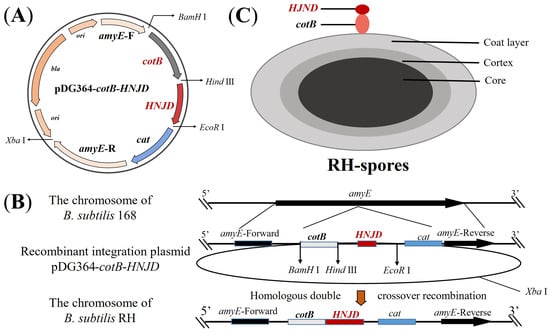
Figure 1.
Schematic representation of homologous recombination and the structure of RH spores. (A) Diagram of the integrative plasmid pDG364-cotB-HNJD. (B) Homologous double-crossover recombination between the recombinant plasmid pDG364-cotB-HNJD and the genome of B. subtilis 168 occurs, leading to the integration of the target gene into the amyE locus. (C) Structure depiction of RH spores showing the fusion expression of the anchor protein cotB and the HNJD protein on the surface of the spores.
2.5. Sporulation and Immunofluorescence Microscopy
Sporulation was induced using Difco sporulation medium (DSM) according to the method described by Stasiłojć et al. [35]. After approximately 24 h of sporulating culture, spores were treated with 2.0 mg/L of lysozyme at 37 °C for 2 h to eliminate any residual vegetative cells. The purified spores were then obtained by sequential washing with 1 M of NaCl, 1 M of MgCl2, and distilled water, as detailed in reference [7]. Finally, the spread plate technique was employed to enumerate the spore colonies.
Immunofluorescence microscopy was used to verify the successful display of the HNJD protein on the spore surface [36], as indicated in Figure 1C. A purified spore suspension was prepared and affixed to microscope slides according to the method described previously [37]. Rabbit anti-NDV hyperimmune serum (diluted 1:200 in PBST) served as the primary antibody, while Cy3-conjugated goat anti-rabbit IgG (diluted 1:200 in PBST, BOSTER, Wuhan, China) was used as the secondary antibody. Additionally, serum from non-immunized mice was applied as the primary antibody to establish a negative control. The immunofluorescence images were captured using a fluorescence microscope (DMi8, Leica, Tokyo, Japan).
2.6. Immunization of Mice and Collection of Samples
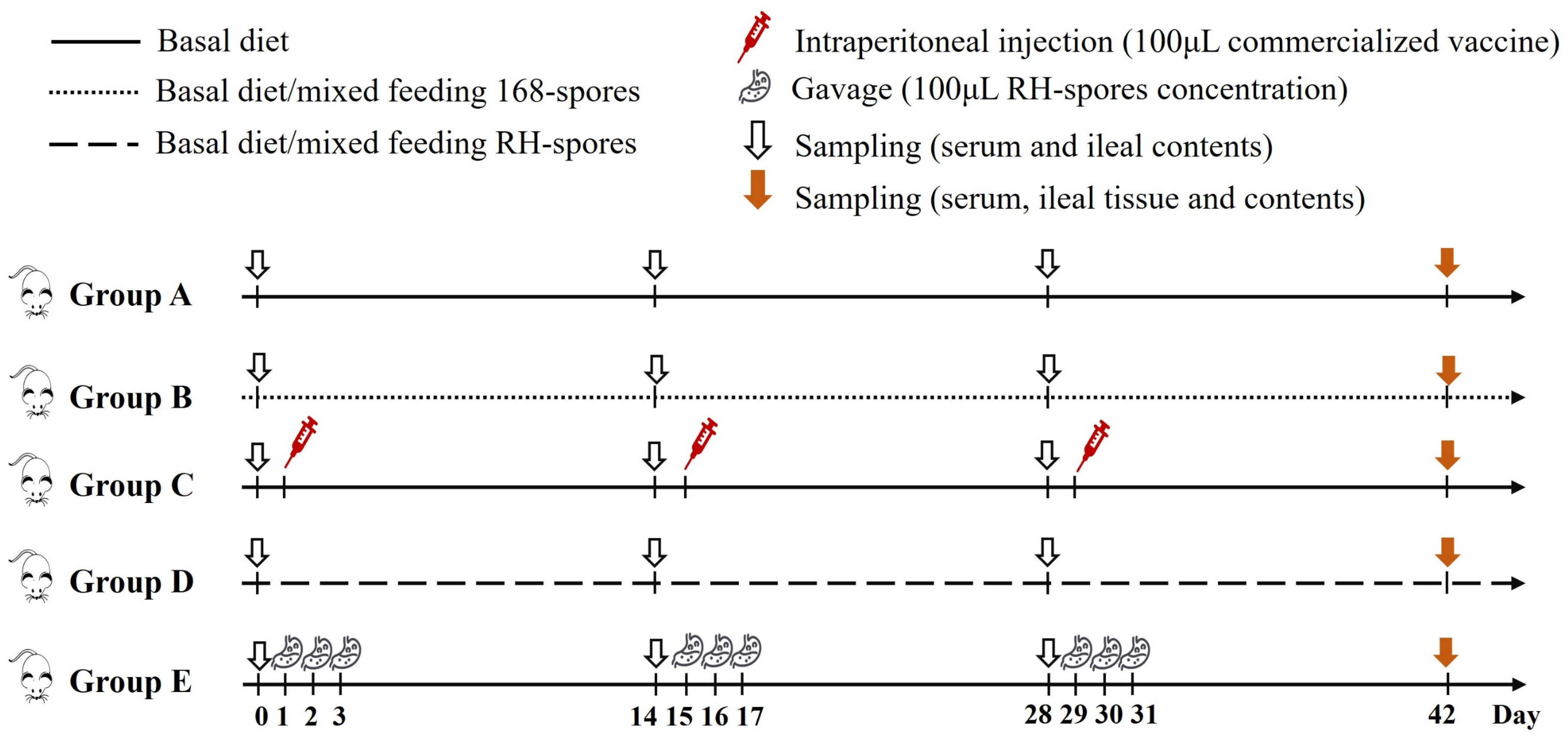
A total of 100 three-week-old female BALB/c mice underwent a seven-day acclimatization period before being randomly distributed into five groups, each comprising 20 individuals. Each group was then subdivided into five cages, with four mice per cage. The feeding regimen and immunization schedule for the mice are detailed in Figure 2. The mice in group A served as the control group and were provided with a basal diet. Group B received a diet mixed with 2.0 × 106 CFU per gram of wild-type B. subtilis 168 spores (168-spores). Group C was intraperitoneally administered 100 μL of inactivated vaccine (strain LaSota, with viral content equal to or exceeding 1.0 × 108 ELD50 prior to inactivation) on days 1, 15, and 29. Group D was fed a diet containing 2.0 × 106 CFU per gram of B. subtilis RH spores (RH-spores). Group E received an RH-spore suspension (2.0 × 1010 CFU/mL) via oral gavage. The mice in group E received 0.1 mL daily on days 1–3, 15–17, and 29–31, as previously described [38]. Each mouse was given 3–5 g of this diet daily.
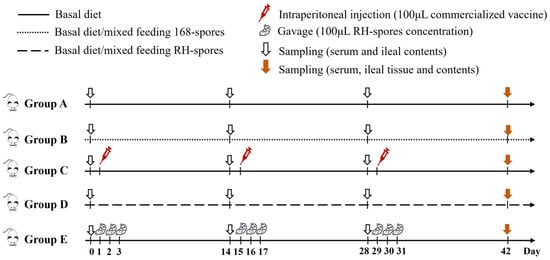
Figure 2.
Schematic representation of the vaccination and feeding trail. Group A: untreated control; Group B: mixed diet with 168-spores; Group C: inactivated vaccine administered; Group D: mixed diet with RH-spores; Group E: RH-spores administered via gavage.
The experimental period spanned 42 days. On days 0, 14, 28, and 42, five mice from each group were randomly selected, and their serum and small intestinal contents were harvested and preserved at −80 °C. The body weight of the mice was recorded on day 42. Following euthanasia, ileal tissues and contents were gathered, and the ileal tissues were preserved in 4% paraformaldehyde. The spleens and thymuses were excised, and organ indices were calculated as follows:
2.7. Detection of NDV-Specific Serum and Mucosal Antibodies
Serum IgG antibody levels were quantified using a competitive enzyme-linked immunosorbent assay (ELISA) with the Serum Antibodies to Newcastle Disease Virus ELISA Kit (Zhenrui Bio, Shenzhen, China). The serum IgG levels were expressed as S/N (sample-to-negative control) values:
The indirect ELISA method was used to detect the anti-NDV sIgA in the small intestinal contents. The ELISA plates were coated with NDV antigen, provided by Shenzhen Zhenrui Biotech Co., Ltd., Shenzhen, China. The intestinal contents were diluted to 1:100 with PBST, and serum from unimmunized mice served as the negative control. The secondary antibody used was horseradish peroxidase (HRP)-conjugated goat anti-mouse IgA (diluted to 1:1200 in PBST, Abcam, Cambridge, UK). The levels of sIgA in the small intestinal contents were expressed as P/N (positive-to-negative control) values:
2.8. Microneutralization Test
The serum-neutralizing antibody (NA) titers were detected with a microneutralization assay, as previously described [39]. CEF cells were seeded in 96-well plates and used for virus neutralization tests when they reached a confluency rate of 50–60%. All serum samples underwent heat inactivation at 56 °C for 30 min and were then serially diluted two-fold from 1:50 to 1:1600 in Dulbecco’s Modified Eagle Medium (DMEM). Subsequently, 50 μL of the NDV CS2 vaccine strain (200 TCID50) was combined with an equal volume of the diluted serum and incubated together at 37 °C for 1 h. This mixture was then added to a 96-well plate containing CEF cells, with three replicate wells for each sample. The cells were maintained in DMEM supplemented with 2% fetal bovine serum (FBS) at 37 °C and 5% CO2. It is crucial to note that if the virus is neutralized by serum antibodies, it will be unable to infect the cells. After incubation for 72 h at 37 °C and 5% CO2, the NA titer for each group was calculated as the geometric mean titer (GMT), which was established by identifying the lowest serum dilution that prevented cytopathic effects in over 50% of the CEF cells.
