Limosilactobacillus reuteri HCS02-001 Attenuates Hyperuricemia through Gut Microbiota-Dependent Regulation of Uric Acid Biosynthesis and Excretion
Abstract
:1. Introduction
2. Materials and Methods
2.1. Probiotic Screening
2.1.1. Strain Cultivation
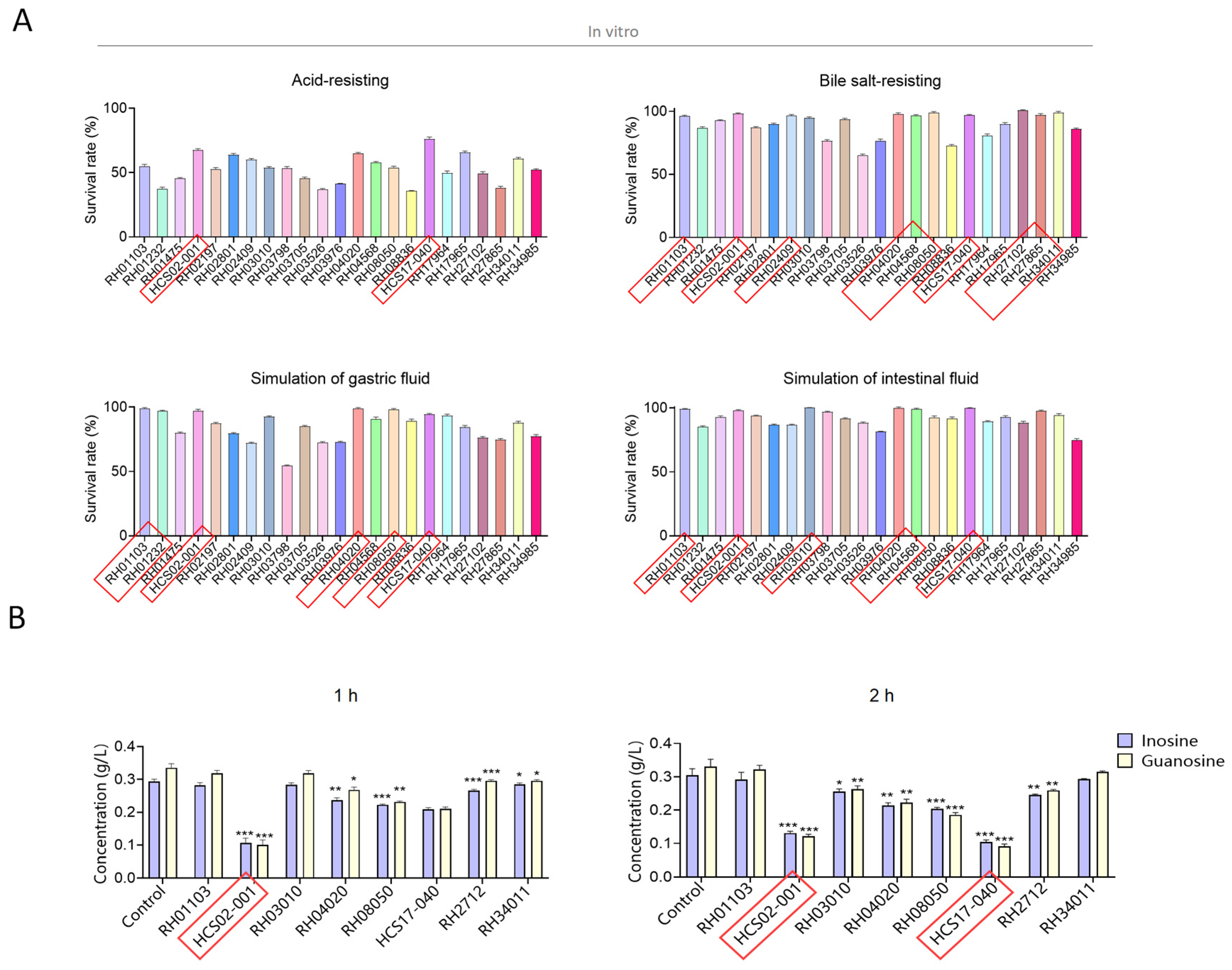
2.1.2. Tolerance to Acid, Bile, Artificial Gastric and Intestinal Fluids
2.1.3. Determination of Nucleoside Degradation Activity In Vitro
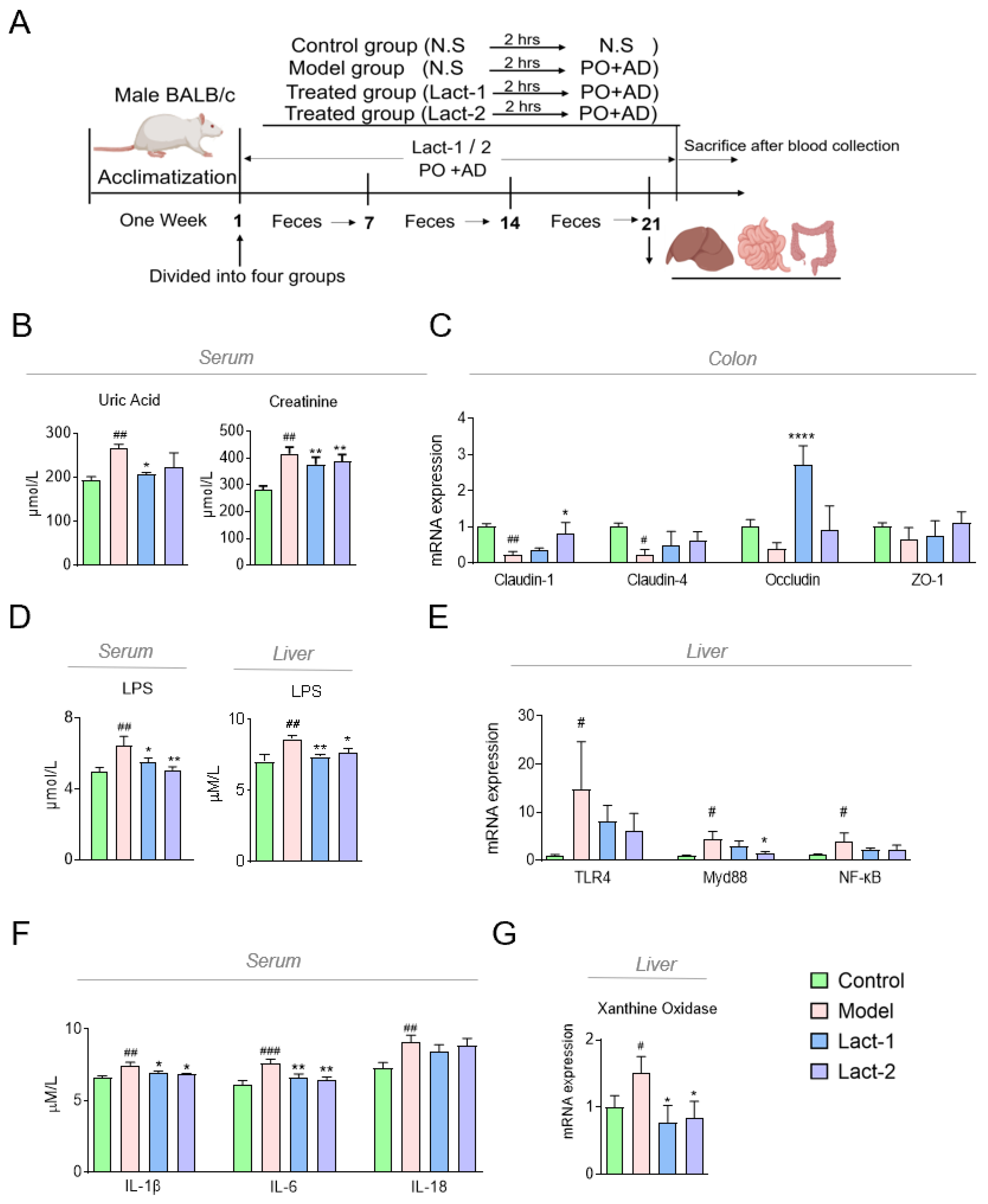
2.2. Animals and Study Design
2.3. Molecular Biology Experiment Design
2.3.1. Biochemical Parameters Analysis
2.3.2. RNA Isolation, cDNA Synthesis, and Quantitative Real-Time PCR (qPCR)
2.3.3. Western Blotting
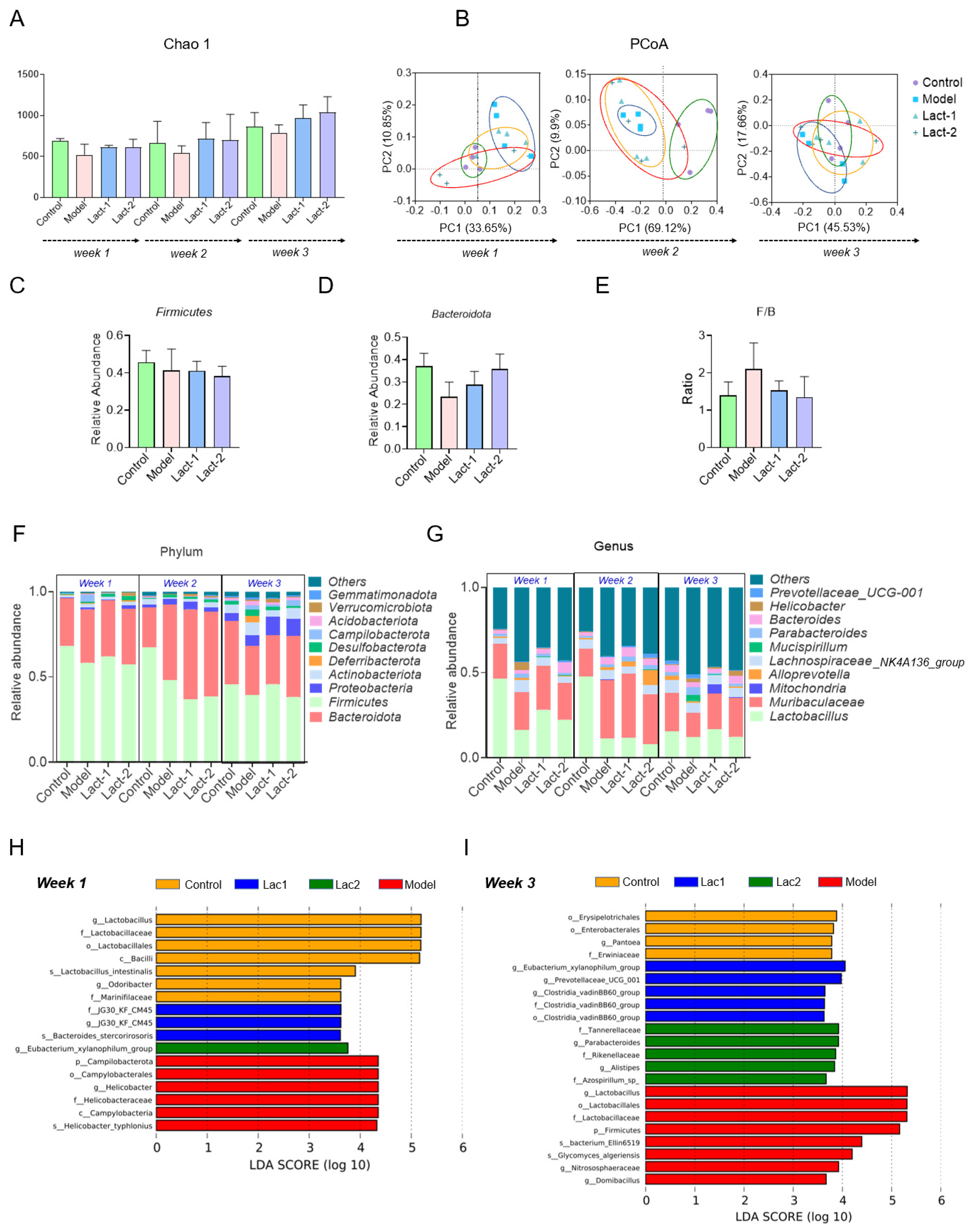
2.4. Genomic DNA Extraction and 16S rRNA Sequencing and Data Analysis
2.5. Cell Lines Culturing
2.6. Transcriptomic Analysis of Metabolites- and Probiotics-Treated HT-29 by RNA Sequencing
2.7. Statistical Analysis
3. Results
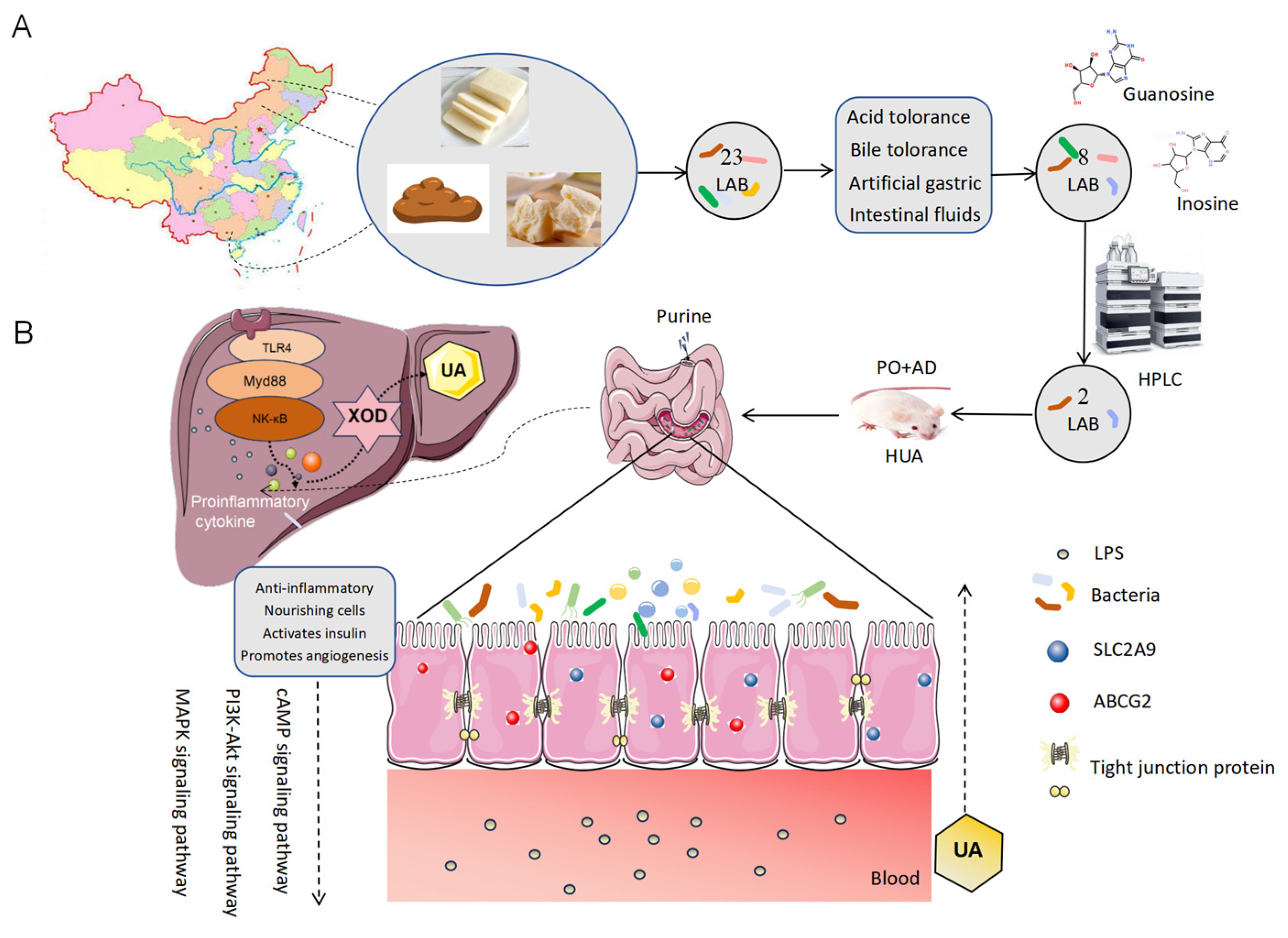
3.1. Screening of Inosine and Guanosine Degrading Lactic Acid Bacteria
3.2. Lact-1 Treatment Alleviated Hyperuricemia in Mice
3.3. Effect of Lact-1 and Lact-2 on Gut Microbiota of Hyperuricemia Mice
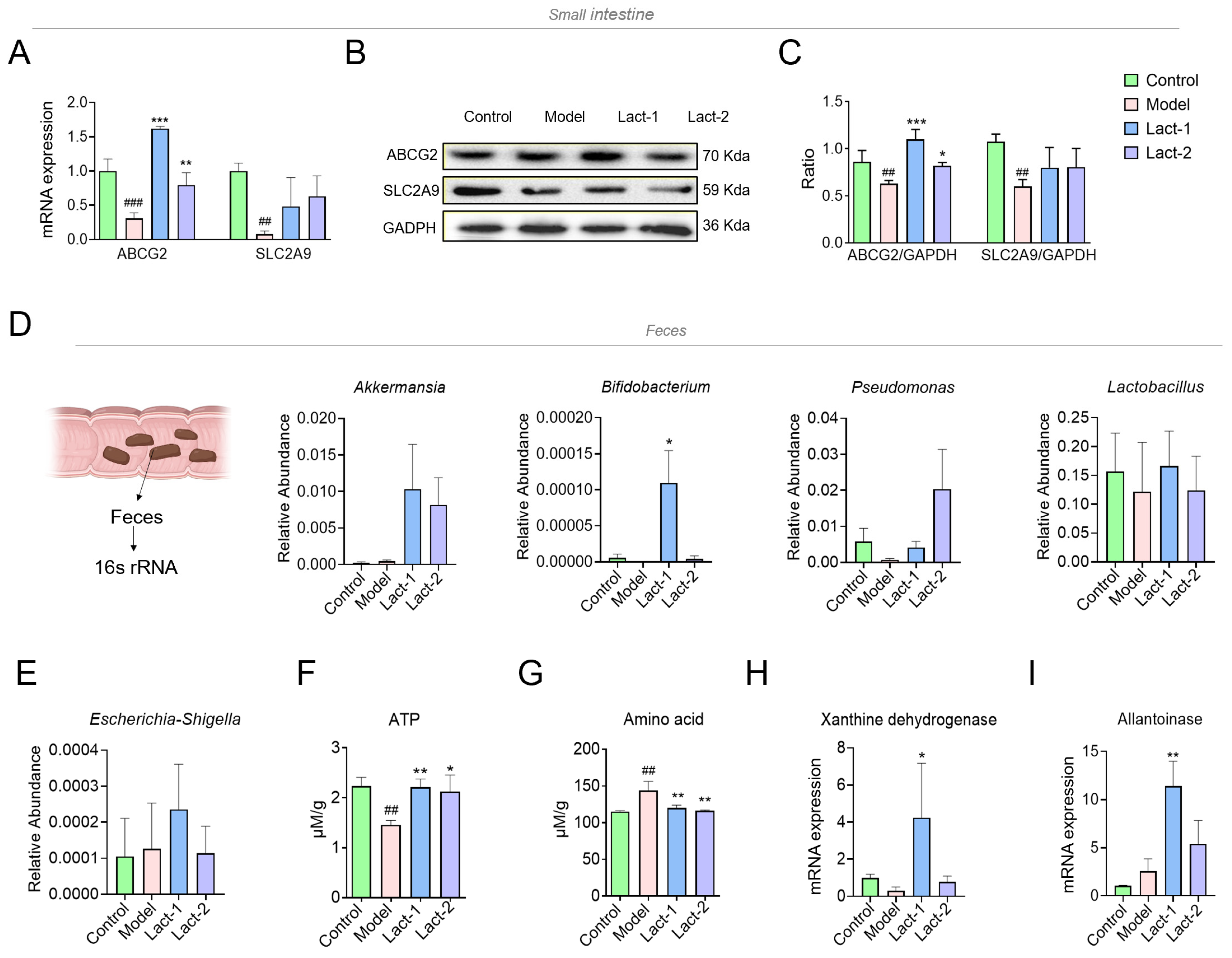
3.4. Effect of Lact-1 and Lact-2 on UA Transporter in Small Intestine
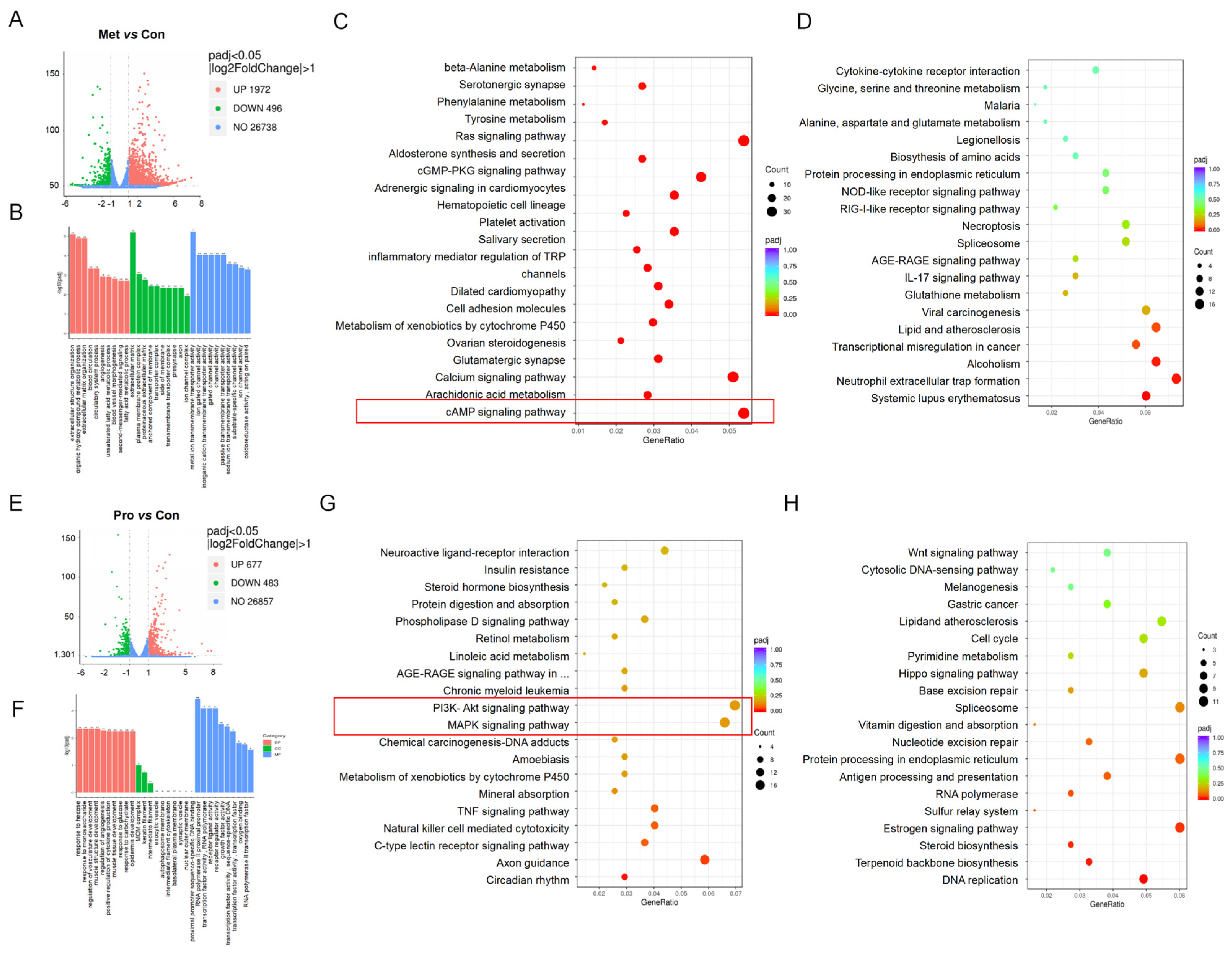
3.5. Transcriptome Sequencing of Lact-1 on HT-29 Cells In Vitro
3.5.1. GO Functional Enrichment Analysis of Metabolites- and Probiotics-Treated HT-29 Cells
3.5.2. KEGG Functional Enrichment Analysis of Metabolites and Probiotics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, W.; Luan, R. Early-life exposure to the Chinese famine of 1959-61 and risk of Hyperuricemia: Results from the China health and retirement longitudinal study. BMC Public Health 2020, 20, 15. [Google Scholar] [CrossRef]
- Joosten, L.A.B.; Crisan, T.O.; Bjornstad, P.; Johnson, R.J. Asymptomatic hyperuricaemia: A silent activator of the innate immune system. Nat. Rev. Rheumatol. 2020, 16, 75–86. [Google Scholar] [CrossRef]
- Bardin, T.; Richette, P. Impact of comorbidities on gout and hyperuricaemia: An update on prevalence and treatment options. BMC Med. 2017, 15, 123. [Google Scholar] [CrossRef]
- Sanikidze, Q.; Mamacashvili, I.; Petriashvili, S. Prevalence of hyperuricemia in patients with chronic heart failure. Georgian Med. News 2021, 311, 85–88. [Google Scholar]
- Chen-Xu, M.; Yokose, C.; Rai, S.K.; Pillinger, M.H.; Choi, H.K. Contemporary prevalence of gout and hyperuricemia in the United states and decadal trends: The national health and nutrition examination survey, 2007–2016. Arthritis Rheumatol. 2019, 71, 991–999. [Google Scholar] [CrossRef]
- Dehlin, M.; Jacobsson, L.; Roddy, E. Global epidemiology of gout: Prevalence, incidence, treatment patterns and risk factors. Nat. Rev. Rheumatol. 2020, 16, 380–390. [Google Scholar] [CrossRef]
