Influence of Age of Infection on the Gut Microbiota in Worker Honey Bees (Apis mellifera iberiensis) Experimentally Infected with Nosema ceranae
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection
2.2. Molecular Analysis
2.3. Statistical Analysis
3. Results
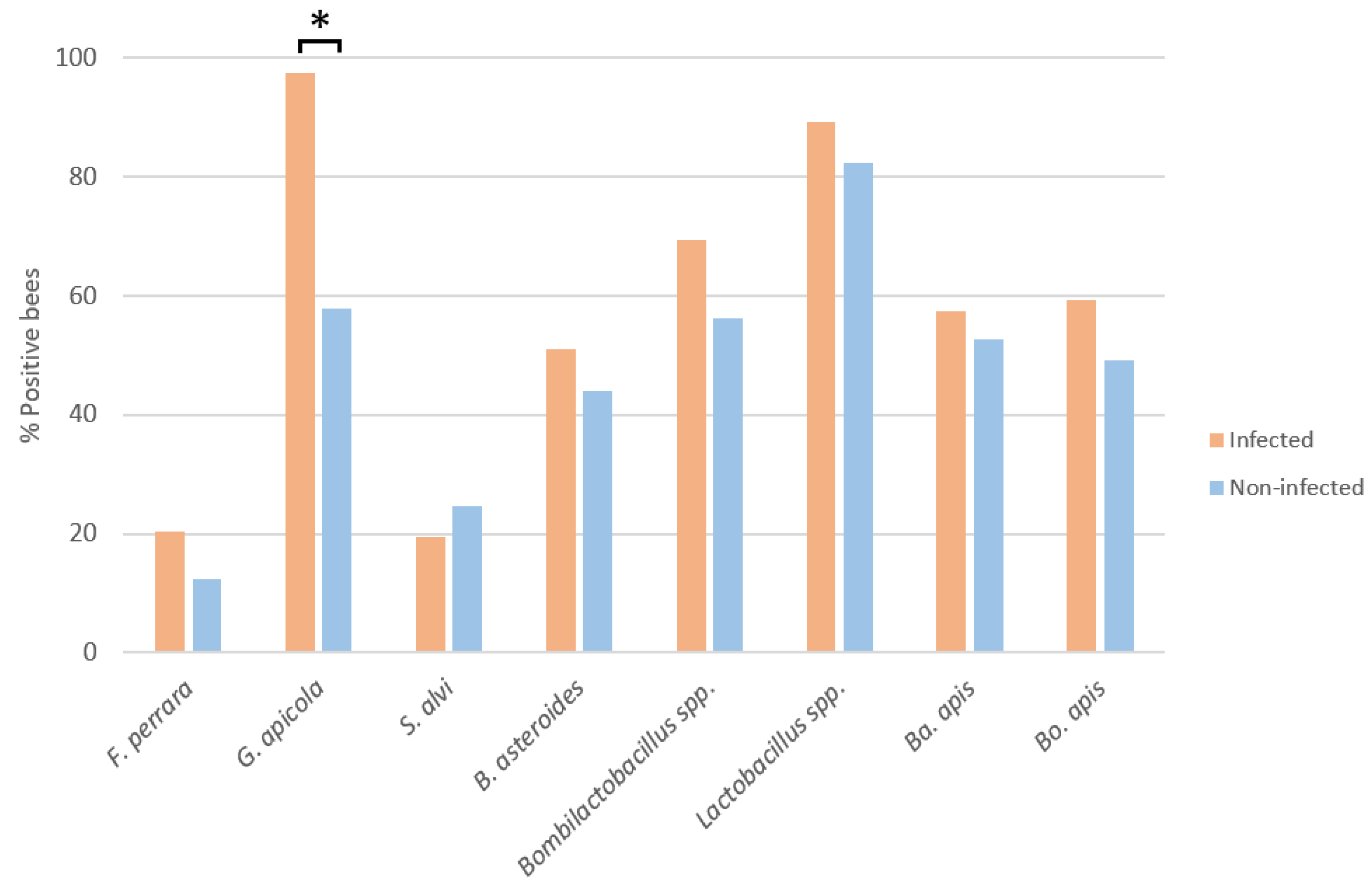
3.1. Influence of N. ceranae Infection on the Honey Bee Gut Bacteria
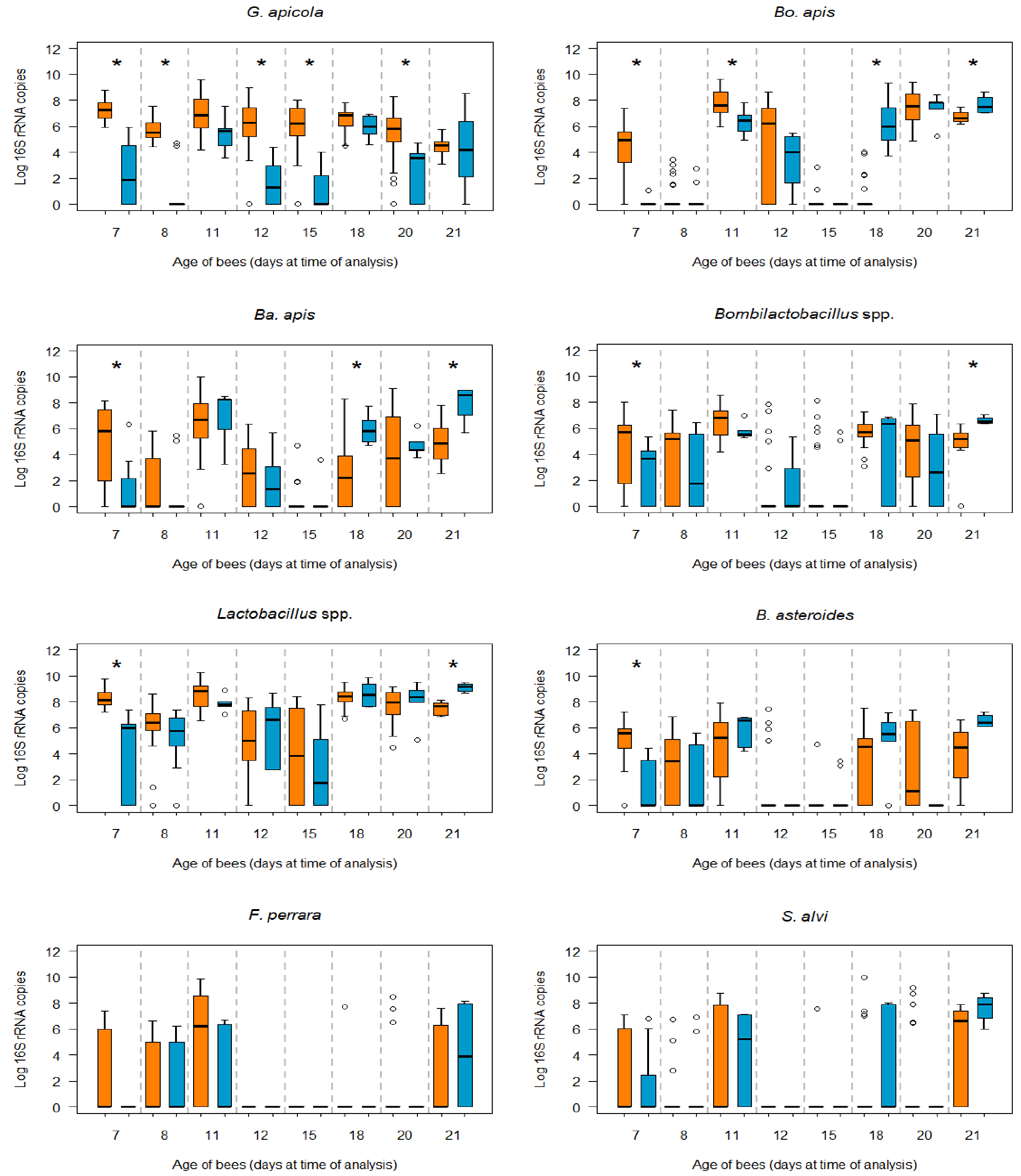
3.2. Bacterial Loads in Infected and Non-Infected Honey Bees of the Same Age
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kostic, A.D.; Howitt, M.R.; Garrett, W.S. Exploring Host-Microbiota Interactions in Animal Models and Humans. Genes Dev. 2013, 27, 701–718. [Google Scholar] [CrossRef] [PubMed]
- Cox-Foster, D.L.; Conlan, S.; Holmes, E.C.; Palacios, G.; Evans, J.D.; Moran, N.A.; Quan, P.L.; Briese, T.; Hornig, M.; Geiser, D.M.; et al. A Metagenomic Survey of Microbes in Honey Bee Colony Collapse Disorder. Science 2007, 318, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Spivak, M. Socialized Medicine: Individual and Communal Disease Barriers in Honey Bees. J. Invertebr. Pathol. 2010, 103, S62–S72. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Kwong, W.K.; McFrederick, Q.; Anderson, K.E.; Barribeau, S.M.; Chandler, J.A.; Cornman, R.S.; Dainat, J.; De Miranda, J.R.; Doublet, V.; et al. The Bee Microbiome: Impact on Bee Health and Model for Evolution and Ecology of Host-Microbe Interactions. mBio 2016, 7, e02164-15. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Steele, M.I.; Leonard, S.P.; Motta, E.V.S.; Moran, N.A. Honey Bees as Models for Gut Microbiota Research. Lab Anim. 2018, 47, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.-W.; Soh, E.J.Y.; Ascher, J.S.; Jaffé, R.; Moran, N.A. Dynamic Microbiome Evolution in Social Bees. Sci. Adv. 2017, 3, e1600513. [Google Scholar] [CrossRef] [PubMed]
- Martinson, V.G.; Danforth, B.N.; Minckley, R.L.; Rueppell, O.; Tingek, S.; Moran, N.A. A Simple and Distinctive Microbiota Associated with Honey Bees and Bumble Bees. Mol. Ecol. 2011, 20, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A. Genomics of the Honey Bee Microbiome. Curr. Opin. Insect Sci. 2015, 10, 22–28. [Google Scholar] [CrossRef]
- Su, Q.; Tang, M.; Hu, J.; Tang, J.; Zhang, X.; Li, X.; Niu, Q.; Zhou, X.; Luo, S.; Zhou, X. Significant Compositional and Functional Variation Reveals the Patterns of Gut Microbiota Evolution among the Widespread Asian Honeybee Populations. Front. Microbiol. 2022, 13, 934459. [Google Scholar] [CrossRef]
- Anderson, K.E.; Sheehan, T.H.; Mott, B.M.; Maes, P.; Snyder, L.; Schwan, M.R.; Walton, A.; Jones, B.M.; Corby-Harris, V. Microbial Ecology of the Hive and Pollination Landscape: Bacterial Associates from Floral Nectar, the Alimentary Tract and Stored Food of Honey Bees (Apis mellifera). PLoS ONE 2013, 8, e83125. [Google Scholar] [CrossRef]