2.9. Hemagglutination Inhibition Test
The HI assay was performed in accordance with the National Standard of the People’s Republic of China [40]. Serum samples were twofold diluted in a 96-well plate. A total of 25 μL of NDV antigen containing four hemagglutination units was introduced into each well, including a positive control. After 30 min of incubation, 25 μL of a 1% suspension of chicken red blood cells was added to each well and incubated for an additional 30 min. The results were observed, and the HI titers were defined as the highest dilution of serum that completely inhibited NDV hemagglutination.
2.10. Gene Expression of Cytokine in Ileum
Total RNA from ileal tissues samples was extracted using the Animal Total Isolation Kit (Foregene Co., Ltd., Chengdu, China). The extracted RNA was subsequently reverse-transcribed into cDNA using the RT-Easy™ II (Foregene, Chengdu, China). The expression levels of cytokine-related mRNAs, including interleukin 1 beta (IL-1β), interleukin 6 (IL-6), interleukin 10 (IL-10), interferon gamma (IFN-γ), and tumor necrosis factor alpha (TNF-α), were quantified by real-time quantitative PCR (RT-qPCR) according to the methodology described by Xin et al. [41]. Primer sequences are listed in Supplementary Table S2. Beta-actin (β-actin) served as the reference gene for data normalization. The relative expression of the target genes was calculate using the 2−ΔΔCT method:
2.11. The Morphology of Ileum
Tissue sections were prepared from ileal tissue fixed in 4% paraformaldehyde (ServiceBio Technology Co., Ltd., Wuhan, China). The imaging of the target areas was performed using an Eclipse Ci-L photographic microscope (Nikon, Tokyo, Japan), following the method described previously [42]. Image analysis was conducted using Image-Pro Plus 6.0 software, with results reported in millimeters. For each intestinal sample, five random fields were selected to measure the villus height (VH) and crypt depth (CD) and to determine the VH/CD ratio. A 100× magnification field was used, and five intact villi were chosen from each tissue section for measurement. The height of the intestinal villi and the number of intraepithelial lymphocytes (IELs) were measured. The number of IELs per unit length of villus was then calculated as follows:
2.12. Statistical Analysis
Data were analyzed using one-way ANOVA followed by the Friedman test for multiple comparisons. Statistical analyses were conducted with IBM SPSS Statistics 26 (IBM Corporation, Armonk, NY, USA), and results are presented as mean ± standard deviation (SD). Statistical significance was denoted by letters, where different letters indicate significant differences (p < 0.05) between groups. Similar letters indicate no significant difference among groups (p > 0.05). Data visualization was performed using Origin 2021 (OriginLab, Northampton, MA, USA).
3. Results
3.1. Construction and Transformation of Recombinant Plasmid
Figure 3 illustrates the construction of recombinant plasmids and the outcomes of the transformation. The HNJD gene was approximately 642 bp in size (Figure 3A). Sequencing revealed that the HNJD gene contained an EcoR I restriction site, with the original and mutated sequences detailed in Supplementary Table S3. The HI titer of the rabbit anti-NDV hyperimmune serum reached a value of 13log2. An analysis via 12% SDS-PAGE and Western blotting (shown in Figure 3B,C) confirmed that the E. coli BL21/pET-32a-HNJD expression product exhibited a protein band around 42 kDa, signifying successful recombinant protein expression with reactivity to NDV-specific antibodies. Following double-enzyme digestion validation, the recombinant plasmid pDG364-cotB-NHJD was introduced into B. subtilis-competent cells (Figure 3D). PCR identification was performed using four primer sets, with the wild-type B. subtilis genome serving as the control. The results presented in Figure 3E demonstrated that all DNA bands were congruent with their theoretical sizes. Furthermore, Figure 3F indicated that the recombinant B. subtilis lost its ability to break down starch due to the insertion of the fusion gene cotB-HNJD into the amylase-encoding locus. In conclusion, we successfully engineered a strain of B. subtilis RH.
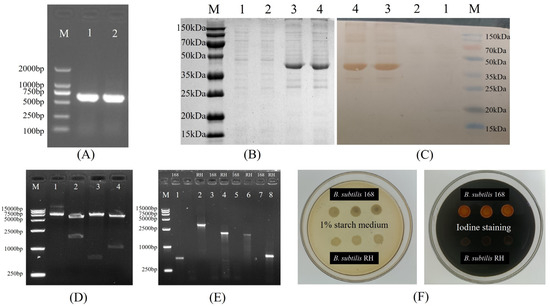
Figure 3.
Construction of B. subtilis RH. (A) Cloning of the truncated HNJD gene. M: DNA marker (100–2000 bp); Lanes 1 and 2: PCR products of the HNJD gene. (B) SDS-PAGE analysis of protein expression on a 12% gel. M: Protein marker (15–150 kDa); Lanes 1 and 2: proteins expressed with the empty pET32a plasmid vector; Lanes 3 and 4: expression products after induction with IPTG for 6 h. (C) Western blot analysis of transmembrane proteins. (D) Double digestion verification of pDG364-cotB-HNJD. M: DNA marker (250–15,000 bp); Lanes 1–4: products from the recombinant plasmid digested with Xba I, BamH I/EcoR I, Hind III/EcoR I, and BamH I/Hind III, respectively. (E) PCR identification of B. subtilis 168 and B. subtilis RH. M: DNA marker (250–10,000 bp); Lanes 1 and 2: amyE-F/R; Lanes 3 and 4: amyE-F/HNJD-R1; Lanes 5 and 6: cotB-F/HNJD-R1; Lanes 7 and 8: HNJD-F2/R2. (F) Identification of B. subtilis RH with amylase activity assay. B. subtilis RH was cultivated on a medium containing 1% starch and then stained with iodine solution.
3.2. Sporulation and Immunofluorescence Microscopy
The results of the indirect immunofluorescence assay for the recombinant B. subtilis RH are presented in Figure 4. Red fluorescent signals were exclusively observed in B. subtilis RH when viewed under a microscope, using anti-NDV hyperimmune serum as the primary antibody. This observation confirms that the HNJD protein was successfully expressed on the surface of B. subtilis RH spores and was capable of binding to NDV antibodies.
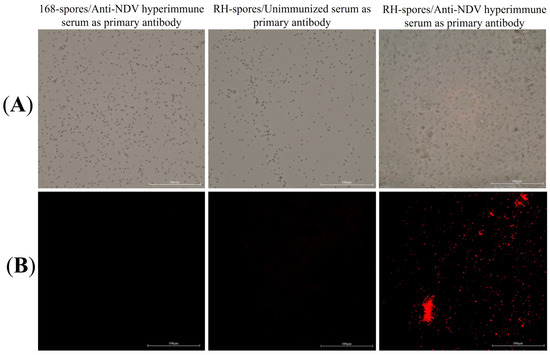
Figure 4.
Indirect immunofluorescence assay. (A) Spores of B. subtilis 168 and RH observed under 40× magnification using bright-field microscopy; (B) Spores of B. subtilis 168 and RH under 40× magnification using fluorescence microscopy, with different serums serving as the primary antibody and Cy3-conjugated goat anti-rabbit IgG employed as the secondary antibody.
3.3. Detection of NDV-Specific Serum and Mucosal Antibodies
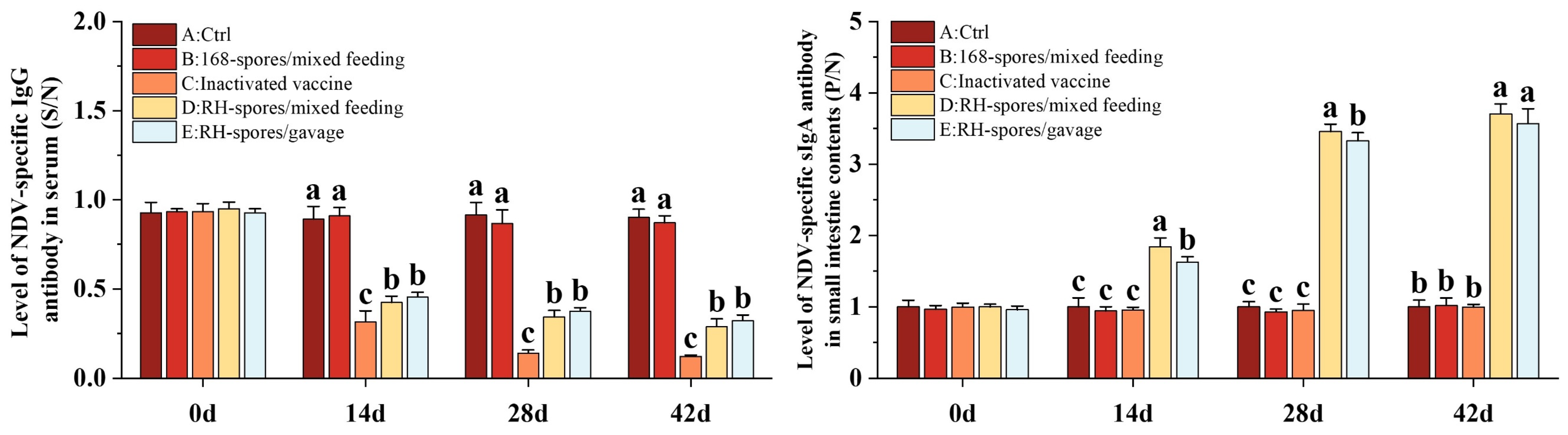
The ELISA results for serum IgG and mucosal sIgA are depicted in Figure 5. The serum IgG levels were quantified using a competitive ELISA approach. The S/N value is inversely related to the concentration of IgG, with lower S/N values indicating higher IgG concentrations. Throughout the study, Groups A and B exhibited consistently lower IgG levels, whereas Groups C, D, and E had significantly elevated serum IgG levels (p < 0.05), with Group C demonstrating the highest concentration of serum IgG. The serum IgG level was higher in Group D compared to Group E. The sIgA levels in the ileal content were determined using an indirect ELISA method. Higher P/N values are indicative of increased sIgA levels. It was observed that Groups A, B, and C did not generate substantial levels of sIgA antibodies. In contrast, both Groups D and E showed a marked increase in sIgA levels (p < 0.05), which progressively rose over time, reaching a plateau on day 28. Notably, Group D achieved higher levels of sIgA than Group E. This suggests that B. subtilis RH effectively triggered an immune response in the mice through both mixed feeding and gavage administration methods. The mixed feeding method elicited a stronger mucosal immune response, potentially due to an extended period of stimulation of the mucous membranes.