- Mehmood, A.; Zhao, L.; Wang, C.; Nadeem, M.; Raza, A.; Ali, N.; Shah, A.A. Management of hyperuricemia through dietary polyphenols as a natural medicament: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2019, 59, 1433–1455. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Qiu, L.; Cheng, X.Q.; Xu, T.; Wu, W.; Zeng, X.J.; Ye, Y.C.; Guo, X.Z.; Cheng, Q.; Liu, Q.; et al. Hyperuricemia and clustering of cardiovascular risk factors in the Chinese adult population. Sci. Rep. 2017, 7, 5456. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Sandoval, J.C.; Madero, M. Treatment of hyperuricemia in chronic kidney disease. Contrib. Nephrol. 2018, 192, 135–146. [Google Scholar] [CrossRef]
- Wilson, L.; Saseen, J.J. Gouty arthritis: A review of acute management and prevention. Pharmacotherapy 2016, 36, 906–922. [Google Scholar] [CrossRef]
- Alvarez-Lario, B.; Alonso-Valdivielso, J.L. Hyperuricemia and gout; the role of diet. Nutr. Hosp. 2014, 29, 760–770. [Google Scholar] [CrossRef]
- Stamp, L.K.; Barclay, M.L. How to prevent allopurinol hypersensitivity reactions? Rheumatology 2018, 57, i35–i41. [Google Scholar] [CrossRef]
- Stamp, L.K.; Chapman, P.T.; Palmer, S.C. Allopurinol and kidney function: An update. Jt. Bone Spine 2016, 83, 19–24. [Google Scholar] [CrossRef]
- Li, L.; Teng, M.; Liu, Y.; Qu, Y.; Zhang, Y.; Lin, F.; Wang, D. Anti-gouty arthritis and antihyperuricemia effects of sunflower (Helianthus annuus) head extract in gouty and hyperuricemia animal models. BioMed Res. Int. 2017, 2017, 5852076. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.T.; Xiu, W.J.; Liu, J.K.; Yang, Y.; Hou, X.G.; Zheng, Y.Y.; Wu, T.T.; Wu, C.X.; Xie, X. Gut microbiota characterization in patients with asymptomatic hyperuricemia: Probiotics increased. Bioengineered 2021, 12, 7263–7275. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liu, Q.; Li, H.; Wen, C.; He, Z. Alterations of the gut microbiome associated with the treatment of hyperuricaemia in male rats. Front. Microbiol. 2018, 9, 2233. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ye, Z.; Feng, P.; Li, R.; Chen, X.; Tian, X.; Han, R.; Kakade, A.; Liu, P.; Li, X. Limosilactobacillus fermentum JL-3 isolated from “Jiangshui” ameliorates hyperuricemia by degrading uric acid. Gut Microbes 2021, 13, 1897211. [Google Scholar] [CrossRef] [PubMed]
- Yamada, N.; Saito, C.; Kano, H.; Fukuuchi, T.; Yamaoka, N.; Kaneko, K.; Asami, Y. Lactobacillus gasseri PA-3 directly incorporates purine mononucleotides and utilizes them for growth. Nucleosides Nucleotides Nucleic Acids 2022, 41, 221–230. [Google Scholar] [CrossRef]
- Yamada, N.; Saito-Iwamoto, C.; Nakamura, M.; Soeda, M.; Chiba, Y.; Kano, H.; Asami, Y. Lactobacillus gasseri PA-3 uses the purines IMP, inosine and hypoxanthine and reduces their absorption in rats. Microorganisms 2017, 5, 10. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, J.; Lin, G.; Gao, K.; Yu, Y.; Chen, S.; Chen, L.; Chen, Z.; Li, L. Probiotic effects of Lacticaseibacillus rhamnosus 1155 and Limosilactobacillus fermentum 2644 on hyperuricemic rats. Front. Nutr. 2022, 9, 993951. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhang, C.; Zeng, X.; Yuan, Z. Microecological treatment of hyperuricemia using Lactobacillus from pickles. BMC Microbiol. 2020, 20, 195. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Liu, Q.; Hao, H.; Bu, Y.; Tian, X.; Wang, T.; Yi, H. Lactobacillus paracasei X11 ameliorates hyperuricemia and modulates gut microbiota in mice. Front. Immunol. 2022, 13, 940228. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Werlinger, P.; Suh, J.W.; Cheng, J. Potential probiotic Lacticaseibacillus paracasei MJM60396 prevents hyperuricemia in a multiple way by absorbing purine, suppressing xanthine oxidase and regulating urate excretion in mice. Microorganisms 2022, 10, 851. [Google Scholar] [CrossRef]
- Kuo, Y.W.; Hsieh, S.H.; Chen, J.F.; Liu, C.R.; Chen, C.W.; Huang, Y.F.; Ho, H.H. Lactobacillus reuteri TSR332 and Lactobacillus fermentum TSF331 stabilize serum uric acid levels and prevent hyperuricemia in rats. PeerJ 2021, 9, e11209. [Google Scholar] [CrossRef]
- Dong, J.; Pang, H.; Zhao, L. Hyperuricemia among mongolian adults and the related factors in inner mongolia autonomous region from 2018 to 2020. Wei Sheng Yan Jiu 2022, 51, 940–946. [Google Scholar] [CrossRef]
- Zhao, S.; Feng, P.; Hu, X.; Cao, W.; Liu, P.; Han, H.; Jin, W.; Li, X. Probiotic Limosilactobacillus fermentum GR-3 ameliorates human hyperuricemia via degrading and promoting excretion of uric acid. iScience 2022, 25, 105198. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.E.; Aldis, H.W. Lymphangitic ureteral metastases from prostatic carcinoma. J. Can. Assoc. Radiol. 1980, 31, 158–162. [Google Scholar]
- Berg, D.; Clemente, J.C.; Colombel, J.F. Can inflammatory bowel disease be permanently treated with short-term interventions on the microbiome? Expert. Rev. Gastroenterol. Hepatol. 2015, 9, 781–795. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, Y.; Dalbeth, N.; Terkeltaub, R.; Yang, T.; Wang, Y.; Yang, Z.; Li, J.; Wu, Z.; Zeng, C.; et al. Association between gut microbiota and elevated serum urate in two independent cohorts. Arthritis Rheumatol. 2022, 74, 682–691. [Google Scholar] [CrossRef]
- Liu, S.; Perez-Ruiz, F.; Miner, J.N. Patients with gout differ from healthy subjects in renal response to changes in serum uric acid. Jt. Bone Spine 2017, 84, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Nieuwdorp, M.; Gilijamse, P.W.; Pai, N.; Kaplan, L.M. Role of the microbiome in energy regulation and metabolism. Gastroenterology 2014, 146, 1525–1533. [Google Scholar] [CrossRef] [PubMed]
- Oshima, S.; Shiiya, S.; Nakamura, Y. Serum uric acid-lowering effects of combined glycine and tryptophan treatments in subjects with mild hyperuricemia: A randomized, double-blind, placebo-controlled, crossover study. Nutrients 2019, 11, 564. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, Y.; Zhong, H.; Chen, F.; Regenstein, J.; Hu, X.; Cai, L.; Feng, F. The gut microbiota as a target to control hyperuricemia pathogenesis: Potential mechanisms and therapeutic strategies. Crit. Rev. Food Sci. Nutr. 2022, 62, 3979–3989. [Google Scholar] [CrossRef] [PubMed]