- Corby-Harris, V.; Maes, P.; Anderson, K.E. The Bacterial Communities Associated with Honey Bee (Apis mellifera) Foragers. PLoS ONE 2014, 9, e95056. [Google Scholar] [CrossRef] [PubMed]
- Ludvigsen, J.; Rangberg, A.; Avershina, E.; Sekelja, M.; Kreibich, C.; Amdam, G.; Rudi, K. Shifts in the Midgut/Pyloric Microbiota Composition within a Honey Bee Apiary throughout a Season. Microbes Environ. 2015, 30, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Bartlett, K.D.; Moran, N.A. The Bacterium Frischella Perrara Causes Scab Formation in the Gut of Its Honeybee Host. mBio 2015, 6, e00193-15. [Google Scholar] [CrossRef] [PubMed]
- Martinson, V.G.; Moy, J.; Moran, N.A. Establishment of Characteristic Gut Bacteria during Development of the Honeybee Worker. Appl. Environ. Microbiol. 2012, 78, 2830–2840. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.E.; Leonard, S.P.; Kwong, W.K.; Engel, P.; Moran, N.A. Genome-Wide Screen Identifies Host Colonization Determinants in a Bacterial Gut Symbiont. Proc. Natl. Acad. Sci. USA 2016, 113, 13887–13892. [Google Scholar] [CrossRef] [PubMed]
- Maes, P.W.; Rodrigues, P.A.P.; Oliver, R.; Mott, B.M.; Anderson, K.E. Diet-Related Gut Bacterial Dysbiosis Correlates with Impaired Development, Increased Mortality and Nosema Disease in the Honeybee (Apis mellifera). Mol. Ecol. 2016, 25, 5439–5450. [Google Scholar] [CrossRef]
- Anderson, K.E.; Ricigliano, V.A. Honey Bee Gut Dysbiosis: A Novel Context of Disease Ecology. Curr. Opin. Insect Sci. 2017, 22, 125–132. [Google Scholar] [CrossRef]
- Vojvodic, S.; Rehan, S.M.; Anderson, K.E. Microbial Gut Diversity of Africanized and European Honey Bee Larval Instars. PLoS ONE 2013, 8, e72106. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. Functional and Evolutionary Insights into the Simple yet Specific Gut Microbiota of the Honey Bee from Metagenomic Analysis. Gut Microbes 2013, 4, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.; Abrol, D.P.; Li, J.; Schmid-Hempel, P. Diversity and Evolutionary Patterns of Bacterial Gut Associates of Corbiculate Bees. Mol. Ecol. 2013, 22, 2028–2044. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.E.; Martinson, V.G.; Urban-Mead, K.; Moran, N.A. Routes of Acquisition of the Gut Microbiota of the Honey Bee Apis mellifera. Appl. Environ. Microbiol. 2014, 80, 7378–7387. [Google Scholar] [CrossRef] [PubMed]
- Kapheim, K.M.; Rao, V.D.; Yeoman, C.J.; Wilson, B.A.; White, B.A.; Goldenfeld, N.; Robinson, G.E. Caste-Specific Differences in Hindgut Microbial Communities of Honey Bees (Apis mellifera). PLoS ONE 2015, 10, e0123911. [Google Scholar] [CrossRef] [PubMed]