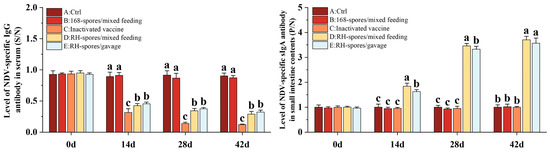
Figure 5.
Immune responses in mice following immunization using different methods. Serum IgG levels are depicted as S/N values, while sIgA levels in the mucosa are represented by P/N values. The data are reported as mean values with SD, based on a sample size of n = 5. Statistical analysis involved one-way ANOVA, followed by a Friedman test. Bars labeled with distinct lowercase letters indicate a significant difference at the p < 0.05 level.
3.4. Detection of NA and HI Antibody Titers
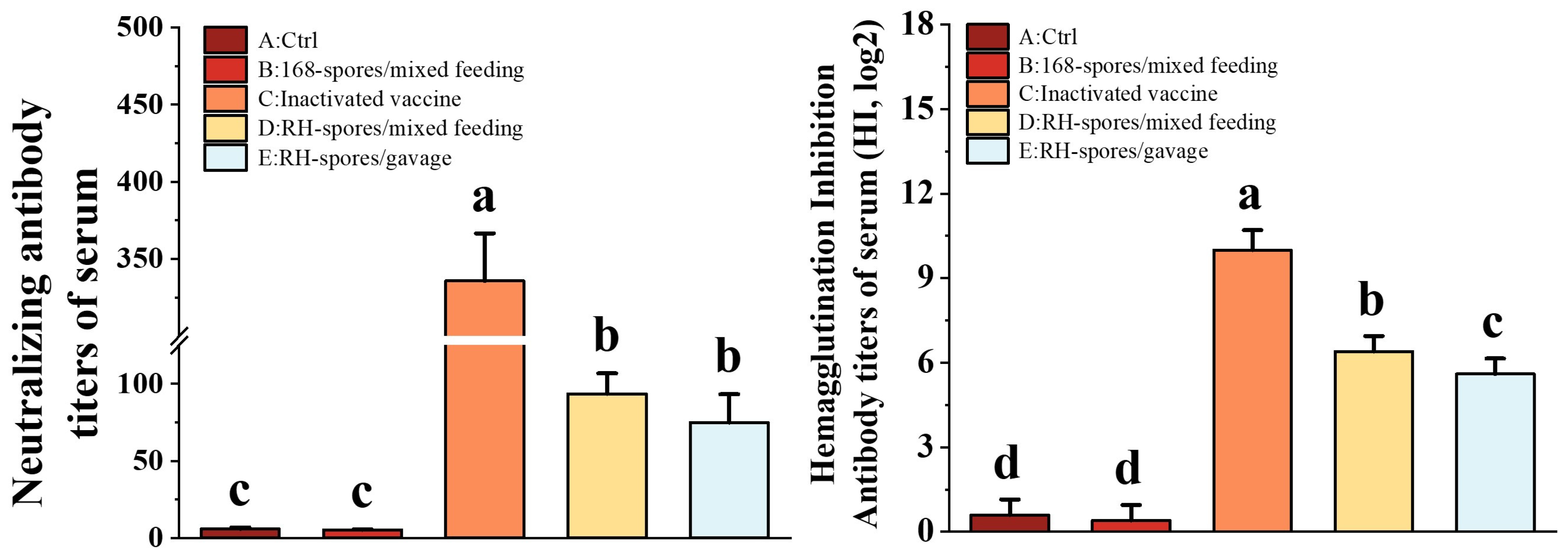
Using the Reed–Muench method, the TCID50 for the NDV CS2 vaccine strain was determined to be 10−6.2/0.1 mL. As depicted in Figure 6, Groups C, D, and E exhibited significantly elevated NA and HI titers compared to Groups A and B (p < 0.05). Group C achieved the highest NA and HI titers, and Group D exhibited significantly higher HI titers than Group E (p < 0.05). These findings indicate that mucosal immunization can elicit the robust production of neutralizing antibodies in mice and that the levels of NA and HI antibodies are correlated with the route of mucosal administration.
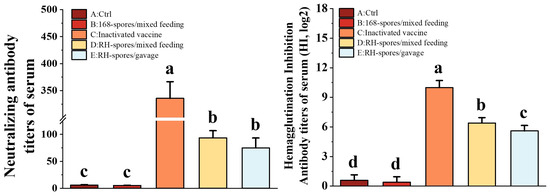
Figure 6.
Determination of NA and HI levels in the serum of each group. The data are presented as mean ± SD, with a sample size of n = 5. Statistical analysis was conducted using one-way ANOVA, followed by a Friedman test. Bars labeled with different lowercase letters indicate a statistically significant difference at the p < 0.05 level.
3.5. Gene Expression of Cytokine in the Ileum
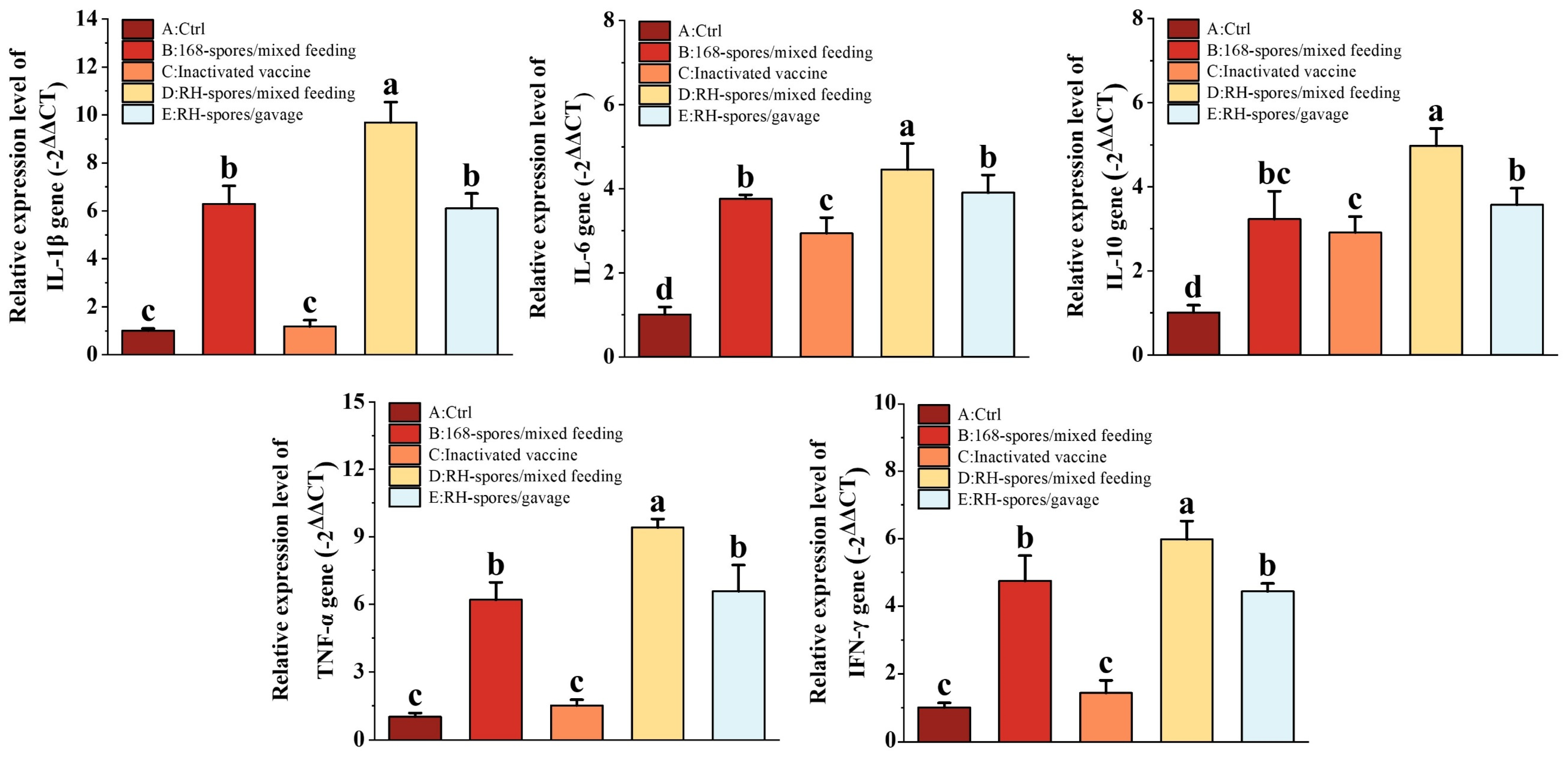
Cytokines related to immunity were quantified using RT-qPCR, and the results are shown in Figure 7. We observed a significant upregulation in the relative mRNA expression of IL-1β, IL-6, IL-10, TNF-α, and IFN-γ in the spore-immunized groups (Group B, D, and E). Notably, all cytokine mRNA expression levels were higher in Group D compared to Group E (p < 0.05), indicating that mixed feeding was more effective than gavage administration. While the expression of IL-6 and IL-10 was significantly elevated in Group C compared to Group A (p < 0.05), their levels remained lower than those in the spore-immunized groups. This indicates that inactivated vaccines can enhance cytokine expression in the intestinal mucosa by modulating the systemic immune response, but this effect is less pronounced than the direct action of the spores on the mucosal surface.