- Akintunde, J.K.; Falomo, I.M.; Akinbohun, O.M.; Erinoso, S.O.; Ugwor, E.; Folayan, A.D.; Ateate, A.D. Naringin corrects renal failure related to Lesch-Nyhan disease in a rat model via NOS-cAMP-PKA and BDNF/TrkB pathways. J. Biochem. Mol. Toxicol. 2023, 38, e23558. [Google Scholar] [CrossRef]
- Xu, L.; Cheng, J.; Lu, J.; Lin, G.; Yu, Q.; Li, Y.; Chen, J.; Xie, J.; Su, Z.; Zhou, Q. Integrating network pharmacology and experimental validation to clarify the anti-hyperuricemia mechanism of cortex phellodendri in mice. Front. Pharmacol. 2022, 13, 964593. [Google Scholar] [CrossRef]
- Lu, J.; Dalbeth, N.; Yin, H.; Li, C.; Merriman, T.R.; Wei, W.H. Mouse models for human hyperuricaemia: A critical review. Nat. Rev. Rheumatol. 2019, 15, 413–426. [Google Scholar] [CrossRef]
- Zhu, J.; Li, Y.; Chen, Z.; Gao, K.; Lin, G.; Chen, S.; Li, L.; Ge, H. Screening of lactic acid bacteria strains with urate-lowering effect from fermented dairy products. J. Food Sci. 2022, 87, 5118–5127. [Google Scholar] [CrossRef]
- Chelakkot, C.; Ghim, J.; Ryu, S.H. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 2018, 50, 1–9. [Google Scholar] [CrossRef]
- Kuo, W.T.; Odenwald, M.A.; Turner, J.R.; Zuo, L. Tight junction proteins occludin and ZO-1 as regulators of epithelial proliferation and survival. Ann. N. Y. Acad. Sci. 2022, 1514, 21–33. [Google Scholar] [CrossRef]
- Liang, L.; Meng, Z.; Zhang, F.; Jianguo, Z.; Fang, S.; Hu, Q.; Tang, X.; Li, Y. Lactobacillus gasseri LG08 and Leuconostoc mesenteroides LM58 exert preventive effect on the development of hyperuricemia by repairing antioxidant system and intestinal flora balance. Front. Microbiol. 2023, 14, 1211831. [Google Scholar] [CrossRef]
- Li, Y.; Li, H.; Wang, R.; Yu, Y.; Liu, X.; Tian, Z. Protective effect of sodium butyrate on intestinal barrier damage and uric acid reduction in hyperuricemia mice. Biomed. Pharmacother. 2023, 161, 114568. [Google Scholar] [CrossRef]
- Xi, Y.; Yan, J.; Li, M.; Ying, S.; Shi, Z. Gut microbiota dysbiosis increases the risk of visceral gout in goslings through translocation of gut-derived lipopolysaccharide. Poult. Sci. 2019, 98, 5361–5373. [Google Scholar] [CrossRef]
- Malesza, I.J.; Malesza, M.; Walkowiak, J.; Mussin, N.; Walkowiak, D.; Aringazina, R.; Bartkowiak-Wieczorek, J.; Madry, E. High-fat, western-style diet, systemic inflammation, and gut microbiota: A narrative review. Cells 2021, 10, 3164. [Google Scholar] [CrossRef]
- Han, J.; Wang, X.; Tang, S.; Lu, C.; Wan, H.; Zhou, J.; Li, Y.; Ming, T.; Wang, Z.J.; Su, X. Protective effects of tuna meat oligopeptides (TMOP) supplementation on hyperuricemia and associated renal inflammation mediated by gut microbiota. FASEB J. 2020, 34, 5061–5076. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, B.; Zhao, X.; Lin, Y.; Wang, J.; Wang, X.; Hu, N.; Wang, S. Chlorogenic acid supplementation ameliorates hyperuricemia, relieves renal inflammation, and modulates intestinal homeostasis. Food Funct. 2021, 12, 5637–5649. [Google Scholar] [CrossRef]