- Kešnerová, L.; Mars, R.A.T.; Ellegaard, K.M.; Troilo, M.; Sauer, U.; Engel, P. Disentangling Metabolic Functions of Bacteria in the Honey Bee Gut. PLoS Biol. 2017, 15, e2003467. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Perreau, J.; Powell, J.E.; Han, B.; Zhang, Z.; Kwong, W.K.; Tringe, S.G.; Moran, N.A. Division of Labor in Honey Bee Gut Microbiota for Plant Polysaccharide Digestion. Proc. Natl. Acad. Sci. USA 2019, 116, 25909–25916. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Tang, M.; Li, X.; Zhou, X. Community Dynamics in Structure and Function of Honey Bee Gut Bacteria in Response to Winter Dietary Shift. mBio 2022, 13, e01131-22. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Powell, J.E.; Steele, M.I.; Dietrich, C.; Moran, N.A. Honeybee Gut Microbiota Promotes Host Weight Gain via Bacterial Metabolism and Hormonal Signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 4775–4780. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.C.; Fruciano, C.; Marchant, J.; Hildebrand, F.; Forslund, S.; Bork, P.; Engel, P.; Hughes, W.O.H. The Gut Microbiome Is Associated with Behavioural Task in Honey Bees. Insectes Soc. 2018, 65, 419–429. [Google Scholar] [CrossRef]
- Wu, Y.; Zheng, Y.; Chen, Y.; Wang, S.; Chen, Y.; Hu, F.; Zheng, H. Honey Bee (Apis mellifera) Gut Microbiota Promotes Host Endogenous Detoxification Capability via Regulation of P450 Gene Expression in the Digestive Tract. Microb. Biotechnol. 2020, 13, 1201–1212. [Google Scholar] [CrossRef]
- Wang, K.; Li, J.; Zhao, L.; Mu, X.; Wang, C.; Wang, M.; Xue, X.; Qi, S.; Wu, L. Gut Microbiota Protects Honey Bees (Apis mellifera L.) against Polystyrene Microplastics Exposure Risks. J. Hazard. Mater. 2021, 402, 123828. [Google Scholar] [CrossRef]
- Engel, P.; Martinson, V.G.; Moran, N.A. Functional Diversity within the Simple Gut Microbiota of the Honey Bee. Proc. Natl. Acad. Sci. USA 2012, 109, 11002–11007. [Google Scholar] [CrossRef]
- Schwarz, R.S.; Huang, Q.; Evans, J.D. Hologenome Theory and the Honey Bee Pathosphere. Curr. Opin. Insect Sci. 2015, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Steele, M.I.; Motta, E.V.S.; Gattu, T.; Martinez, D.; Moran, N.A. The Gut Microbiota Protects Bees from Invasion by a Bacterial Pathogen. Microbiol. Spectr. 2021, 9, e00394-21. [Google Scholar] [CrossRef] [PubMed]
- Higes, M.; Martín-Hernández, R.; Garrido-Bailón, E.; Botías, C.; García-Palencia, P.; Meana, A. Regurgitated Pellets of Merops apiaster as Fomites of Infective Nosema ceranae (Microsporidia) Spores. Environ. Microbiol. 2008, 10, 1374–1379. [Google Scholar] [CrossRef] [PubMed]
- Stevanovic, J.; Stanimirovic, Z.; Genersch, E.; Kovacevic, S.R.; Ljubenkovic, J.; Radakovic, M.; Aleksic, N. Dominance of Nosema ceranae in Honey Bees in the Balkan Countries in the Absence of Symptoms of Colony Collapse Disorder. Apidologie 2011, 42, 49–58. [Google Scholar] [CrossRef]
- Yoshiyama, M.; Kimura, K. Distribution of Nosema ceranae in the European Honeybee, Apis mellifera in Japan. J. Invertebr. Pathol. 2011, 106, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.; Leal, G.; Conget, P. Nosema ceranae an Emergent Pathogen of Apis mellifera in Chile. Parasitol. Res. 2012, 111, 601–607. [Google Scholar] [CrossRef]