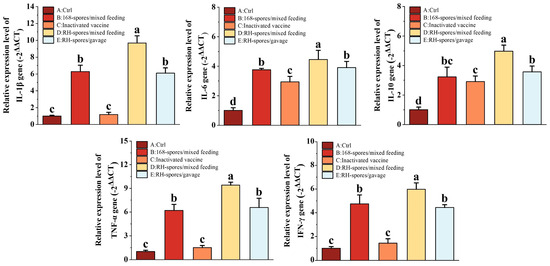
Figure 7.
Expression levels of cytokines IL-1β, IL-6, IL-10, TNF-α, and IFN-γ in the small intestine were quantified using RT-qPCR. The data were normalized using the −2ΔΔCT method. The results are presented as mean ± SD, with a sample size of n = 5. Statistical analysis was conducted using one-way ANOVA, followed by a Friedman test. Bars labeled with distinct lowercase letters denote a statistically significant difference at the p < 0.05 level.
3.6. Histomorphology and Intraepithelial Lymphocytes of the Mouse Ileum
Figure 8A illustrates the ileum morphology of the mice from each group, revealing a clear and intact histological structure of the ileum. The measurements of VH and CD in the mouse ileum are shown in Figure 8B. The mice from the spore-immunized groups exhibited a significant increase in VH and the VH/CD ratio and a decrease in CD compared to Groups A and C (p < 0.05). However, no significant differences were observed among the spore-immunized groups (p > 0.05). The number of IELs per unit length of the intestinal villi was quantified and is also presented in Figure 8B. The spore-immunized groups exhibited a significantly higher number of IELs per unit length compared to Groups A and C (p < 0.05), with Group D demonstrating the highest count. These findings suggest that the administration of B. subtilis 168 and RH can effectively enhance intestinal mucosa development and promote the maturation of intestinal structures in mice. Notably, B. subtilis RH appears to be more effective in stimulating the proliferation of IELs and modulating the immune response within the intestinal mucosa.
Figure 8.
Histomorphologic analysis and enumeration of IELs in ileal tissue. (A) Representative images of ileal tissue from each group are shown, with intestinal crypt (1) and villi (2) indicated by black arrows. IELs are localized within the mucosal epithelial layer of the small intestine, characterized by their irregularly shaped nuclei and darker staining relative to the surrounding epithelium. Microscopic magnification is set at 100×. (B) Quantitative measurements of intestinal CD, VH, VH/CD ratio, and IELs per unit length are presented. Data are presented as the mean ± SD, with a sample size of n = 5. Statistical analysis involved one-way ANOVA, followed by a Friedman test, where bars labeled with distinct lowercase letters signify a significant difference at the p < 0.05 level. VH refers to the height of the villus, CD denotes the depth of the crypts, and the VH/CD ratio represents the metric of the villus height relative to the crypt depth. IELs are defined as intestinal intraepithelial lymphocytes.
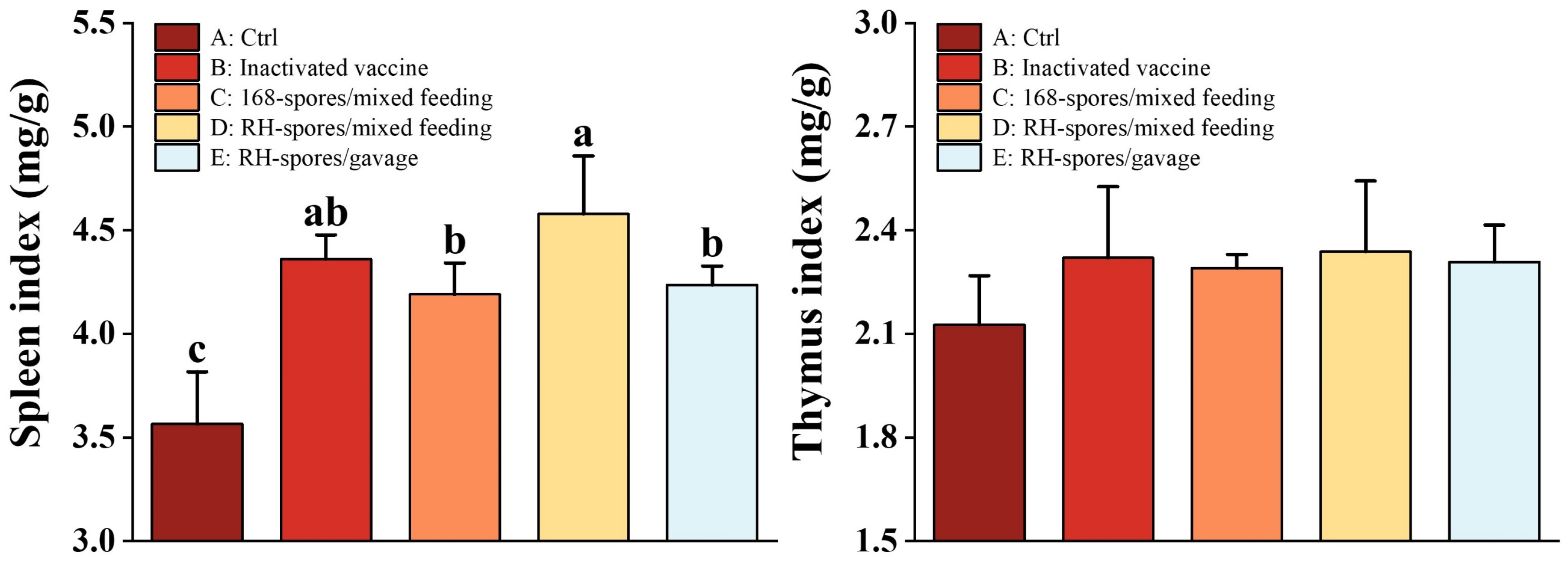
3.7. Mouse Immune Organ Index
Figure 9 demonstrates that the spleen index was increased in all groups except for Group A. Notably, Group D exhibited a significantly higher spleen index than both Group C and Group E (p < 0.05). In contrast, no significant differences were found in the thymus index across all the groups (p > 0.05).
Figure 9.
Organ indices of the thymus and spleen. Data are presented as mean ± SD, with a sample sized of n = 5. Statistical analysis was conducted using one-way ANOVA, followed by a Friedman test. Bars labeled with different lowercase letters indicate a statistically significant difference at the p < 0.05 level.
4. Discussion
In the realm of livestock and poultry production, probiotics are increasingly being adopted as a viable alternative to antibiotics [43]. B. subtilis, a probiotic species, has been acknowledged by the FDA as safe and is commonly incorporated into functional feed additives due to its robust probiotic characteristics and the convenience of its isolation and cultivation. This bacterium exhibits remarkable resistance to the harsh conditions within the gastrointestinal tract, enabling it to maintain potent biological activity in the digestive system [44,45]. It contributes to enhanced digestion by providing enzymes, antimicrobial peptides, and other advantageous metabolic byproducts, which collectively improve feed efficiency, growth performance, and the integrity of the gut barrier [46,47]. B. subtilis also plays a pivotal role in modulating the intestinal microbiota and the immune response, thereby warding off pathogenic microorganisms and preventing diseases [48,49,50]. In addition, B. subtilis is often employed as an engineering bacterium for the efficient expression of exogenous proteins. A technique for displaying proteins on the surface of B. subtilis spores was first proposed by Isticato et al. [51] in 2001. They successfully expressed a 459-amino-acid C-terminal fragment of the tetanus toxin (TTFC) on a spore surface, demonstrating its reactivity. With the elucidation of the B. subtilis genome and advances in proteomic research, this technique has been effectively utilized across various domains, including mucosal immunization, industrial enzyme production, environmental remediation, pharmaceutical development, and more [3]. Owing to its probiotic attributes and stability, B. subtilis holds promising advantages as a mucosal adjuvant for the delivery of antigenic proteins. Recombinant probiotics displaying the HN protein can elicit specific immune responses when used as mucosal vaccines, offering substantial potential in research and application areas. Our laboratory has successfully constructed several recombinant B. subtilis strains that display heterologous proteins, including Salmonella OmpC [52], Rotavirus VP8 [33], and Porcine circovirus type 2 Cap proteins [53]. In the current study, we excised a portion of the antigenic region from the HN protein and spliced it into smaller fragments following Chen’s methodology [32]. This approach preserved the native hemagglutinin–neuraminidase active sites of the HN protein, along with the majority of its antigenic sites and all the cysteine residues crucial for its tertiary structure. The resulting truncated HN protein is efficiently expressed in prokaryotic systems as a monomer, in contrast to the wild-type tetramer form. The prokaryotically expressed HNJD protein is readily recognized by NDV-specific antibodies, demonstrating robust reactivity. Immunofluorescence assays corroborated the accurate expression and correct anchoring of the HNJD protein to the spore surface during sporulation.