- Shi, R.; Ye, J.; Fan, H.; Xiao, C.; Wang, D.; Xia, B.; Zhao, Z.; Zhao, B.; Dai, X.; Liu, X. Lactobacillus plantarum LLY-606 supplementation ameliorates hyperuricemia via modulating intestinal homeostasis and relieving inflammation. Food Funct. 2023, 14, 5663–5677. [Google Scholar] [CrossRef]
- Wan, H.; Han, J.; Tang, S.; Bao, W.; Lu, C.; Zhou, J.; Ming, T.; Li, Y.; Su, X. Comparisons of protective effects between two sea cucumber hydrolysates against diet induced hyperuricemia and renal inflammation in mice. Food Funct. 2020, 11, 1074–1086. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Kan, J.; Cai, H.; Hong, J.; Jin, C.; Zhang, M. In vitro and in vivo ameliorative effects of polyphenols from purple potato leaves on renal injury and associated inflammation induced by hyperuricemia. J. Food Biochem. 2022, 46, e14049. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Li, H.; Liu, Z.; Li, C.; Chen, Y.; Jiang, C.; Yu, Y.; Tian, Z. Impaired intestinal barrier function in a mouse model of hyperuricemia. Mol. Med. Rep. 2019, 20, 3292–3300. [Google Scholar] [CrossRef] [PubMed]
- Ni, C.; Li, X.; Wang, L.; Li, X.; Zhao, J.; Zhang, H.; Wang, G.; Chen, W. Lactic acid bacteria strains relieve hyperuricaemia by suppressing xanthine oxidase activity via a short-chain fatty acid-dependent mechanism. Food Funct. 2021, 12, 7054–7067. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.; Xu, D.; Zhang, X.; Yang, X.; Zhao, P.; Cui, X.; Liu, X.; Yang, W.; Yang, G.; Xing, S. Association of hyperuricemia with immune disorders and intestinal barrier dysfunction. Front. Physiol. 2020, 11, 524236. [Google Scholar] [CrossRef] [PubMed]
- Stojanov, S.; Berlec, A.; Strukelj, B. The Influence of probiotics on the Firmicutes/Bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Ren, P.; Yang, R.; Gao, Y.; Tang, Q.; Xue, C.; Wang, Y. Ameliorative effect of mannuronate oligosaccharides on hyperuricemic mice via promoting uric acid excretion and modulating gut microbiota. Nutrients 2023, 15, 417. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wu, X.; Guo, Z.; Gao, R.; Ni, Z.; Cui, H.; Zong, M.; Van Bockstaele, F.; Lou, W. Lactiplantibacillus plantarum enables blood urate control in mice through degradation of nucleosides in gastrointestinal tract. Microbiome 2023, 11, 153. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, X.; Zhang, L.; Meng, F.; Zhou, L.; Pang, X.; Lu, Z.; Lu, Y. Lacticaseibacillus rhamnosus Fmb14 prevents purine induced hyperuricemia and alleviate renal fibrosis through gut-kidney axis. Pharmacol. Res. 2022, 182, 106350. [Google Scholar] [CrossRef]
- Eckenstaler, R.; Benndorf, R.A. The role of ABCG2 in the pathogenesis of primary hyperuricemia and gout-an update. Int. J. Mol. Sci. 2021, 22, 6678. [Google Scholar] [CrossRef]
- Xu, Y.; Cao, X.; Zhao, H.; Yang, E.; Wang, Y.; Cheng, N.; Cao, W. Impact of Camellia japonica bee pollen polyphenols on hyperuricemia and gut microbiota in potassium oxonate-induced mice. Nutrients 2021, 13, 2665. [Google Scholar] [CrossRef]