- Guerrero-Molina, C.; Correa-Benítez, A.; Hamiduzzaman, M.M.; Guzman-Novoa, E. Nosema ceranae Is an Old Resident of Honey Bee (Apis mellifera) Colonies in Mexico, Causing Infection Levels of One Million Spores per Bee or Higher during Summer and Fall. J. Invertebr. Pathol. 2016, 141, 38–40. [Google Scholar] [CrossRef]
- Wang, Q.; Dai, P.; Guzman-Novoa, E.; Wu, Y.; Hou, C.; Diao, Q. Nosema ceranae, the Most Common Microsporidium Infecting Apis mellifera in the Main Beekeeping Regions of China since at Least 2005. J. Apic. Res. 2019, 58, 562–566. [Google Scholar] [CrossRef]
- Dussaubat, C.; Brunet, J.L.; Higes, M.; Colbourne, J.K.; Lopez, J.; Choi, J.H.; Martín-Hernández, R.; Botías, C.; Cousin, M.; McDonnell, C.; et al. Gut Pathology and Responses to the Microsporidium Nosema ceranae in the Honey Bee Apis mellifera. PLoS ONE 2012, 7, e37017. [Google Scholar] [CrossRef]
- Martín-Hernández, R.; Higes, M.; Sagastume, S.; Juarranz, Á.; Dias-Almeida, J.; Budge, G.E.; Meana, A.; Boonham, N. Microsporidia Infection Impacts the Host Cell’s Cycle and Reduces Host Cell Apoptosis. PLoS ONE 2017, 12, e0170183. [Google Scholar] [CrossRef]
- Mayack, C.; Naug, D. Energetic Stress in the Honeybee Apis mellifera from Nosema ceranae Infection. J. Invertebr. Pathol. 2009, 100, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Antúnez, K.; Martín-Hernández, R.; Prieto, L.; Meana, A.; Zunino, P.; Higes, M. Immune Suppression in the Honey Bee (Apis mellifera) Following Infection by Nosema ceranae (Microsporidia). Environ. Microbiol. 2009, 11, 2284–2290. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, R.S.; Evans, J.D. Single and Mixed-Species Trypanosome and Microsporidia Infections Elicit Distinct, Ephemeral Cellular and Humoral Immune Responses in Honey Bees. Dev. Comp. Immunol. 2013, 40, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Gage, S.L.; Kramer, C.; Calle, S.; Carroll, M.; Heien, M.; DeGrandi-Hoffman, G. Nosema ceranae Parasitism Impacts Olfactory Learning and Memory and Neurochemistry in Honey Bees (Apis mellifera). J. Exp. Biol. 2018, 221, jeb161489. [Google Scholar] [CrossRef] [PubMed]
- Ares, A.M.; Nozal, M.J.; Bernal, J.L.; Martín-Hernández, R.; Higes, M.; Bernal, J. Liquid Chromatography Coupled to Ion Trap-Tandem Mass Spectrometry to Evaluate Juvenile Hormone III Levels in Bee Hemolymph from Nosema spp. Infected Colonies. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2012, 899, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; McMahon, D.P.; Lim, K.S.; Pull, C.D.; Clark, S.J.; Paxton, R.J.; Osborne, J.L. So near and yet so Far: Harmonic Radar Reveals Reduced Homing Ability of Nosema Infected Honeybees. PLoS ONE 2014, 9, e103989. [Google Scholar] [CrossRef] [PubMed]
- Botías, C.; Martín-Hernández, R.; Barrios, L.; Meana, A.; Higes, M. Nosema spp. Infection and Its Negative Effects on Honey Bees (Apis mellifera iberiensis) at the Colony Level. Vet. Res. 2013, 44, 25. [Google Scholar] [CrossRef] [PubMed]