Upon ingestion by animals, B. subtilis spores can safely navigate through the stomach and germinate within the upper intestine, where they undergo a brief period of multiplication before sporulating again in the lower intestine [54,55]. Both spores and vegetative cells possess the capacity to interact with intestinal epithelial cells, thereby stimulating the proliferation of intestinal-associated lymphocytes and fortifying the mucosal immune system [56]. The mucosal immune system is pivotal for immune defense, acting as the body’s initial barrier against pathogenic invasion. In the context of preventive vaccination against Newcastle disease, eliciting an effective mucosal immune response can significantly diminish the rates of infection and morbidity, constituting the most efficacious strategy for the prevention and control of the disease [57]. In our investigation, we discovered that B. subtilis RH has the capacity to induce both systemic and local mucosal immune responses. It was observed that supplementing mice with B. subtilis RH resulted in elevated levels of serum IgG and intestinal sIgA. sIgA is the most abundant immunoglobulin produced by the intestinal mucosa, contributing to mucosal homeostasis by inhibiting pathogen adherence to the mucosal surface and playing a vital role in the early prevention of infection [58]. In contrast, commercial vaccines administered via intraperitoneal injection fail to stimulate the production of anti-NDV-specific sIgA in mice. This limitation is due to the fact that most commercial vaccines delivered through non-intestinal routes provide only partial protection against clinical infections and do not completely eliminate infection at the site of initial mucosa invasion. Chicks immunized with commercially inactivated vaccines often require mucosal immune adjuvants to elicit the effective production of specific mucosal antibodies, as indicated in previous studies [59,60]. HI and NA assays are used to measure antibody potency, reflecting their capacity to neutralize the virus and prevent infection. Through microneutralization and HI assays, we have shown that serum antibodies derived from mice vaccinated with RH-spores effectively neutralize NDV, preventing its cellular invasion. This suggests that antigen delivery via spores can effectively penetrate the selectively permeable mucus layer of the intestinal tract, leading to the generation of antibodies and the secretion of cytokines by associated lymphocytes [61,62], which contributes to combating disease infections. Gonçalves et al. [15] immunized zebrafish with spores displaying the Vibrio antigen OmpK, leading to a 50–90% increase in survival rates after infection with Vibrio anguillarum and Vibrio parahaemolyticus. Oh et al. [63] constructed a recombinant B. subtilis strain expressing the protective antigen (PA) of Bacillus anthracis, which successfully elicited a protective immune response in mice, with the level of protection varying according to the immunization method used. In our study, we observed that the immunogenic effect was dependent on the mode of administration, with the mixed feeding group showing superior performance over the intermittent gavage group in all measured parameters, including IgG and sIgA levels, as well as NA and HI titers. This could be attributed to the fact that dietary spores germinate in the jejunum and ileum, whereas spores and vegetative cells do not permanently colonize the intestine and are only in it for a brief duration [54,64]. Our findings suggest that continuous feeding immunization is more effective at inducing intestinal mucosal immunity and probiotic effects than high-dose gavage administered a limited number of times, aligning with previous research conducted in our laboratory. Additionally, gavage may induce stress and disrupt homeostasis in animals, indicating that intermittent gavage is not an effective or suitable immunization method.
In our study, B. subtilis RH modulated the expression levels of cytokines such as IL-1β, IL-6, IL-10, TNF-α, and IFN-γ in mouse ileal tissues. These cytokines are instrumental in orchestrating the Th1/Th2 pathway, which is essential for the regulation of both innate and adaptive immune responses in animals [65]. CD4+ T lymphocytes can differentiate into either Th1 or Th2 cell subsets. The elevated expression of IL-1β, TNF-α, and IFN-γ contributes to cellular immunity by fostering a converging Th1-type immune response. IFN-γ, acting as a pivotal activator of cellular immunity, stimulates CD8+ T lymphocytes and macrophages to eliminate foreign pathogens or infected cells. On the other hand, IL-6 and IL-10 respond to Th2 chemotaxis and also propel the proliferation of T and B cells, thereby enhancing the innate immune response [35]. Unlike many commercially inactivated vaccines that predominantly elicit Th2-type responses [66], B. subtilis RH is capable of promoting both cellular and innate immunity. The spores of the wild-type B. subtilis strain also upregulated cytokine expression, demonstrating their capacity to bolster the immune system. This observation aligns with the findings reported by LEE et al. [67].
Compared to conventional injectable vaccines, mucosal vaccines offer convenience, enhanced safety, and cost-effectiveness. Our investigation has demonstrated that B. subtilis RH can elicit a specific immune response when administered orally, highlighting its potential as an adjuvant for mucosal vaccines, which is particularly advantageous in the livestock and poultry industries. Measures such as gut histomorphometry and immune organ indices are frequently employed in research to assess gastrointestinal development and performance. The morphology of the small intestine directly mirrors the health of the gut, as well as its digestive and absorptive capabilities, which are critical for sustaining the intestinal immune system [68,69,70]. Longer villi correlate with epithelial cell proliferation and can stimulate cell mitosis, whereas shorter villi and deeper crypts are associated with nutrient malabsorption and diminished gastrointestinal function [71,72,73,74]. Our research has determined that dietary supplementation with B. subtilis RH results in an increase in the height of ileal villi and a decrease in crypt depth, thereby enhancing intestinal tissue structure in mice. In related findings, Dong et al. [75] found that B. subtilis BYS2 not only increased the height of avian duodenal and jejunal villi but also increased the villi-to-crypts rates, reduced viral load following NDV infection, and significantly improved survival rates. Jayaraman et al. [76] discovered that B. subtilis PB6 could replace antibiotic growth promoters (AGP) to prevent necrotizing enterocolitis while concurrently improving small intestine organization and the growth performance of broilers through mixed feeding. The intestinal mucosa’s epithelial layer is replete with IELs, which are crucial for preserving intestinal epithelial integrity and play a pivotal role in immune surveillance and cell-mediated mucosal immunity [56,77]. In our study, both strains of B. subtilis effectively stimulated the proliferation of IELs in the mouse ileum, thereby enhancing immunoprotection against virus challenges and potentially blocking the infection route of NDV. However, it remains uncertain if this effect is replicated with commercial vaccines. The organ index serves as an objective measure to assess the growth and development of immune organs and is a crucial parameter for evaluating the micro-ecological impacts of probiotics [78,79]. Our findings indicate that B. subtilis RH promotes splenic growth in mice, yet its influence on thymic development was not evident on day 42. This could be attributed to the fact that the thymus is among the first immune organs to develop and undergoes age-related involution [80], culminating in no significant disparity between the experimental groups. Furthermore, B. subtilis RH has been utilized as a genetically engineered probiotic for animal feed at the recommended dosage (1.0–2.0 × 106 CFU/g), which is consistent with that of most probiotics [81,82]. This approach ensures that it functions as an immune adjuvant while eliciting an effective immune response. In a related study by Pham et al. [83], when the conventional dose (1.0 × 106 CFU/g) of CotB-VP28 spores in feed pellets was increased by 50-fold (5.0 × 107 CFU/g) or 1000-fold (1.0 × 109 CFU/g) for black tiger shrimps, the enhancement in protection against spot syndrome virus infection was marginally increased by only 2.5% and 7.5%, respectively. Thus, routine dosages of spore supplementation can provide effective immune protection, and higher concentrations are not only unnecessary but also impractical in the poultry industry due to cost implications. Nonetheless, we posit that the optimal concentration of various recombinant B. subtilis strains may differ depending on the vectors used, the expression methods, and efficiency. Future poultry experiments could explore the immunoprotective effects across different dosages. In summary, these findings indicate that B. subtilis RH, when used as a feed additive, significantly enhances the development of intestinal and immune organs in animals, promotes the proliferation of immune-related lymphocytes, and shows promise as a mucosal immune adjuvant.
Although B. subtilis RH was capable of inducing an effective specific immune response, the IgG levels and NA titers were not as high as those achieved with commercial vaccines. One plausible explanation for this discrepancy could be the absence of a linker peptide, which might impact the expression of exogenous proteins. The inclusion of a linker peptide can form a stable helical structure that bridges the anchor protein and the target protein, alleviating rigidity issues and enhancing their stability and expression. However, the direct fusion of anchor proteins may lead to undesirable outcomes such as incorrect folding, low yields, and compromised biological activity [84]. Hinc et al. [85] compared the expression efficiency of fusion proteins in cotZ-UreA versus cotB-GGGEAAAKGGGG-UreA, finding that the latter, which incorporates flexible linker peptides, expressed up to 100-fold more recombinant proteins on a spore than the former. Although various types of linker peptides have been designed over time [86,87], the ideal configuration remains elusive. The selection of different anchor proteins [8,85] and ligation strategies [88,89] can also significantly enhance the presentation efficiency of exogenous proteins. Determining how to improve the display efficiency of the HNJD protein on the spore surface may be a focus of our future research endeavors. Looking ahead, omics technologies should be harnessed to uncover the precise mechanisms through which B. subtilis RH induces immune responses and exerts its probiotic effects.
5. Conclusions
In this study, we developed a recombinant B. subtilis RH strain capable of expressing the NDV HN protein on the surface of its spores. This innovation induced both mucosal and systemic immune responses in mice. Additionally, the recombinant B. subtilis enhanced the expression of cytokines in the ileum, fostered the development of immune organs and the intestinal tract, and stimulated the proliferation of intestinal-associated lymphocytes. As such, it also serves as a mucosal immune adjuvant, positioning it as a potential novel probiotic strain engineered to combat NDV. The evaluation of its immunogenicity in mice conducted herein lays the groundwork for future challenge studies in poultry models.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/microorganisms12030439/s1. The supplementary files provided include Table S1, which lists the trains, vaccines, cell lines, plasmids, and primer sequences used; Table S2, detailing the primer sequences for real-time quantitative PCR; and Table S3, presenting the HNJD gene sequences along with the optimized sites.
Author Contributions
J.L. and M.Y. contributed equally to this study. J.L., M.Y. and K.P. contributed to the conceptualization; the methodology was developed by J.L., M.Y., B.C., Z.W., Y.C. and K.P.; the formal analysis was conducted by J.L. and M.Y.; the resources were provided by D.Z. and Y.Z.; data curation involved B.C., Z.W. and Y.C.; the original draft was written by J.L. and M.Y.; X.N., Y.Z. and K.P. reviewed and edited the draft; visualization was created by Y.Y. and M.Z.; supervision was provided by X.N., Y.Z. and K.P.; project administration was managed by K.P.; funding acquisition was secured by K.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Technology Innovation Research Team in the University of Sichuan Province (Grant No. KM406183.1), the Sichuan Province Science and Technology Plan Key Research and Development Project (Grant No. 2021FYS0002), the Sichuan Agricultural University Double Branch Program (Grant No. 03571146), and the Chengdu Agricultural College Science and Technology Fund (Grant No. 20QN01).