- Li, X.; Gao, X.; Zhang, H.; Liu, Y.; Sarker, M.M.R.; Wu, Y.; Chen, X.; Zhao, C. The anti-hyperuricemic effects of green alga Enteromorpha prolifera polysaccharide via regulation of the uric acid transporters in vivo. Food Chem. Toxicol. 2021, 158, 112630. [Google Scholar] [CrossRef]
- Shan, B.; Wu, M.; Chen, T.; Tang, W.; Li, P.; Chen, J. Berberine attenuates hyperuricemia by regulating urate transporters and gut microbiota. Am. J. Chin. Med. 2022, 50, 2199–2221. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Yu, Y.; Chen, Z.; Wang, L.; Ma, J.; Bai, X.; Sun, Y.; Wang, D. Effects of beta-carotin and green tea powder diets on alleviating the symptoms of gouty arthritis and improving gut microbiota in C57BL/6 mice. Front. Microbiol. 2022, 13, 837182. [Google Scholar] [CrossRef]
- McFarlane, C.; Krishnasamy, R.; Stanton, T.; Savill, E.; Snelson, M.; Mihala, G.; Morrison, M.; Johnson, D.W.; Campbell, K.L. Diet quality and protein-bound uraemic toxins: Investigation of novel risk factors and the role of microbiome in chronic kidney disease. J. Ren. Nutr. 2022, 32, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhong, J.; Yang, H.; Wang, D.; Zhang, Y.; Yang, Y.; Xing, G.; Kon, V. Biotic supplements in patients with chronic kidney disease: Meta-analysis of randomized controlled trials. J. Ren. Nutr. 2022, 32, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Maske, B.L.; de Melo Pereira, G.V.; Vale, A.D.S.; de Carvalho Neto, D.P.; Karp, S.G.; Viesser, J.A.; De Dea Lindner, J.; Pagnoncelli, M.G.; Soccol, V.T.; Soccol, C.R. A review on enzyme-producing lactobacilli associated with the human digestive process: From metabolism to application. Enzyme Microb. Technol. 2021, 149, 109836. [Google Scholar] [CrossRef] [PubMed]
- Fushinobu, S.; Abou Hachem, M. Structure and evolution of the bifidobacterial carbohydrate metabolism proteins and enzymes. Biochem. Soc. Trans. 2021, 49, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, K.; Wagener, J.; Ames, R.M.; Christou, S.; MacCallum, D.M.; Bates, S.; Gow, N.A.R. Three related enzymes in candida albicans achieve arginine- and agmatine-dependent metabolism that is essential for growth and fungal virulence. mBio 2020, 11, e01845-20. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.H.; Chang, Y.P.; Li, T.; Han, F.; Li, C.J.; Li, X.Y.; Xue, M.; Cheng, Y.; Meng, Z.Y.; Han, Z.; et al. Empagliflozin attenuates hyperuricemia by upregulation of ABCG2 via ampk/akt/creb signaling pathway in type 2 diabetic mice. Int. J. Biol. Sci. 2020, 16, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhong, Y.; Ouyang, X.; Wang, C.; Yin, L.; Huang, J.; Li, Y.; Wang, Q.; Xie, J.; Huang, P.; et al. Effects of beta-alanine on intestinal development and immune performance of weaned piglets. Anim. Nutr. 2023, 12, 398–408. [Google Scholar] [CrossRef] [PubMed]
- Qin, N.; Qin, M.; Shi, W.; Kong, L.; Wang, L.; Xu, G.; Guo, Y.; Zhang, J.; Ma, Q. Investigation of pathogenesis of hyperuricemia based on untargeted and targeted metabolomics. Sci. Rep. 2022, 12, 13980. [Google Scholar] [CrossRef]