- Benvau, L.R.; Nieh, J.C. Larval Honey Bees Infected with Nosema ceranae Have Increased Vitellogenin Titers as Young Adults. Sci. Rep. 2017, 7, 14144. [Google Scholar] [CrossRef]
- Urbieta-Magro, A.; Higes, M.; Meana, A.; Gómez-Moracho, T.; Rodríguez-García, C.; Barrios, L.; Martín-Hernández, R. The Levels of Natural Nosema Spp. Infection in Apis mellifera iberiensis Brood Stages. Int. J. Parasitol. 2019, 49, 657–667. [Google Scholar] [CrossRef]
- Jabal-Uriel, C.; Albarracín, V.N.; Calatayud, J.; Higes, M.; Martín-Hernández, R. Age and Season Effect the Timing of Adult Worker Honeybee Infection by Nosema ceranae. Front. Cell. Infect. Microbiol. 2022, 11, 1–12. [Google Scholar] [CrossRef]
- Smart, M.D.; Sheppard, W.S. Nosema ceranae in Age Cohorts of the Western Honey Bee (Apis mellifera). J. Invertebr. Pathol. 2012, 109, 148–151. [Google Scholar] [CrossRef]
- Jack, C.J.; Lucas, H.M.; Webster, T.C.; Sagili, R.R. Colony Level Prevalence and Intensity of Nosema ceranae in Honey Bees (Apis mellifera L.). PLoS ONE 2016, 11, e0163522. [Google Scholar] [CrossRef] [PubMed]
- Chaimanee, V.; Chantawannakul, P.; Chen, Y.; Evans, J.D.; Pettis, J.S. Effects of Host Age on Susceptibility to Infection and Immune Gene Expression in Honey Bee Queens (Apis mellifera) Inoculated with Nosema ceranae. Apidologie 2014, 45, 451–463. [Google Scholar] [CrossRef]
- Urbieta-Magro, A.; Higes, M.; Meana, A.; Barrios, L.; Martín-Hernández, R. Age and Method of Inoculation Influence the Infection of Worker Honey Bees (Apis mellifera) by Nosema ceranae. Insects 2019, 10, 417. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.K.; Ye, K.T.; Huang, W.F.; Ying, B.H.; Su, X.; Lin, L.H.; Li, J.H.; Chen, Y.P.; Li, J.L.; Bao, X.L.; et al. Influence of Feeding Type and Nosema ceranae Infection on the Gut Microbiota of Apis cerana Workers. mSystems 2018, 3, e00177-18. [Google Scholar] [CrossRef] [PubMed]
- Rubanov, A.; Russell, K.A.; Rothman, J.A.; Nieh, J.C.; McFrederick, Q.S. Intensity of Nosema ceranae Infection Is Associated with Specific Honey Bee Gut Bacteria and Weakly Associated with Gut Microbiome Structure. Sci. Rep. 2019, 9, 3820. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, X.; Huang, S.; Zhang, L.; Su, S.; Huang, W.F. Nosema ceranae Infection Enhances Bifidobacterium spp. Abundances in the Honey Bee Hindgut. Apidologie 2019, 50, 353–362. [Google Scholar] [CrossRef]
- Paris, L.; Peghaire, E.; Moné, A.; Diogon, M.; Debroas, D.; Delbac, F.; El Alaoui, H. Honeybee Gut Microbiota Dysbiosis in Pesticide/Parasite Co-Exposures Is Mainly Induced by Nosema ceranae. J. Invertebr. Pathol. 2020, 172, 107348. [Google Scholar] [CrossRef] [PubMed]
- Taupin, V.; Méténier, G.; Vivarès, C.P.; Prensier, G. An Improved Procedure for Percoll Gradient Separation of Sporogonial Stages in Encephalitozoon Cuniculi (Microsporidia). Parasitol. Res. 2006, 99, 708–714. [Google Scholar] [CrossRef]