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Piewngam, P.; Khongthong, S.; Roekngam, N.; Theapparat, Y.; Sunpaweravong, S.; Faroongsarng, D.; Otto, M. Probiotic for Pathogen-Specific Staphylococcus aureus Decolonisation in Thailand: A Phase 2, Double-Blind, Randomised, Placebo-Controlled Trial. Lancet Microbe 2023, 4, e75–e83. [Google Scholar] [CrossRef]
- Brophy, J.A.N.; Triassi, A.J.; Adams, B.L.; Renberg, R.L.; Stratis-Cullum, D.N.; Grossman, A.D.; Voigt, C.A. Engineered Integrative and Conjugative Elements for Efficient and Inducible DNA Transfer to Undomesticated Bacteria. Nat. Microbiol. 2018, 3, 1043–1053. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Y.; Yang, R. Recent Progress in Bacillus subtilis Spore-Surface Display: Concept, Progress, and Future. Appl. Microbiol. Biotechnol. 2017, 101, 933–949. [Google Scholar] [CrossRef]
- Sarao, L.K.; Arora, M. Probiotics, Prebiotics, and Microencapsulation: A Review. Crit. Rev. Food Sci. Nutr. 2017, 57, 344–371. [Google Scholar] [CrossRef]
- Setlow, P. Spores of Bacillus subtilis: Their Resistance to and Killing by Radiation, Heat and Chemicals. J. Appl. Microbiol. 2006, 101, 514–525. [Google Scholar] [CrossRef]
- Saggese, A.; Baccigalupi, L.; Donadio, G.; Ricca, E.; Isticato, R. The Bacterial Spore as a Mucosal Vaccine Delivery System. Int. J. Mol. Sci. 2023, 24, 10880. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Lin, Z.; Zhao, L.; Chen, T.; Shang, M.; Jiang, H.; Tang, Z.; Zhou, X.; Shi, M.; Zhou, L.; et al. Bacillus subtilis Spore with Surface Display of Paramyosin from Clonorchis sinensis Potentializes a Promising Oral Vaccine Candidate. Parasites Vectors 2018, 11, 156. [Google Scholar] [CrossRef]
- Negri, A.; Potocki, W.; Iwanicki, A.; Obuchowski, M.; Hinc, K. Expression and Display of Clostridium difficile Protein FliD on the Surface of Bacillus subtilis Spores. J. Med. Microbiol. 2013, 62, 1379–1385. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, Y.L.; Lv, F.L.; Xu, L.D.; Huang, Y.W. Surface Display of Peptides Corresponding to the Heptad Repeat 2 Domain of the Feline Enteric Coronavirus Spike Protein on Bacillus subtilis Spores Elicits Protective Immune Responses against Homologous Infection in a Feline Aminopeptidase-N-Transduced Mouse Model. Front. Immunol. 2022, 13, 925922. [Google Scholar]
- Barnes, A.G.C.; Cerovic, V.; Hobson, P.S.; Klavinskis, L.S. Bacillus subtilis Spores: A Novel Microparticle Adjuvant Which Can Instruct a Balanced Th1 and Th2 Immune Response to Specific Antigen. Eur. J. Immunol. 2007, 37, 1538–1547. [Google Scholar] [CrossRef] [PubMed]
- Amuguni, H.; Lee, S.; Kerstein, K.; Brown, D.; Belitsky, B.; Herrmann, J.; Keusch, G.; Sonenshein, A.; Tzipori, S. Sublingual Immunization with an Engineered Bacillus subtilis Strain Expressing Tetanus Toxin Fragment C Induces Systemic and Mucosal Immune Responses in Piglets. Microbes Infect. 2012, 14, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Cuburu, N.; Kweon, M.N.; Song, J.H.; Hervouet, C.; Luci, C.; Sun, J.B.; Hofman, P.; Holmgren, J.; Anjuère, F.; Czerkinsky, C. Sublingual Immunization Induces Broad-Based Systemic and Mucosal Immune Responses in Mice. Vaccine 2007, 25, 8598–8610. [Google Scholar] [CrossRef] [PubMed]
- Mou, C.; Zhu, L.; Xing, X.; Lin, J.; Yang, Q. Immune Responses Induced by Recombinant Bacillus subtilis Expressing the Spike Protein of Transmissible Gastroenteritis Virus in Pigs. Antivir. Res. 2016, 131, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Mou, C.; Zhu, L.; Yang, J.; Xu, W.; Cheng, X.; Yang, Q. Immune Responses Induced by Recombinant Bacillus subtilis Expressing the Hemagglutinin Protein of H5N1 in Chickens. Sci. Rep. 2016, 6, 38403. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, G.; Santos, R.A.; Coutinho, F.; Pedrosa, N.; Curado, M.; Machado, M.; Costas, B.; Bonneville, L.; Serrano, M.; Carvalho, A.P.; et al. Oral Vaccination of Fish against Vibriosis Using Spore-Display Technology. Front. Immunol. 2022, 13, 1012301. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.S.; Alvarez, J.; Bicout, D.J.; Calistri, P.; Canali, E.; Drewe, J.A.; Garin-Bastuji, B.; Gonzales Rojas, J.L.; Gortázar Schmidt, C.; Herskin, M.; et al. Assessment of the Control Measures of the Category a Diseases of Animal Health Law: Newcastle Disease. EFSA J. 2021, 19, e06946. [Google Scholar]
- Absalón, A.E.; Cortés-Espinosa, D.V.; Lucio, E.; Miller, P.J.; Afonso, C.L. Epidemiology, Control, and Prevention of Newcastle Disease in Endemic Regions: Latin America. Trop. Anim. Health Prod. 2019, 51, 1033–1048. [Google Scholar] [CrossRef]
- Dimitrov, K.M.; Afonso, C.L.; Yu, Q.; Miller, P.J. Newcastle Disease Vaccines-A Solved Problem or a Continuous Challenge? Vet. Microbiol. 2017, 206, 126–136. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, J.; Xu, H.; Li, J.; Hu, Z.; Hu, S.; And, X.W.; Liu, X. Effects of the HN Antigenic Difference between the Vaccine Strain and the Challenge Strain of Newcastle Disease Virus on Virus Shedding and Transmission. Viruses 2017, 9, 225. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Kwon, H.J.; Kim, T.E.; Kim, J.H.; Yoo, H.S.; Kim, S.J. Variation of a Newcastle Disease Virus Hemagglutinin-Neuraminidase Linear Epitope. J. Clin. Microbiol. 2008, 46, 1541–1544. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Iorio, R.M.; Syddall, R.J.; Sheehan, J.P.; Bratt, M.A.; Glickman, R.L.; Riel, A.M. Neutralization Map of the Hemagglutinin-Neuraminidase Glycoprotein of Newcastle Disease Virus: Domains Recognized by Monoclonal Antibodies That Prevent Receptor Recognition. J. Virol. 1991, 65, 4999–5006. [Google Scholar] [CrossRef] [PubMed]
- Boursnell, M.E.; Green, P.F.; Samson, A.C.; Campbell, J.I.; Deuter, A.; Peters, R.W.; Millar, N.S.; Emmerson, P.T.; Binns, M.M. A Recombinant Fowlpox Virus Expressing the Hemagglutinin-Neuraminidase Gene of Newcastle Disease Virus (NDV) Protects Chickens against Challenge by NDV. Virology 1990, 178, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Mayahi, V.; Esmaelizad, M.; Harzandi, N. Designing a Novel Recombinant HN Protein with Multi Neutralizing Antigenic Sites and Auto Tag Removal Ability Based on NDV-VIIj for Diagnosis and Vaccination Application. Indian J. Microbiol. 2018, 58, 326–331. [Google Scholar] [CrossRef]
- Hu, S.; Wang, T.; Liu, Y.; Meng, C.; Wang, X.; Wu, Y.; Liu, X. Identification of a Variable Epitope on the Newcastle Disease Virus Hemagglutinin-Neuraminidase Protein. Vet. Microbiol. 2010, 140, 92–97. [Google Scholar] [CrossRef]
- Pedersen, K.; Marks, D.R.; Arsnoe, D.M.; Afonso, C.L.; Bevins, S.N.; Miller, P.J.; Randall, A.R.; DeLiberto, T.J. Avian Paramyxovirus Serotype 1 (Newcastle Disease Virus), Avian Influenza Virus, and Salmonella spp. In Mute Swans (Cygnus olor) in the Great Lakes Region and Atlantic Coast of the United States. Avian Dis. 2014, 58, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Young, J.K.; Li, D.; Abramowitz, M.C.; Morrison, T.G. Interaction of Peptides with Sequences from the Newcastle Disease Virus Fusion Protein Heptad Repeat Regions. J. Virol. 1999, 73, 5945–5956. [Google Scholar] [CrossRef]
- Ju, A.; Duan, A.; Zhang, Y.; Qin, Y.; Xue, L.; Ma, X.; Luan, W.; Yang, S. The Construction of Recombinant Lactobacillus casei Expressing Hemagglutinin-Neuraminidase Protein and Its Immune Response in Chickens. Microb. Pathog. 2021, 158, 105091. [Google Scholar] [CrossRef]
- Kaiser, A.; Willer, T.; Sid, H.; Petersen, H.; Baumgärtner, W.; Steinberg, P.; Rautenschlein, S. Susceptibility of Primary Chicken Intestinal Epithelial Cells for Low Pathogenic Avian Influenza Virus and Velogenic Viscerotropic Newcastle Disease Virus. Virus Res. 2016, 225, 50–63. [Google Scholar] [CrossRef]
- Dong, B.; Tang, N.; Guan, Y.; Qu, G.; Miao, L.; Han, W.; Shen, Z. Type and Abundance of Sialic Acid Receptors on Host Cell Membrane Affect Infectivity and Viral Titer of Different Strains of Newcastle Disease Virus. J. Virol. Methods 2022, 302, 114488. [Google Scholar] [CrossRef]