- Mehmood, A.; Zhao, L.; Ishaq, M.; Usman, M.; Zad, O.D.; Hossain, I.; Raka, R.N.; Naveed, M.; Zhao, L.; Wang, C.; et al. Uricostatic and uricosuric effect of grapefruit juice in potassium oxonate-induced hyperuricemic mice. J. Food Biochem. 2020, 44, e13213. [Google Scholar] [CrossRef]
- Guo, X.L.; Gao, Y.Y.; Yang, Y.X.; Zhu, Q.F.; Guan, H.Y.; He, X.; Zhang, C.L.; Wang, Y.; Xu, G.B.; Zou, S.H.; et al. Amelioration effects of alpha-viniferin on hyperuricemia and hyperuricemia-induced kidney injury in mice. Phytomedicine 2023, 116, 154868. [Google Scholar] [CrossRef]
- Li, Z.; Guo, X.; Liu, Y.; Chang, Y.; Sun, Y.; Zhu, G.; Abraham, M.R. The relation of moderate alcohol consumption to hyperuricemia in a rural general population. Int. J. Environ. Res. Public. Health 2016, 13, 732. [Google Scholar] [CrossRef]
- Ponticelli, C.; Podesta, M.A.; Moroni, G. Hyperuricemia as a trigger of immune response in hypertension and chronic kidney disease. Kidney Int. 2020, 98, 1149–1159. [Google Scholar] [CrossRef]
- Li, H.; Zhang, H.; Yan, F.; He, Y.; Ji, A.; Liu, Z.; Li, M.; Ji, X.; Li, C. Kidney and plasma metabolomics provide insights into the molecular mechanisms of urate nephropathy in a mouse model of hyperuricemia. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166374. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, M.; Veronese, F.V.; Moresco, R.N. Uric acid and kidney damage in systemic lupus erythematosus. Clin. Chim. Acta 2020, 508, 197–205. [Google Scholar] [CrossRef]
- Wang, X.; Liu, X.; Gao, Q.; Gu, X.; Zhang, G.; Sheng, Z.; Wu, T.; Su, Z.; Wang, W.; Ye, M. Gegen Qinlian decoction treatment of asymptomatic hyperuricemia by targeting circadian immune function. Chin. Med. 2023, 18, 77. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Wan, L.; Chen, X.; Li, X.; Hao, X.; Li, X.; Li, J.; Ding, H. Conjugated linoleic acid ameliorates high fructose-induced hyperuricemia and renal inflammation in rats via NLRP3 inflammasome and tlr4 signaling pathway. Mol. Nutr. Food Res. 2019, 63, e1801402. [Google Scholar] [CrossRef]
- Qi, X.; Guan, K.; Liu, C.; Chen, H.; Ma, Y.; Wang, R. Whey protein peptides PEW and LLW synergistically ameliorate hyperuricemia and modulate gut microbiota in potassium oxonate and hypoxanthine-induced hyperuricemic rats. J. Dairy Sci. 2023, 106, 7367–7381. [Google Scholar] [CrossRef]
- Wang, Z.; Song, L.; Li, X.; Xiao, Y.; Huang, Y.; Zhang, Y.; Li, J.; Li, M.; Ren, Z. Lactiplantibacillus pentosus P2020 protects the hyperuricemia and renal inflammation in mice. Front. Nutr. 2023, 10, 1094483. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.S.; Lu, T.M.; Chen, C.H.; Guo, B.C.; Hsu, C.P. Hyperuricemia induces endothelial dysfunction and accelerates atherosclerosis by disturbing the asymmetric dimethylarginine/dimethylarginine dimethylaminotransferase 2 pathway. Redox Biol. 2021, 46, 102108. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, Z.; Luo, J.; Jiang, Y.; Li, L.; Chen, Y.; Zhang, L.; Huang, Q.; Cao, Y.; Zhou, P.; et al. Apigenin ameliorates hyperuricemic nephropathy by inhibiting URAT1 and GLUT9 and relieving renal fibrosis via the Wnt/beta-catenin pathway. Phytomedicine 2021, 87, 153585. [Google Scholar] [CrossRef] [PubMed]