- Martín-Hernández, R.; Botías, C.; Bailón, E.G.; Martínez-Salvador, A.; Prieto, L.; Meana, A.; Higes, M. Microsporidia Infecting Apis mellifera: Coexistence or Competition. Is Nosema ceranae Replacing Nosema apis? Environ. Microbiol. 2012, 14, 2127–2138. [Google Scholar] [CrossRef]
- Smith, E.A.; Newton, I.L.G. Genomic Signatures of Honey Bee Association in an Acetic Acid Symbiont. Genome Biol. Evol. 2020, 12, 1882–1894. [Google Scholar] [CrossRef]
- Castelli, L.; Branchiccela, B.; Garrido, M.; Invernizzi, C.; Porrini, M.; Romero, H.; Santos, E.; Zunino, P.; Antúnez, K. Impact of Nutritional Stress on Honeybee Gut Microbiota, Immunity, and Nosema ceranae Infection. Microb. Ecol. 2020, 80, 908–919. [Google Scholar] [CrossRef]
- Zhang, Y.; Su, M.; Wang, L.; Huang, S.; Su, S.; Huang, W.-F. Vairimorpha (Nosema) ceranae Infection Alters Honey Bee Microbiota Composition and Sustains the Survival of Adult Honey Bees. Biology 2021, 10, 905. [Google Scholar] [CrossRef]
- Rouzé, R.; Moné, A.; Delbac, F.; Belzunces, L.; Blot, N. The Honeybee Gut Microbiota Is Altered after Chronic Exposure to Different Families of Insecticides and Infection by Nosema ceranae. Microbes Environ. 2019, 34, 226–233. [Google Scholar] [CrossRef]
- Li, J.H.; Evans, J.D.; Li, W.F.; Zhao, Y.Z.; DeGrandi-Hoffman, G.; Huang, S.K.; Li, Z.G.; Hamilton, M.; Chen, Y.P. New Evidence Showing That the Destruction of Gut Bacteria by Antibiotic Treatment Could Increase the Honey Bee’s Vulnerability to Nosema Infection. PLoS ONE 2017, 12, e0187505. [Google Scholar] [CrossRef]
- Panjad, P.; Yongsawas, R.; Sinpoo, C.; Pakwan, C.; Subta, P.; Krongdang, S.; In-On, A.; Chomdej, S.; Chantawannakul, P.; Disayathanoowat, T. Impact of Nosema Disease and American Foulbrood on Gut Bacterial Communities of Honeybees Apis mellifera. Insects 2021, 12, 525. [Google Scholar] [CrossRef]
- Naree, S.; Ellis, J.D.; Benbow, M.E.; Suwannapong, G. Experimental Nosema ceranae Infection Is Associated with Microbiome Changes in the Midguts of Four Species of Apis (Honey Bees). J. Apic. Res. 2022, 61, 435–447. [Google Scholar] [CrossRef]
- de Oliveira, A.H.; Souza, A.M.D.C.; de Resende, M.T.C.S.; Carneiro, L.S.; de Oliveira, J.F.; Serra, R.S.; Serrão, J.E. The Peritrophic Matrix Delays Nosema ceranae Infection in the Honey Bee Apis mellifera Midgut. Physiol. Entomol. 2023, 48, 61–67. [Google Scholar] [CrossRef]
- Horton, M.A.; Oliver, R.; Newton, I.L. No Apparent Correlation between Honey Bee Forager Gut Microbiota and Honey Production. PeerJ 2015, 2015, e1329. [Google Scholar] [CrossRef] [PubMed]
- Raymann, K.; Moran, N.A. The Role of the Gut Microbiome in Health and Disease of Adult Honey Bee Workers. Curr. Opin. Insect Sci. 2018, 26, 97–104. [Google Scholar] [CrossRef]
- Kakumanu, M.L.; Reeves, A.M.; Anderson, T.D.; Rodrigues, R.R.; Williams, M.A. Honey Bee Gut Microbiome Is Altered by In-Hive Pesticide Exposures. Front. Microbiol. 2016, 7, 1255. [Google Scholar] [CrossRef]