- Zhang, D.; Ding, Z.; Xu, X. Pathologic Mechanisms of the Newcastle Disease Virus. Viruses 2023, 15, 864. [Google Scholar] [CrossRef]
- Mao, Q.; Ma, S.; Schrickel, P.L.; Zhao, P.; Wang, J.; Zhang, Y.; Li, S.; Wang, C. Review Detection of Newcastle Disease Virus. Front. Vet. Sci. 2022, 9, 936251. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, C.; Zhang, O. Prokaryotic Expression of Structural Domains of the Newcastle Disease Virus Hemagglutinin-Neuraminidase. Chin. J. Prev. Vet. Med. 2007, 29, 32–35. [Google Scholar]
- Li, W.; Feng, J.; Li, J.; Li, J.; Wang, Z.; Khalique, A.; Yang, M.; Ni, X.; Zeng, D.; Zhang, D.; et al. Surface Display of Antigen Protein VP8* of Porcine Rotavirus on Bacillus subtilis Spores Using CotB as a Fusion Partner. Molecules 2019, 24, 3793. [Google Scholar] [CrossRef]
- Julkowska, D.; Obuchowski, M.; Holland, I.B.; Séror, S.J. Comparative Analysis of the Development of Swarming Communities of Bacillus subtilis 168 and a Natural Wild Type: Critical Effects of Surfactin and the Composition of the Medium. J. Bacteriol. 2005, 187, 65–76. [Google Scholar] [CrossRef]
- Stasiłojć, M.; Hinc, K.; Peszyńska-Sularz, G.; Obuchowski, M.; Iwanicki, A. Recombinant Bacillus subtilis Spores Elicit Th1/Th17-Polarized Immune Response in a Murine Model of Helicobacter pylori Vaccination. Mol. Biotechnol. 2015, 57, 685–691. [Google Scholar] [CrossRef]
- Wang, H.; Yang, R.; Hua, X.; Zhao, W.; Zhang, W. Functional Display of Active β-Galactosidase on Bacillus subtilis Spores Using Crust Proteins as Carriers. Food Sci. Biotechnol. 2015, 24, 1755–1759. [Google Scholar] [CrossRef]
- Hinc, K.; Isticato, R.; Dembek, M.; Karczewska, J.; Iwanicki, A.; Peszyńska-Sularz, G.; De Felice, M.; Obuchowski, M.; Ricca, E. Expression and Display of UreA of Helicobacter acinonychis on the Surface of Bacillus subtilis Spores. Microb. Cell Fact. 2010, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Duc, L.H.; Hong, H.A.; Fairweather, N.; Ricca, E.; Cutting, S.M. Bacterial Spores as Vaccine Vehicles. Infect. Immun. 2003, 71, 2810–2818. [Google Scholar] [CrossRef] [PubMed]
- Chumbe, A.; Izquierdo-Lara, R.; Calderón, K.; Fernández-Díaz, M.; Vakharia, V.N. Development of a Novel Newcastle Disease Virus (NDV) Neutralization Test Based on Recombinant NDV Expressing Enhanced Green Fluorescent Protein. Virol. J. 2017, 14, 232. [Google Scholar] [CrossRef] [PubMed]
- GB/T 16550-2020: Diagnostic Techniques for Newcastle Disease. Available online: https://openstd.samr.gov.cn/bzgk/gb/newGbInfo?hcno=DAA58A6FCC696F91339EED5BF8460CAB (accessed on 14 December 2020).
- Xin, J.; Wang, H.; Sun, N.; Bughio, S.; Zeng, D.; Li, L.; Wang, Y.; Khalique, A.; Zeng, Y.; Pan, K.; et al. Probiotic Alleviate Fluoride-Induced Memory Impairment by Reconstructing Gut Microbiota in Mice. Ecotox. Environ. Safe. 2021, 215, 112108. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Dai, X.; Wang, Z.; Ni, X.; Zeng, D.; Zeng, Y.; Zhang, D.; Pan, K. Effects of Antimicrobial Peptides Gal-13 on the Growth Performance, Intestinal Microbiota, Digestive Enzyme Activities, Intestinal Morphology, Antioxidative Activities, and Immunity of Broilers. Probiotics Antimicrob. Proteins 2023, 15, 694–705. [Google Scholar] [CrossRef]
- de Melo Pereira, G.V.; de Oliveira Coelho, B.; Magalhães Júnior, A.I.; Thomaz-Soccol, V.; Soccol, C.R. How to Select a Probiotic? A Review and Update of Methods and Criteria. Biotechnol. Adv. 2018, 36, 2060–2076. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.A.; Huang, J.M.; Khaneja, R.; Hiep, L.V.; Urdaci, M.C.; Cutting, S.M. The Safety of Bacillus subtilis and Bacillus indicus as Food Probiotics. J. Appl. Microbiol. 2008, 105, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Kovács, Á.T. Bacillus subtilis . Trends Microbiol. 2019, 27, 724–725. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Xu, D.; Xie, D.; Wang, M.; Li, Z.; Guo, X. Effects of Antibacterial Peptide-Producing Bacillus subtilis and Lactobacillus buchneri on Fermentation, Aerobic Stability, and Microbial Community of Alfalfa Silage. Bioresour. Technol. 2020, 315, 123881. [Google Scholar] [CrossRef] [PubMed]
- Ramlucken, U.; Ramchuran, S.O.; Moonsamy, G.; Lalloo, R.; Thantsha, M.S.; Jansen van Rensburg, C. A Novel Bacillus Based Multi-Strain Probiotic Improves Growth Performance and Intestinal Properties of Clostridium perfringens Challenged Broilers. Poult. Sci. 2020, 99, 331–341. [Google Scholar] [CrossRef]
- Azimirad, M.; Alebouyeh, M.; Naji, T. Inhibition of Lipopolysaccharide-Induced Interleukin 8 in Human Adenocarcinoma Cell Line HT-29 by Spore Probiotics: B. coagulans and B. subtilis (natto). Probiotics Antimicrob. Proteins 2017, 9, 56–63. [Google Scholar] [CrossRef]
- Pi, X.; Teng, W.; Fei, D.; Zhao, G.; Liu, W. Effects of Live Combined Bacillus subtilis and Enterococcus faecium on Gut Microbiota Composition in C57BL/6 Mice and in Humans. Front. Cell. Infect. Microbiol. 2022, 12, 821662. [Google Scholar] [CrossRef]
- Clemente, J.C.; Manasson, J.; Scher, J.U. The Role of the Gut Microbiome in Systemic Inflammatory Disease. BMJ 2018, 360, j5145. [Google Scholar] [CrossRef]
- Isticato, R.; Cangiano, G.; Tran, H.T.; Ciabattini, A.; Medaglini, D.; Oggioni, M.R.; De Felice, M.; Pozzi, G.; Ricca, E. Surface Display of Recombinant Proteins on Bacillus subtilis Spores. J. Bacteriol. 2001, 183, 6294–6301. [Google Scholar] [CrossRef]
- Dai, X.; Liu, M.; Pan, K.; Yang, J. Surface Display of OmpC of Salmonella Serovar Pullorum on Bacillus subtilis Spores. PLoS ONE 2018, 13, e0191627. [Google Scholar] [CrossRef]
- Li, W.; Li, J.; Dai, X.; Liu, M.; Khalique, A.; Wang, Z.; Zeng, Y.; Zhang, D.; Ni, X.; Zeng, D.; et al. Surface Display of Porcine Circovirus Type 2 Antigen Protein Cap on the Spores of Bacillus subtilis 168: An Effective Mucosal Vaccine Candidate. Front. Immunol. 2022, 13, 1007202. [Google Scholar] [CrossRef]
- Hoa, T.T.; Duc, L.H.; Isticato, R.; Baccigalupi, L.; Ricca, E.; Van, P.H.; Cutting, S.M. Fate and Dissemination of Bacillus subtilis Spores in a Murine Model. Appl. Environ. Microbiol. 2001, 67, 3819–3823. [Google Scholar] [CrossRef]
- Hoa, N.T.; Baccigalupi, L.; Huxham, A.; Smertenko, A.; Van, P.H.; Ammendola, S.; Ricca, E.; Cutting, A.S. Characterization of Bacillus Species Used for Oral Bacteriotherapy and Bacterioprophylaxis of Gastrointestinal Disorders. Appl. Environ. Microbiol. 2000, 66, 5241–5247. [Google Scholar] [CrossRef]
- Olivares-Villagómez, D.; Van Kaer, L. Intestinal Intraepithelial Lymphocytes: Sentinels of the Mucosal Barrier. Trends Immunol. 2018, 39, 264–275. [Google Scholar] [CrossRef]
- Shan, C.; Sun, B.; Dalloul, R.A.; Zhai, Z.; Sun, P.; Li, M.; Yang, S.; Luan, W. Effect of the Oral Administration of Astragalus Polysaccharides on Jejunum Mucosal Immunity in Chickens Vaccinated against Newcastle Disease. Microb. Pathog. 2019, 135, 103621. [Google Scholar] [CrossRef]
- Cerutti, A.; Chen, K.; Chorny, A. Immunoglobulin Responses at the Mucosal Interface. Annu. Rev. Immunol. 2011, 29, 273–293. [Google Scholar] [CrossRef]
- Rahman, M.M.; Uyangaa, E.; Han, Y.W.; Hur, J.; Park, S.Y.; Lee, J.H.; Kim, K.; Eo, S.K. Modulation of Systemic and Mucosal Immunity against an Inactivated Vaccine of Newcastle Disease Virus by Oral Co-Administration of Live Attenuated Salmonella enterica Serovar Typhimurium Expressing Chicken Interleukin-18 and Interferon-α. J. Vet. Med. Sci. 2015, 77, 395–403. [Google Scholar] [CrossRef]
- Xu, X.; Qian, J.; Qin, L.; Li, J.; Xue, C.; Ding, J.; Wang, W.; Ding, W.; Yin, R.; Jin, N.; et al. Chimeric Newcastle Disease Virus-Like Particles Containing DC-Binding Peptide-Fused Haemagglutinin Protect Chickens from Virulent Newcastle Disease Virus and H9N2 Avian Influenza Virus Challenge. Virol. Sin. 2020, 35, 455–467. [Google Scholar] [CrossRef]