| Sample Number | Sampling Spot | Strain Source |
|---|---|---|
| 1 | Bayin Irangacha, Ewenke Banner, Hulunbuir, Inner Mongolia | Feces of healthy infant |
| 2 | Tongda Ranch in Manzhouli, Hulunbuir, Inner Mongolia | Fermented milk curd |
| 3 | West Ujur Sumu, Chenbarhu Banner, Hulunbuir, Inner Mongolia | Feces of healthy infant |
| 4 | Xilinhot, Xilingol League, Inner Mongolia | Feces of healthy infant |
| 5 | Bama Longevity Village, Guangxi | Feces of healthy infant |
| 6 | Dashimo, New Baerhuyou Banner, Hulunbuir, Inner Mongolia | Feces of healthy infant |
| 7 | Sanhe Hui Township, Erguna city, Hulunbuir, Inner Mongolia | Fermented milk lump |
| 8 | Chenbarhu Banner, Hulunbuir, Inner Mongolia | Fermented milk lump |
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| XOD | ATGACGAGGACAACGGTAGAT | TCATACTTGGAGATCATCACGGT |
| TLR4 | CCTTTTCATCTCTGCCTTCACTAC | GGGACACCACGACAATAACCT |
| MyD88 | GATGGTAGCGTTGTCTCTGAT | GATGCTGGGGAACTCTTTCTTC |
| NF-κB | CCCATGTAGACAGCACCACCTATGAT | ACAGAGGCTCAAAGTTCTCCACCA |
| Claudin-1 | CCAGGTACGAATTTGGTCAGG | TGGTGTTGGGTAAGAGGTTGT |
| Claudin-4 | TGGGGCTACAGGTAATGGG | GGTCTGCGAGGTGACAATGTT |
| Occludin | CTTCCAATGGCAAAGTGAATG | TACCACCGCTGCTGTAACGAG |
| ZO-1 | GAGCCTAATCTGACCTATGAACC | TGAGGACTCGTATCTGTATGTGG |
| ABCG2 | AAATGCTGTTCAGGTTATGTGGT | TCCGACCTTAGAATCTGCTACTT |
| SLC2A9 | TTGCTTTAGCTTCCCTGATGTG | GAGAGGTTGTACCCGTAGAGG |
| XHD | CATGGGGAAGACAACCACAGG | ATGGTCCTGATCCTGGCATCC |
| Allantoinase | GCTTACGATATGGTGATA | TCAATGTCAGAACCTATC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, A.; Rui, B.; Ullah, H.; Dai, P.; Ahmad, K.; Yuan, J.; Liu, Y.; Li, M. Limosilactobacillus reuteri HCS02-001 Attenuates Hyperuricemia through Gut Microbiota-Dependent Regulation of Uric Acid Biosynthesis and Excretion. Microorganisms 2024, 12, 637. https://doi.org/10.3390/microorganisms12040637
Hussain A, Rui B, Ullah H, Dai P, Ahmad K, Yuan J, Liu Y, Li M. Limosilactobacillus reuteri HCS02-001 Attenuates Hyperuricemia through Gut Microbiota-Dependent Regulation of Uric Acid Biosynthesis and Excretion. Microorganisms. 2024; 12(4):637. https://doi.org/10.3390/microorganisms12040637
Chicago/Turabian StyleHussain, Akbar, Binqi Rui, Hayan Ullah, Panpan Dai, Kabir Ahmad, Jieli Yuan, Yinhui Liu, and Ming Li. 2024. "Limosilactobacillus reuteri HCS02-001 Attenuates Hyperuricemia through Gut Microbiota-Dependent Regulation of Uric Acid Biosynthesis and Excretion" Microorganisms 12, no. 4: 637. https://doi.org/10.3390/microorganisms12040637
APA StyleHussain, A., Rui, B., Ullah, H., Dai, P., Ahmad, K., Yuan, J., Liu, Y., & Li, M. (2024). Limosilactobacillus reuteri HCS02-001 Attenuates Hyperuricemia through Gut Microbiota-Dependent Regulation of Uric Acid Biosynthesis and Excretion. Microorganisms, 12(4), 637. https://doi.org/10.3390/microorganisms12040637