- Higes, M.; García-Palencia, P.; Martín-Hernández, R.; Meana, A. Experimental Infection of Apis mellifera Honeybees with Nosema ceranae (Microsporidia). J. Invertebr. Pathol. 2007, 94, 211–217. [Google Scholar] [CrossRef]
- Crailsheim, K. Regulation of Food Passage in the Intestine of the Honeybee (Apis mellifera L.). J. Insect Physiol. 1988, 34, 85–90. [Google Scholar] [CrossRef]
- Crailsheim, K. The Protein Balance of the Honey Bee Worker. Apidologie 1990, 21, 417–429. [Google Scholar] [CrossRef]
- Gillott, C. Entomology; Springer: Dordrecht, The Netherlands, 1995; ISBN 978-0-306-44967-3. [Google Scholar]
- Terra, W.R.; Ferreira, C.; Baker, J.E. Compartmentalization of Digestion. In Biology of the Insect Midgut; Lehane, M.J., Billingsley, P.F., Eds.; Springer: Dordrecht, The Netherlands, 1996; ISBN 978-94-010-7179-6. [Google Scholar]
- Lee, F.J.; Miller, K.I.; McKinlay, J.B.; Newton, I.L.G. Differential Carbohydrate Utilization and Organic Acid Production by Honey Bee Symbionts. FEMS Microbiol. Ecol. 2018, 94, fiy113. [Google Scholar] [CrossRef] [PubMed]
- Callegari, M.; Crotti, E.; Fusi, M.; Marasco, R.; Gonella, E.; De Noni, I.; Romano, D.; Borin, S.; Tsiamis, G.; Cherif, A.; et al. Compartmentalization of Bacterial and Fungal Microbiomes in the Gut of Adult Honeybees. NPJ Biofilms Microbiomes 2021, 7, 42. [Google Scholar] [CrossRef]
- Vidau, C.; Panek, J.; Texier, C.; Biron, D.G.; Belzunces, L.P.; Le Gall, M.; Broussard, C.; Delbac, F.; El Alaoui, H. Differential Proteomic Analysis of Midguts from Nosema ceranae-Infected Honeybees Reveals Manipulation of Key Host Functions. J. Invertebr. Pathol. 2014, 121, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Paris, L.; Roussel, M.; Pereira, B.; Delbac, F.; Diogon, M. Disruption of Oxidative Balance in the Gut of the Western Honeybee Apis mellifera Exposed to the Intracellular Parasite Nosema ceranae and to the Insecticide Fipronil. Microb. Biotechnol. 2017, 10, 1702–1717. [Google Scholar] [CrossRef] [PubMed]
- Aufauvre, J.; Misme-Aucouturier, B.; Viguès, B.; Texier, C.; Delbac, F.; Blot, N. Transcriptome Analyses of the Honeybee Response to Nosema ceranae and Insecticides. PLoS ONE 2014, 9, e91686. [Google Scholar] [CrossRef]
- Tan, S.Q.; Zhang, K.Q.; Chen, H.X.; Ge, Y.; Ji, R.; Shi, W.P. The Mechanism for Microsporidian Parasite Suppression of the Hindgut Bacteria of the Migratory Locust Locusta migratoria manilensis. Sci. Rep. 2015, 5, 17365. [Google Scholar] [CrossRef]
- Zheng, H.; Nishida, A.; Kwong, W.K.; Koch, H.; Engel, P.; Steele, M.I.; Moran, N.A. Metabolism of Toxic Sugars by Strains of the Bee Gut Symbiont Gilliamella apicola. mBio 2016, 7, e01326-16. [Google Scholar] [CrossRef]
- Alberoni, D.; Di Gioia, D.; Baffoni, L. Alterations in the Microbiota of Caged Honeybees in the Presence of Nosema ceranae Infection and Related Changes in Functionality. Microb. Ecol. 2022, 86, 601–616. [Google Scholar] [CrossRef] [PubMed]