- Marasini, N.; Skwarczynski, M.; Toth, I. Oral Delivery of Nanoparticle-Based Vaccines. Expert Rev. Vaccines 2014, 13, 1361–1376. [Google Scholar] [CrossRef]
- Zhao, K.; Xie, Y.; Lin, X.; Xu, W. The Mucoadhesive Nanoparticle-Based Delivery System in the Development of Mucosal Vaccines. Int. J. Nanomed. 2022, 17, 4579–4598. [Google Scholar] [CrossRef]
- Oh, Y.; Kim, J.A.; Kim, C.H.; Choi, S.K.; Pan, J.G. Bacillus subtilis Spore Vaccines Displaying Protective Antigen Induce Functional Antibodies and Protective Potency. BMC Vet. Res. 2020, 16, 259. [Google Scholar] [CrossRef]
- Latorre, J.D.; Hernandez-Velasco, X.; Kallapura, G.; Menconi, A.; Pumford, N.R.; Morgan, M.J.; Layton, S.L.; Bielke, L.R.; Hargis, B.M.; Téllez, G. Evaluation of Germination, Distribution, and Persistence of Bacillus subtilis Spores through the Gastrointestinal Tract of Chickens. Poult. Sci. 2014, 93, 1793–1800. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Chen, T.; Sun, H.; Tang, Z.; Yu, J.; Lin, Z.; Ren, P.; Zhou, X.; Huang, Y.; Li, X.; et al. Immune Response Induced by Oral Delivery of Bacillus subtilis Spores Expressing Enolase of Clonorchis sinensis in Grass Carps (Ctenopharyngodon idellus). Fish Shellfish Immunol. 2017, 60, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Rosales-Mendoza, S.; Angulo, C. Bacillus subtilis Comes of Age as a Vaccine Production Host and Delivery Vehicle. Expert Rev. Vaccines 2015, 14, 1135–1148. [Google Scholar] [PubMed]
- Lee, J.E.; Kye, Y.C.; Park, S.M.; Shim, B.S.; Yoo, S.; Hwang, E.; Kim, H.; Kim, S.J.; Han, S.H.; Park, T.S.; et al. Bacillus subtilis Spores as Adjuvants against Avian Influenza H9N2 Induce Antigen-Specific Antibody and T Cell Responses in White Leghorn Chickens. Vet. Res. 2020, 51, 68. [Google Scholar] [CrossRef] [PubMed]
- Xun, W.; Shi, L.; Zhou, H.; Hou, G.; Cao, T.; Zhao, C. Effects of Curcumin on Growth Performance, Jejunal Mucosal Membrane Integrity, Morphology and Immune Status in Weaned Piglets Challenged with Enterotoxigenic Escherichia coli. Int. Immunopharmacol. 2015, 27, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Awad, W.A.; Ghareeb, K.; Abdel-Raheem, S.; Böhm, J. Effects of Dietary Inclusion of Probiotic and Synbiotic on Growth Performance, Organ Weights, and Intestinal Histomorphology of Broiler Chickens. Poult. Sci. 2009, 88, 49–56. [Google Scholar] [CrossRef] [PubMed]
- da Silva, B.P.; Martino, H.S.D.; Tako, E. Plant Origin Prebiotics Affect Duodenal Brush Border Membrane Functionality and Morphology, in Vivo (Gallus Gallus). Food Funct. 2021, 12, 6157–6166. [Google Scholar] [CrossRef] [PubMed]
- Aliakbarpour, H.R.; Chamani, M.; Rahimi, G.; Sadeghi, A.A.; Qujeq, D. The Bacillus subtilis and Lactic Acid Bacteria Probiotics Influences Intestinal Mucin Gene Expression, Histomorphology and Growth Performance in Broilers. Asian Australas. J. Anim. Sci. 2012, 25, 1285–1293. [Google Scholar] [CrossRef]
- Ji, L.; Zhang, L.; Liu, H.; Shen, J.; Zhang, Y.; Lu, L.; Zhang, X.; Ma, X. Bacillus subtilis M6 Improves Intestinal Barrier, Antioxidant Capacity and Gut Microbial Composition in AA Broiler. Front. Nutr. 2022, 9, 965310. [Google Scholar] [CrossRef] [PubMed]
- Qiu, K.; Li, C.L.; Wang, J.; Qi, G.H.; Gao, J.; Zhang, H.J.; Wu, S.G. Effects of Dietary Supplementation with Bacillus subtilis, as an Alternative to Antibiotics, on Growth Performance, Serum Immunity, and Intestinal Health in Broiler Chickens. Front. Nutr. 2021, 8, 786878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, Z.; Gu, X.; Zhao, J.; Guo, T.; Kong, J. Probiotic Bacillus subtilis LF11 Protects Intestinal Epithelium against Salmonella Infection. Front. Cell. Infect. Microbiol. 2022, 12, 837886. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Li, R.; Liu, Y.; Ma, L.; Zha, J.; Qiao, X.; Chai, T.; Wu, B. Benefit of Dietary Supplementation with Bacillus subtilis BYS2 on Growth Performance, Immune Response, and Disease Resistance of Broilers. Probiotics Antimicrob. Proteins 2020, 12, 1385–1397. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, S.; Das, P.P.; Saini, P.C.; Roy, B.; Chatterjee, P.N. Use of Bacillus subtilis PB6 as a Potential Antibiotic Growth Promoter Replacement in Improving Performance of Broiler Birds. Poult. Sci. 2017, 96, 2614–2622. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.K.; Peng, V.; Sudan, R.; Ulezko Antonova, A.; Di Luccia, B.; Ohara, T.E.; Fachi, J.L.; Grajales-Reyes, G.E.; Jaeger, N.; Trsan, T.; et al. Repression of the Aryl-Hydrocarbon Receptor Prevents Oxidative Stress and Ferroptosis of Intestinal Intraepithelial Lymphocytes. Immunity 2023, 56, 797–812. [Google Scholar] [CrossRef]
- Villagrán-de la Mora, Z.; Vázquez-Paulino, O.; Avalos, H.; Ascencio, F.; Nuño, K.; Villarruel-López, A. Effect of a Synbiotic Mix on Lymphoid Organs of Broilers Infected with Salmonella typhimurium and Clostridium perfringens. Animals 2020, 10, 886. [Google Scholar] [CrossRef]
- Sun, S.; Li, B.; Wu, M.; Deng, Y.; Li, J.; Xiong, Y.; He, S. Effect of Dietary Supplemental Vitamin C and Betaine on the Growth Performance, Humoral Immunity, Immune Organ Index, and Antioxidant Status of Broilers under Heat Stress. Trop. Anim. Health Prod. 2023, 55, 96. [Google Scholar] [CrossRef]
- Gui, J.; Mustachio, L.M.; Su, D.M.; Craig, R.W. Thymus Size and Age-Related Thymic Involution: Early Programming, Sexual Dimorphism, Progenitors and Stroma. Aging Dis. 2012, 3, 280–290. [Google Scholar]
- Xu, Y.; Yu, Y.; Shen, Y.; Li, Q.; Lan, J.; Wu, Y.; Zhang, R.; Cao, G.; Yang, C. Effects of Bacillus subtilis and Bacillus licheniformis on Growth Performance, Immunity, Short Chain Fatty Acid Production, Antioxidant Capacity, and Cecal Microflora in Broilers. Poult. Sci. 2021, 100, 101358. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, H.; Yu, Y.; Zhang, R.; Wu, Y.; Yue, M.; Yang, C. Effects of Bacillus coagulans on Growth Performance, Antioxidant Capacity, Immunity Function, and Gut Health in Broilers. Poult. Sci. 2021, 100, 101168. [Google Scholar] [CrossRef]
- Pham, K.C.; Tran, H.T.T.; Van Doan, C.; Le, P.H.; Van Nguyen, A.T.; Nguyen, H.A.; Hong, H.A.; Cutting, S.M.; Phan, T.N. Protection of Penaeus monodon against White Spot Syndrome by Continuous Oral Administration of a Low Concentration of Bacillus subtilis Spores Expressing the VP28 Antigen. Lett. Appl. Microbiol. 2017, 64, 184–191. [Google Scholar] [CrossRef]
- Chen, H.; Wu, B.; Zhang, T.; Jia, J.; Lu, J.; Chen, Z.; Ni, Z.; Tan, T. Effect of Linker Length and Flexibility on the Clostridium thermocellum Esterase Displayed on Bacillus subtilis Spores. Appl. Biochem. Biotechnol. 2017, 182, 168–180. [Google Scholar] [CrossRef]
- Hinc, K.; Iwanicki, A.; Obuchowski, M. New Stable Anchor Protein and Peptide Linker Suitable for Successful Spore Surface Display in B. subtilis. Microb. Cell Fact. 2013, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Iwanicki, A.; Piątek, I.; Stasiłojć, M.; Grela, A.; Lęga, T.; Obuchowski, M.; Hinc, K. A System of Vectors for Bacillus subtilis Spore Surface Display. Microb. Cell Fact. 2014, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Ullah, J.; Chen, H.; Vastermark, A.; Jia, J.; Wu, B.; Ni, Z.; Le, Y.; Wang, H. Impact of Orientation and Flexibility of Peptide Linkers on T. maritima Lipase Tm1350 Displayed on Bacillus subtilis Spores Surface Using CotB as Fusion Partner. World J. Microbiol. Biotechnol. 2017, 33, 166. [Google Scholar] [CrossRef] [PubMed]
- Potocki, W.; Negri, A.; Peszyńska-Sularz, G.; Hinc, K.; Obuchowski, M.; Iwanicki, A. The Combination of Recombinant and Non-Recombinant Bacillus subtilis Spore Display Technology for Presentation of Antigen and Adjuvant on Single Spore. Microb. Cell Fact. 2017, 16, 151. [Google Scholar] [CrossRef] [PubMed]
- Ghaedmohammadi, S.; Rigi, G.; Zadmard, R.; Ricca, E.; Ahmadian, G. Immobilization of Bioactive Protein a from Staphylococcus aureus (SpA) on the Surface of Bacillus subtilis Spores. Mol. Biotechnol. 2015, 57, 756–766. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).