- Hubert, J.; Bicianova, M.; Ledvinka, O.; Kamler, M.; Lester, P.J.; Nesvorna, M.; Kopecky, J.; Erban, T. Changes in the Bacteriome of Honey Bees Associated with the Parasite Varroa destructor, and Pathogens Nosema and Lotmaria passim. Microb. Ecol. 2017, 73, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Jabal-Uriel, C.; Alba, C.; Higes, M.; Rodríguez, J.M.; Martín-Hernández, R. Effect of Nosema ceranae Infection and Season on the Gut Bacteriome Composition of the European Honeybee (Apis mellifera). Sci. Rep. 2022, 12, 9326. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.E.; Maes, P. Social Microbiota and Social Gland Gene Expression of Worker Honey Bees by Age and Climate. Sci. Rep. 2022, 12, 10690. [Google Scholar] [CrossRef]


| Age of Infection (Days p.e.) | 0 | 1 | 4 | 5 | 8 | 11 | 13 | 14 | Total |
|---|---|---|---|---|---|---|---|---|---|
| Age of Analysis (Days p.e.) | 7 | 8 | 11 | 12 | 15 | 18 | 20 | 21 | |
| Infected bees (N) | 30 | 30 | 30 | 30 | 28 | 30 | 30 | 7 | 215 |
| Non-infected bees (N) | 11 | 10 | 6 | 6 | 10 | 6 | 6 | 6 | 61 |
| Primers | Sequence (5′-3′) | Size |
|---|---|---|
| ALPHA 2.2-F | CCGAGAGAGGGTTGTGGAATT | 67 pb |
| ALPHA 2.2-R | AGATATTGGGAAGAACACCG | |
| ALPHA 2.2-P | 6FAM-TGTAGA+GG+T+GAA+AT+TC-BHQ1 |
| Age of Infection (Days p.e.) | 0 | 1 | 4 | 5 | 8 | 11 | 13 | 14 | Total |
|---|---|---|---|---|---|---|---|---|---|
| Age of Analysis (Days p.e.) | 7 | 8 | 11 | 12 | 15 | 18 | 20 | 21 | |
| Infected bees (N) | 28 | 30 | 27 | 27 | 27 | 30 | 30 | 7 | 206 |
| Non-infected bees (N) | 11 | 10 | 5 | 6 | 10 | 6 | 5 | 4 | 57 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguado-López, D.; Urbieta Magro, A.; Higes, M.; Rodríguez, J.M.; Martín-Hernández, R. Influence of Age of Infection on the Gut Microbiota in Worker Honey Bees (Apis mellifera iberiensis) Experimentally Infected with Nosema ceranae. Microorganisms 2024, 12, 635. https://doi.org/10.3390/microorganisms12040635
Aguado-López D, Urbieta Magro A, Higes M, Rodríguez JM, Martín-Hernández R. Influence of Age of Infection on the Gut Microbiota in Worker Honey Bees (Apis mellifera iberiensis) Experimentally Infected with Nosema ceranae. Microorganisms. 2024; 12(4):635. https://doi.org/10.3390/microorganisms12040635
Chicago/Turabian StyleAguado-López, Daniel, Almudena Urbieta Magro, Mariano Higes, Juan Miguel Rodríguez, and Raquel Martín-Hernández. 2024. "Influence of Age of Infection on the Gut Microbiota in Worker Honey Bees (Apis mellifera iberiensis) Experimentally Infected with Nosema ceranae" Microorganisms 12, no. 4: 635. https://doi.org/10.3390/microorganisms12040635
APA StyleAguado-López, D., Urbieta Magro, A., Higes, M., Rodríguez, J. M., & Martín-Hernández, R. (2024). Influence of Age of Infection on the Gut Microbiota in Worker Honey Bees (Apis mellifera iberiensis) Experimentally Infected with Nosema ceranae. Microorganisms, 12(4), 635. https://doi.org/10.3390/microorganisms12040635








